Introduction
Cancer outcome is continually improved in recent
years with the introduction of early detection, advances in the
chemotherapy, radiotherapy, and targeted therapy strategies; and
the implementation of multidisciplinary cancer care (1). In light of the prominent role of
chemotherapy, anthracycline especially doxorubicin remains to be
considered the footstone chemotherapy regimen of cancer treatment
for a wide range of malignancies, particularly for breast cancer
and lymphoma (2,3). Over decades from its discovery, the
anti-tumor and cardiotoxic mechanisms of doxorubicin seem to
continuously evoke considerable interest in basic science and
clinical trial research. Despite the great achievement in the
survival of cancer patients, there is a serious alert nowadays that
today's cancer patients may be tomorrow's cardiac patients due to
the increased awareness of the doxorubicin-induced cardiotoxicity
(4,5).
Paradoxically, apparently prolonged survival with
doxorubicin is accompanid by increased risk of cardiac damage in
cancer patients, particularly in children, because the damage may
not manifest for many years and sustains to be a life-long threat
(6). Reportedly, the risk of
cardiotoxicity varies according to the type and intensity of cancer
treatment (7). A devastating
cardiotoxic effect of doxorubicin is principally heart failure,
with incidence rates from 0.14 to 48% (estimated risk ranges from
0.14 to 5% for doses >400 mg/m2, 7 to 26% for 550
mg/m2 and 18 to 48% for 700 mg/m2) (8). Clinically, in terms of the benefit as
well as cardiotoxic effect with doxorubicin, the paradox could
greatly undermine the decision making of clinicians with the lack
of effective and convenient approaches to monitor and quantify the
cardiotoxic effects.
Although recent emphasis on the development of
biomarkers indicative of doxorubicin-induced cardiac damage has
resulted in the identification of several proposed biomarkers, such
as troponins, myoglobin, actate dehydrogenase, creatine
phosphokinase, C-redactive protein, brain-type natriuretic peptide
(BNP), and pro-BNP (6,9–11),
consistent findings focusing on concerns were not always presented.
For instance, administration of doxorubicin was not always
associated with elevated troponin level (12). Moreover, an increase in pro-BNP level
has been found early after doxorubicin use, however, elevated
pro-BNP level does not predict left ventricular dysfunction
(13,14).
Considering the uncertainty about the effectiveness
of cardiac markers to quantify doxorubicin-induced cardiotoxicity,
we therefore conducted a microarray-based study to systematically
identify more potential candidate blood indicators for doxorubicin-
induced heart failure via a bioinformatics analysis.
Materials and methods
Microarray data
The microarray expression profile dataset GSE40447
deposited by McCaffrey et al (15), was downloaded from the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40447)
and was based on the platform of GPL16006[(HG-U133_Plus_2)
Affymetrix Human Genome U133 Plus 2.0 Array]. A total of 15 blood
samples from 5 women with doxorubicin chemotherapy-induced heart
failure (HF) and 10 women with no HF but a history of doxorubicin
chemotherapy, were used for the analysis. The microarray expression
profile dataset GSE9128 including 12 blood samples from chronic
heart failure patients and 12 age- and sex-matched controls was
used for validation analysis (16).
The GSE9128 dataset deposited by Cappuzzello et al (16) was based on the platform of GPL96
[(HG-U133A) Affymetrix Human Genome U133A Array].
Data preprocessing and identification
of differentially expressed genes (DEGs)
For GSE40447 data, the probe-level raw data (CEL
format) were preprocessed by using a robust multiarray average
algorithm with the Affy package in Bioconductor (17). Missing data were imputed by the
KNN-based method and median data normalization was performed using
robust multichip averaging for multiple probes corresponding to a
common gene symbol (18,19). The DEGs following doxorubicin
chemotherapy between HF and non-HF samples were identified by using
the limma package in R/Bioconductor (20). For GSE9128 data, GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/),
a R-based interactive web tool commonly used to compare two
independent groups of samples for identifying DEGs with lack of raw
CEL format data, was used to screen DEGs between HF and control
samples. Similarly, the limma package involving empirical Bayes
statistics was integrated for DEG analysis. |logFC|≥0.585
(|logFC|≥0.585 indicated that fold-change >1.5) and P-value
<0.05 were considered as threshold values for DEGs in Limma and
GEO2R.
Functional enrichment analysis
Gene Ontology (GO) functional enrichment including
biological process and molecular function and Kyoto Encyclopedia of
Gene and Genomes (KEGG) pathway enrichment analyses were performed
for DEGs in GSE40447 by using the Database for Annotation,
Visualization and Integrated Discovery (DAVID) to study
differentially expressed clusters at the functional level
(https://david.ncifcrf.gov/) (21). P<0.05 was considered as the
cut-off criterion for the enrichment analysis.
Screening for cardiovascular disease
(CVD)-related DEGs
CVD-related DEGs were identified by mapping to a
Literature Based Multi-Omics Database for Major Cardiovascular
Diseases: CardioGenBase (http://cardiogenbase.com/), which collected
gene-disease association data from PubMed and MEDLINE and covered
major CVDs such as cerebrovascular disease, coronary artery disease
(CAD), hypertensive heart disease, inflammatory heart disease,
ischemic heart disease and rheumatic heart disease (22). The CardioGenBase database documented
~1,500 CVD genes from ~2,4000 research articles.
Construction of protein-protein
interaction (PPI) network
The DEGs in GSE40447 data were mapped to the overall
PPIs by using the database Search Tool for the Retrieval of
Interacting Genes (STRING, http://string-db.org/), which integrates a variety of
predicted and experimentally validated interactions of proteins
(23). Then, the primary PPI network
was constructed with a threshold value of combined score >0.4
output by the STRING database and visualized by using Cytoscape
(http://cytoscape.org/) (24).
Microarray-based validation for
CVD-related DEGs
The CVD-related DEGs were validated by an
intersection analysis with DEGs in the GSE9128 data. The overlap
with consistent expression pattern of DEGs in GSE40447 and GSE9128
were identified as candidate indicators for doxorubicin- induced
heart failure. Meanwhile, the expression files of candidate
indicators in experiment and control groups in GSE9128 were
obtained from the GEO database, of which the relative expression
corresponded to the results of DEG analysis was graphically
presented.
Results
Identification of DEGs in GSE40447
microarray data
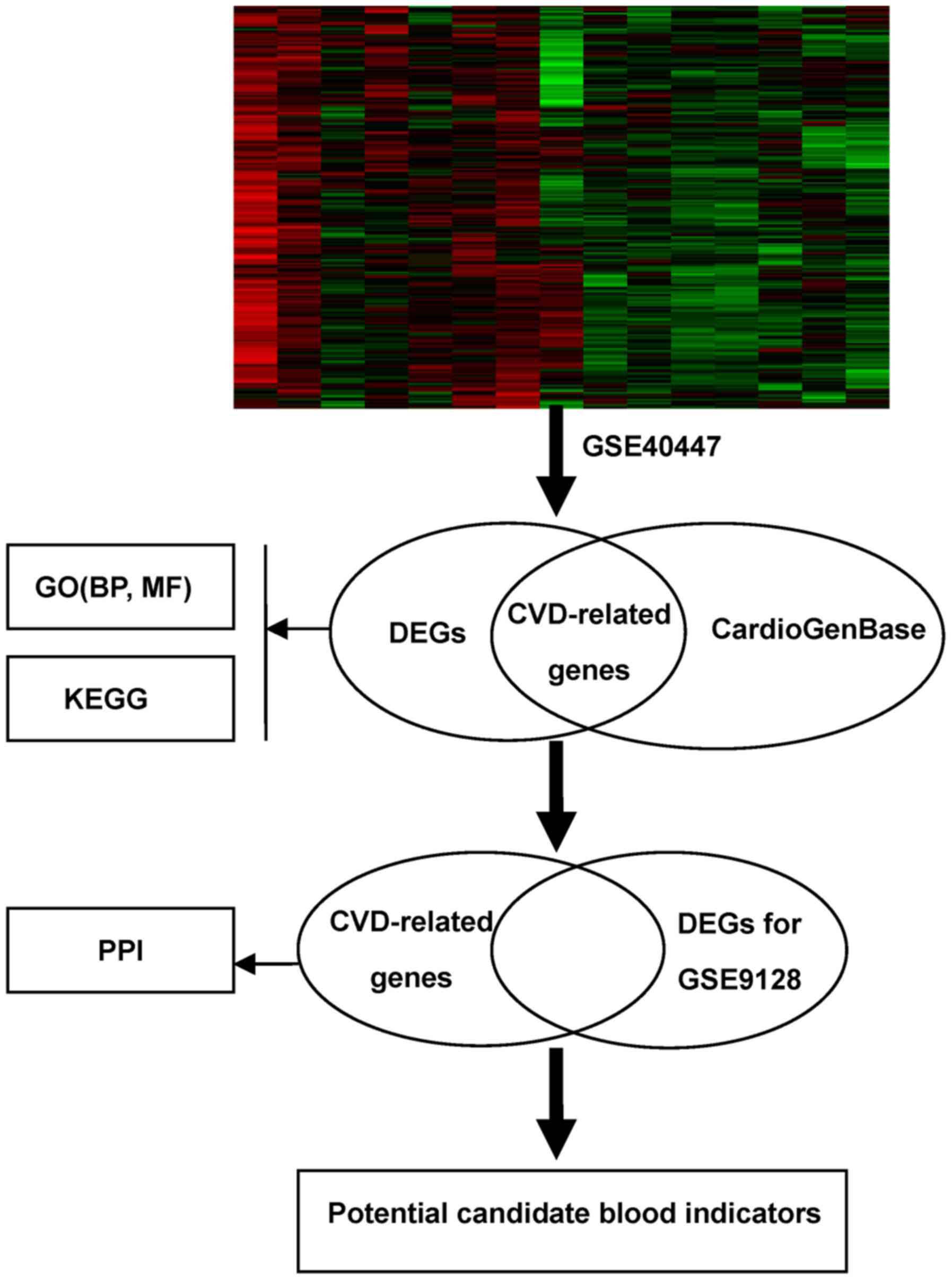
The workflow of the proposed analysis was performed
as Fig. 1. As a result shown in
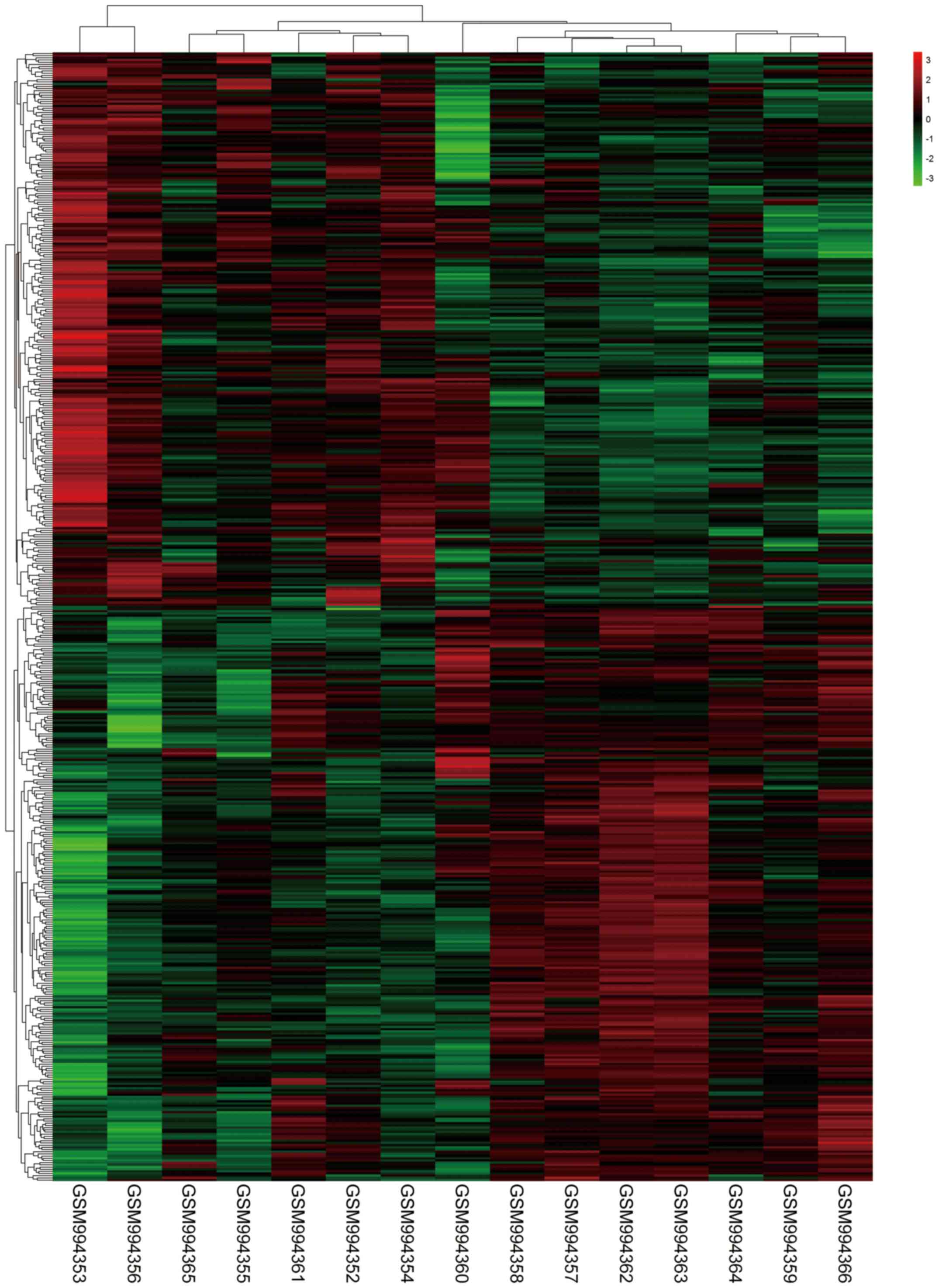
Fig. 2, a total of 516 DEGs in
GSE40447 including 263 down-regulated genes and 253 up-regulated
genes were identified in HF group following doxorubicin
chemotherapy compared to non-HF group with a history of doxorubicin
chemotherapy according to the established cut-off criterion. The
heatmap for DEGs is shown in Fig.
3.
GO and KEGG pathway enrichment
analyses of DEGs in GSE40447 data
DAVID provided comprehensively functional annotation
to understand the biological meaning behind the large list of
genes. In the current study, GO enrichment analysis using DAVID was
performed to explore DEGs functions from two aspects, including
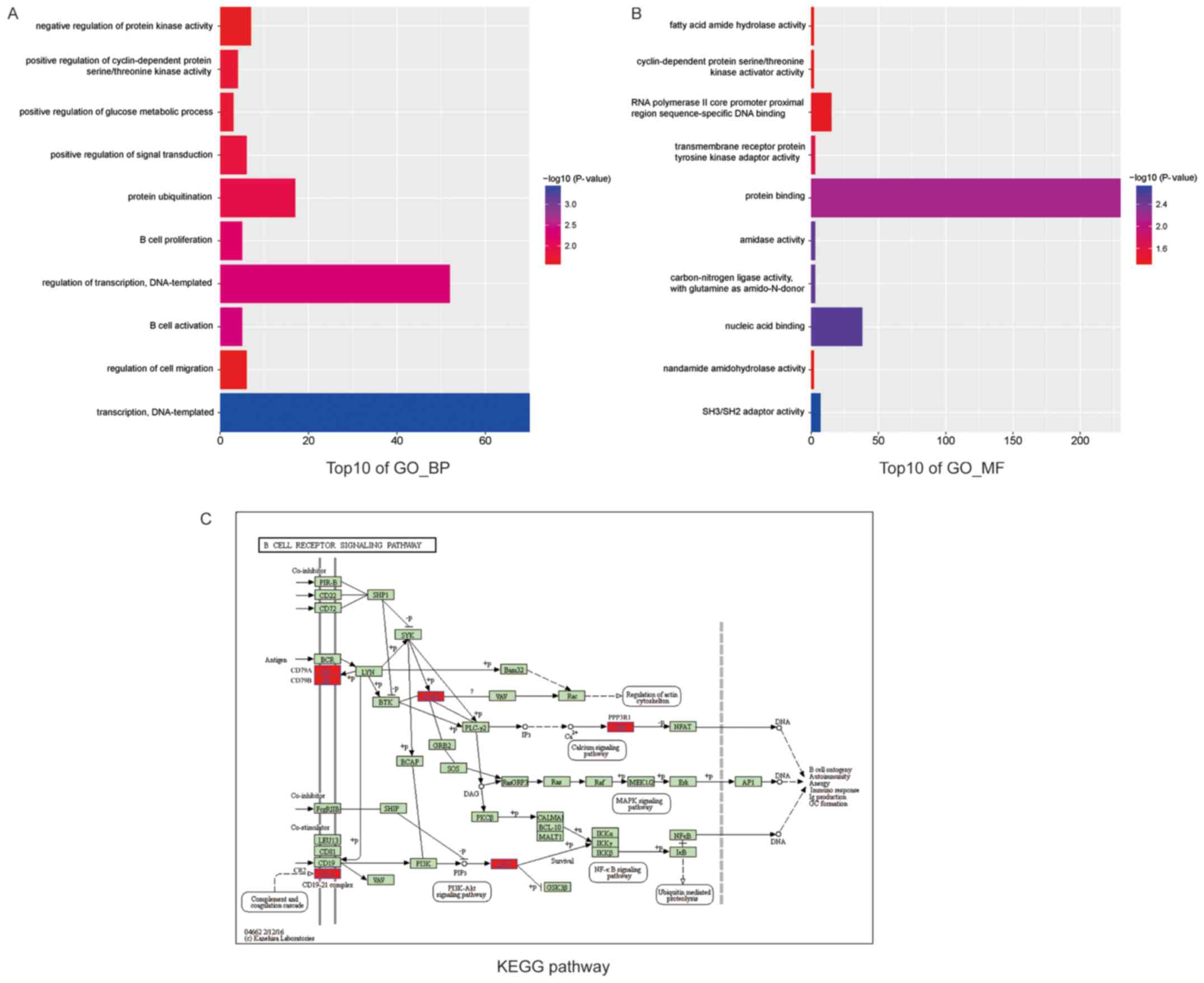
molecular function and biological process. As presented in Fig. 4, the results showed that the top10
overrepresented GO terms in biological processes were enriched in
B-cell activation, leukocyte activation, cell activation,
lymphocyte activation, regulation of transcription, transcription,
negative regulation of B-cell proliferation, inflammatory response,
regulation of lymphocyte proliferation and regulation of
mononuclear cell proliferation. Additionally, the top10 enriched GO
terms in molecular function were protein domain-specific binding,
molecular adaptor activity, carbon-nitrogen ligase activity, with
glutamine as an amido-N-donor, copper ion binding, phospholipid
binding, SH3/SH2 adaptor activity, zinc ion binding,
phosphoinositide binding, UDP-galactose: Glucosylceramide
beta-1,4-galactosyltrans- ferase activity and transcription factor
binding. On the other hand, we adopted KEGG enrichment analysis by
clustering similar genes into a same network to understand the DEGs
action mode. The B-cell receptor signaling pathway containing 6
DEGs (BLNK, CD79A, CD79B, CR2, PPP3R1, AKT1) indicated with red was
enriched (Fig. 4).
Screening for CVD-related DEGs
Many DEGs generated may result from various
responses to doxorubicin chemotherapy with multifarious etiologies
but not just cardiac damage. In order to screen the CVD-related
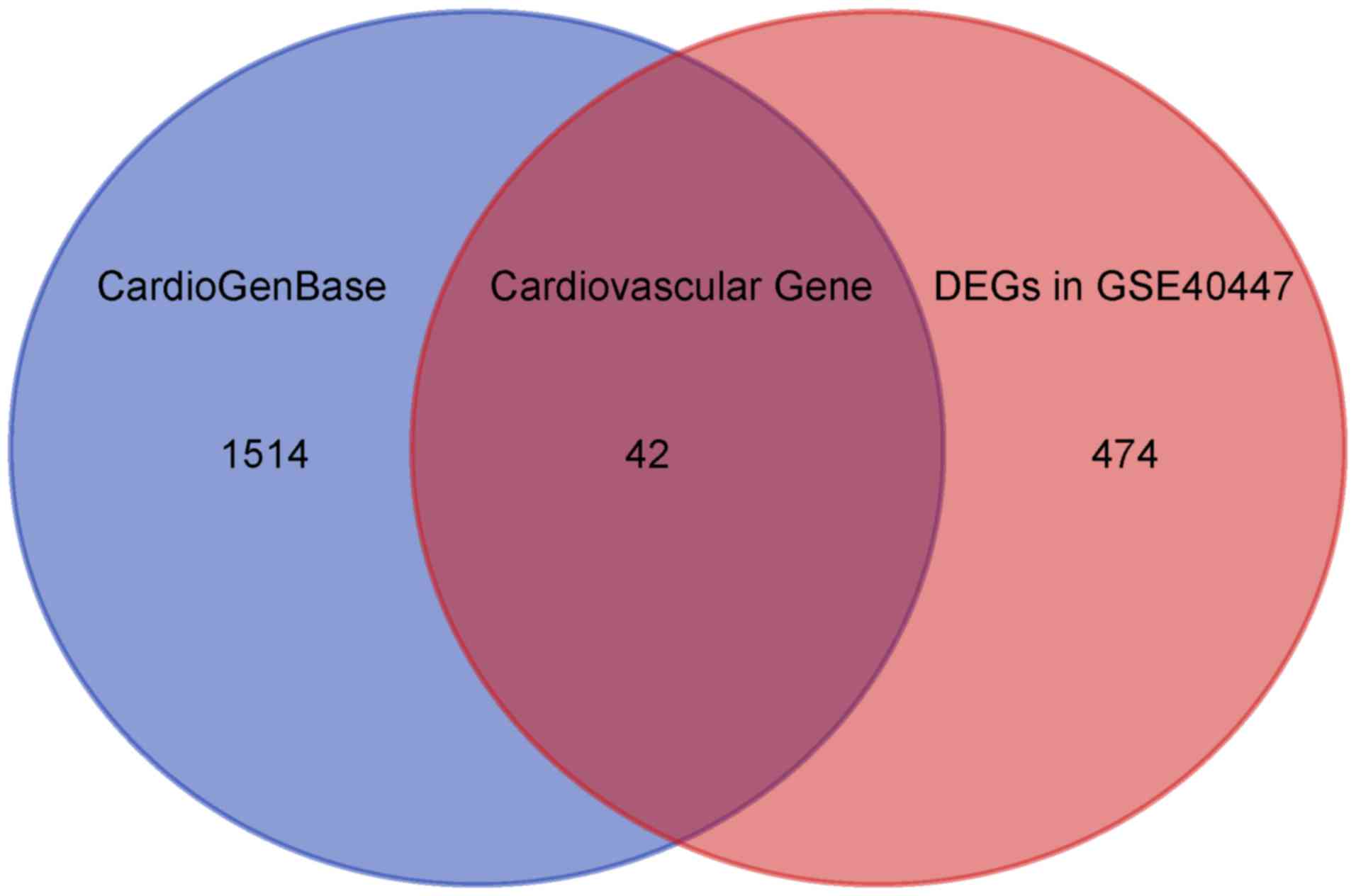
DEGs, the CardioGenBase database containing 1556 CVD-related genes
was applied. Finally, the intersection analysis showed 42 DEGs
associated with CVD (Fig. 2 and
Table I). Of note, all 42 genes were
literarily confirmed with a tight relevance to CVD in MEDLINE.
 | Table I.CVD-related DEGs mapped in the
CardioGenBase database. |
Table I.
CVD-related DEGs mapped in the
CardioGenBase database.
| Gene Symbol | HGNC ID | Gene Location | No. of
articles |
|---|
| ABCB1 | 40 | 7q21.12 | 3 |
| AKT1 | 391 | 14q32.33 | 22 |
| ANPEP | 500 | 15q25-q26 | 1 |
| ARHGAP5 | 675 | 14q12 | 2 |
| BGLAP | 1043 | 1q22 | 11 |
| CD163 | 1631 | 12p13 | 2 |
| CD28 | 1653 | 2q33 | 1 |
| CD40 | 11919 | 20q12-q13.2 | 30 |
| CD79A | 1698 | 19q13.2 | 2 |
| CD83 | 1703 | 6p23 | 1 |
| CDKN2D | 1790 | 19p13 | 1 |
| CELSR2 | 3231 | 1p13.3 | 4 |
| CHML | 1941 | 1q43 | 3 |
| CTNS | 2518 | 17p13 | 1 |
| EBF1 | 3126 | 5q34 | 1 |
| F5 | 3542 | 1q23 | 54 |
| GAA | 4065 | 17q25.2-q25.3 | 3 |
| GABPA | 4071 | 21q21-q22.1 | 1 |
| GATA2 | 4171 | 3q21 | 3 |
| GEN1 | 26881 | 2p24.2 | 1 |
| HDC | 4855 | 15q21.2 | 1 |
| IDUA | 5391 | 4p16.3 | 1 |
| IGFBP4 | 5473 | 17q21.2 | 3 |
| IL3RA | 6012 | Xp22.3 and
Yp13.3 | 1 |
| KLRB1 | 6373 | 12p13 | 1 |
| MACROD2 | 16126 | 20p12.1 | 1 |
| MPO | 7218 | 17q21.3-q23 | 82 |
| NEFL | 7739 | 8p21.2 | 1 |
| OLR1 | 8133 | 12p13.1-p12.3 | 27 |
| PAWR | 8614 | 12q21.2 | 6 |
| PEX6 | 8859 | 6p22-p11 | 1 |
| PPP3R1 | 9317 | 2p14 | 1 |
| PTGDS | 9592 | 9q34.2-q34.3 | 2 |
| SASS6 | 25403 | 1p21.3 | 4 |
| SCN3A | 10590 | 2q24 | 1 |
| SLC22A16 | 20302 | 6q21 | 1 |
| SLC25A20 | 1421 | 3p21.31 | 3 |
| SPRY1 | 11269 | 4q | 1 |
| TBXAS1 | 11609 | 7q34-q35 | 52 |
| TLR5 | 11851 | 1q32.3-q42 | 1 |
| UNC13D | 23147 | 17q25.3 | 1 |
| XRCC1 | 12828 | 19q13.2 | 5 |
PPI network construction for
CVD-related DEGs
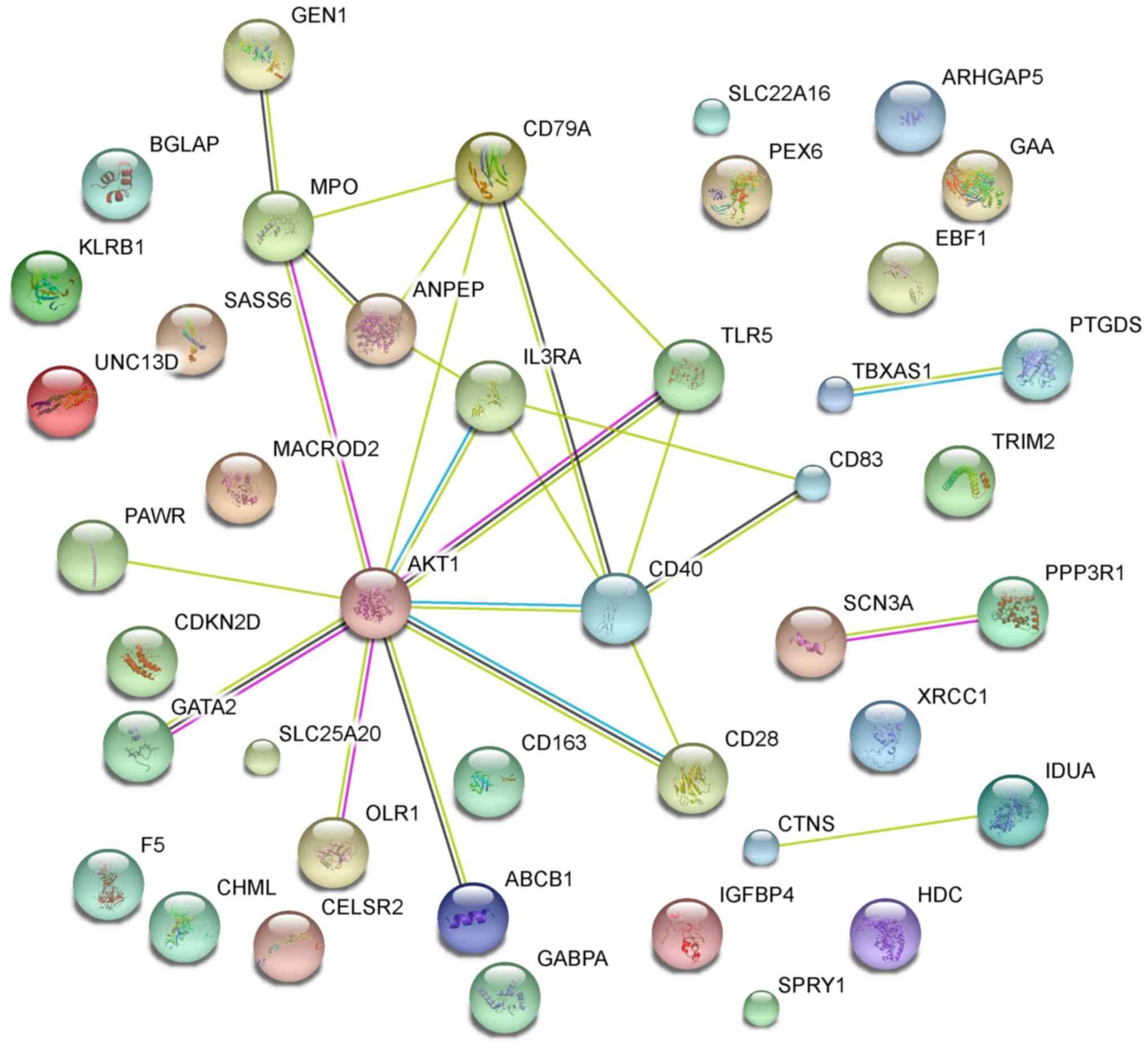
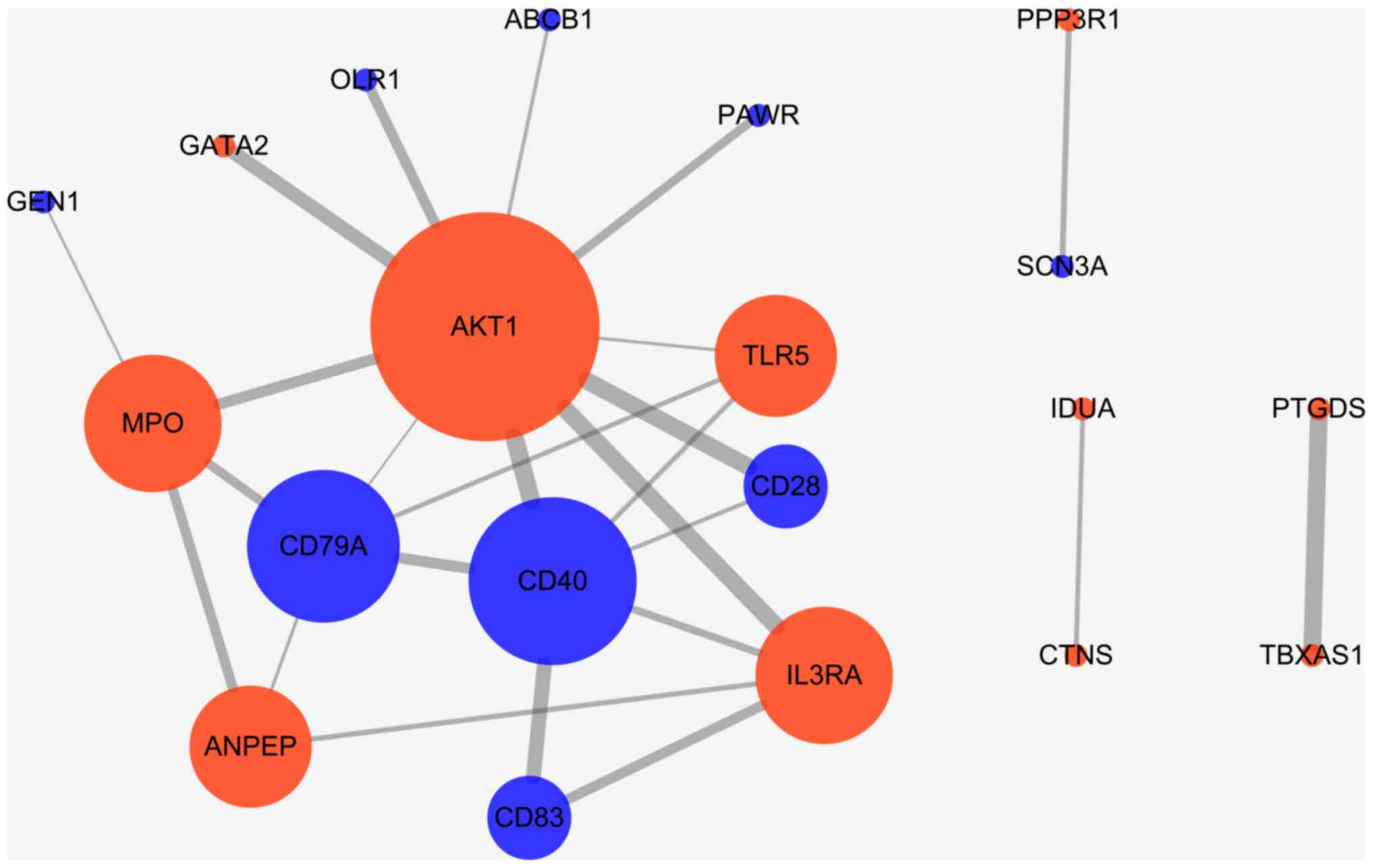
The primary PPI network including all CVD-related
DEGs was constructed using STRING database (Fig. 5) and was further constructed with a
threshold value of a combined score >0.4 and visualized by
Cytoscape. Overall, 20 nodes and 25 edges were mapped in the PPI
network of identified DEGs, including 11 up-regulated genes and 9
down-regulated genes (Fig. 6). The 7
nodes with the higher degrees were screened as hub genes (more than
3 interactions for each hub gene), including AKT1, MPO, ANPEP,
CD79A, CD40, IL3RA and TLR5.
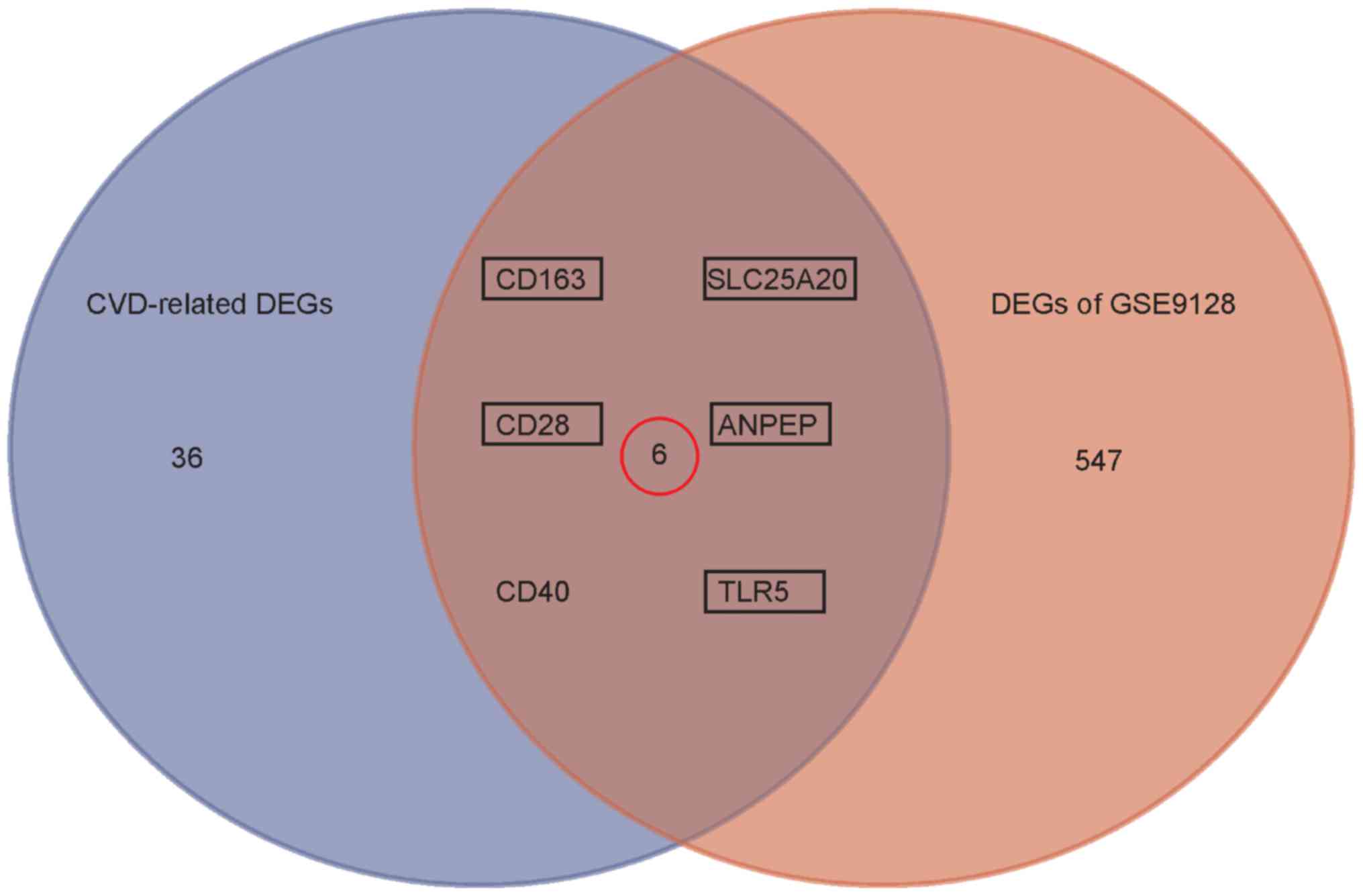
Validation for CVD-related DEGs by
GSE9128
The study involving GSE9128 was designed to identify
diagnostic/prognostic blood markers and gene expression profiles of
chronic heart failure. Based on the established cut-off criterion
for DEGs, 651 DEGs (330 down-regulated and 321 up-regulated) were
identified in HF group compared to healthy control. To validate the
CVD-related DEGs induced by doxorubicin treatment, an intersection
analysis with DEGs in GSE9128 was performed, and 6 genes (CD163,
CD28, SLC25A20, ANPEP, TLR5, CD40) was shown in the overlap
(Fig. 7). However, 5 genes (CD163,
CD28, SLC25A20, ANPEP, TLR5) were highlighted with a consistent
expression pattern in GSE40447 and GSE9128, and were finally
identified as potential candidate blood indicators for
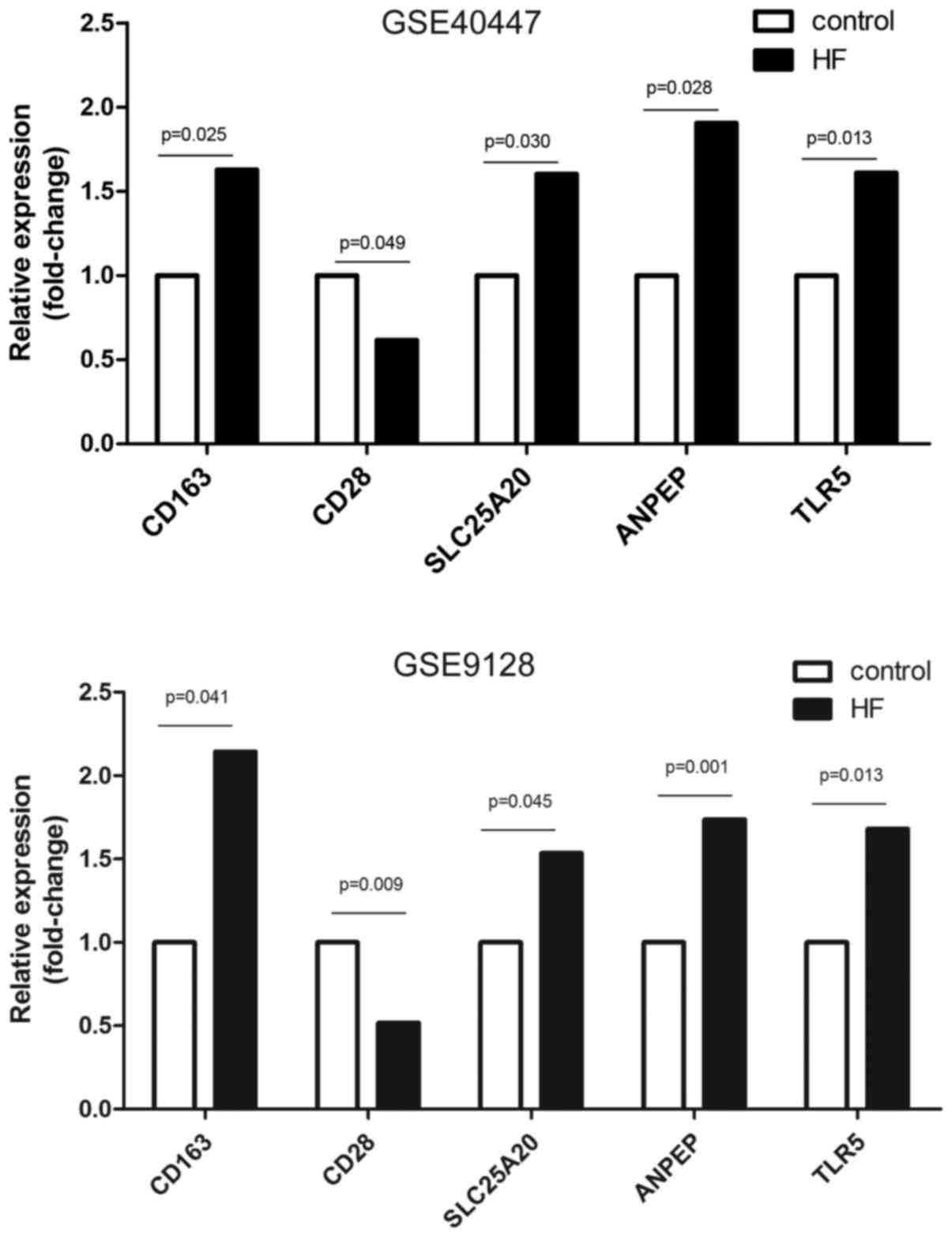
doxorubicin-induced heart failure (Table II). Corresponded to the detailed
results of DEG analysis, the expression files of the candidate
indicators obtained from GSE9128 in experiment and control groups
were graphically presented. As shown in Fig. 8, 2.14, 0.52, 1.53, 1.74 and 1.68
fold-change transcriptional level for CD163, CD28, SLC25A20, ANPEP
and TLR5 were observed compared to the non-HF healthy control,
respectively.
 | Table II.Expression of CVD-related genes in
GSE40447 and GSE9128 datasets. |
Table II.
Expression of CVD-related genes in
GSE40447 and GSE9128 datasets.
|
| LogFC | P-value | Expression
pattern |
|---|
|
|
|
|
|
|---|
| Gene symbol | GSE40447 | GSE9128 | GSE40447 | GSE9128 | GSE40447 | GSE9128 |
|---|
| CD163 | 0.7023137 | 1.0998679 | 0.0254562 | 0.0410781 | Up | Up |
| CD28 | −0.700993 | −0.9513838 | 0.049 | 0.0097194 | Down | Down |
| SLC25A20 | 0.6808281 | 0.6178648 | 0.0299684 | 0.0449655 | Up | Up |
| ANPEP | 0.9295412 | 0.7951052 | 0.0275249 | 0.0012539 | Up | Up |
| TLR5 | 0.6858654 | 0.7478113 | 0.0332472 | 0.0127178 | Up | Up |
| CD40 | −1.36061145 | 0.8154462 | 0.002348725 | 0.04237951 | Down | Up |
Discussion
Currently, bioinformatics analysis with
high-throughput gene expression profiling has provided systematic
insights into the mechanism of doxorubicin-induced cardiotoxic
effects, related to the underlying gene activity changes and more
importantly, has enabled the identification of targets for a
diagnosis and prevention policy. In the present study, a total of
516 blood DEGs potentially related to doxorubicin-induced HF were
initially identified with the GSE40447 microarray data. Functional
enrichment analysis showed that these DEGs were mainly related to B
-cell receptor signaling pathway. Of the DEGs, 42 were literarily
evidenced as CVD-related genes. Importantly, the further validation
analysis revealed that 5 CVD-related DEGs (CD163, CD28, SLC25A20,
ANPEP, TLR5) may served as potential candidate blood indicators for
doxorubicin-induced heart failure.
The pathogenesis of doxorubicin-induced HF is a
complex process driven by specific genetic alterations and
susceptible drug toxicity. Hemoglobin scavenger receptor CD163 is a
macrophage-specific protein and its up-regulation is one of the
major changes in the macrophage switch to alternative activated
phenotypes in inflammation (25).
This upregulation may echo the activation of inflammation triggered
by doxorubicin-related cardiac damage in that enhanced inflammation
has been confirmed as a hallmark of cardiac injury (26). In support of this, several previous
studies have revealed increased serum level of CD163 along with
elevated inflammatory cytokines in patients with atrial
fibrillation (27) and coronary
heart disease (28). In addition,
elevated serum CD163 level was also found in patients with chronic
heart failure with reduced ejection fraction (29). Hence, the results of the current
study are in line with previous findings, suggesting the potential
role of increased CD163 expression in doxorubicin-induced
cardiotoxicity. Moreover, the CD28 molecule is a type I
transmembrane protein expressed on the surface of 80% of human CD4+
T cells and 50% of human CD8+ T cells (30), which is well known as one of the most
important co-stimulatory receptors for T-lymphocyte activation and
T-cell receptor signal transduction, thereby regulating the
production of inflammatory cytokine/chemokines (31). Several separate studies have reported
that CD28 might act as an accomplice but not always a defender by
demonstrating the deleterious effect of CD4+CD28 null T-lymphocytes
in patients with atrial fibrillation, chronic heart failure,
coronary artery disease, atherosclerosis and other CVDs (32–34).
Investigators have uncovered partial mechanisms responsible for the
deleterious effect of activation of cytotoxic lymphocytes secreting
pro-inflammatory cytokines to promote inflammation and the
development of cardiovascular inflammatory diseases (35). Therefore, it is not difficult to
speculate the detrimental role of CD28 down-regulation in
doxorubicin-induced cardiotoxicity.
The present study also suggested a role for
Toll-like receptor 5 (TLR5) up-regulation in doxorubicin-induced
cardiotoxicity. In the heart, previous research found that TLR5
activation may condition vascular dendritic cells (DCs) to support
perivasculitic infiltrates possibly by increasing the suppressive
activity of Treg cells and inducing the synthesis of
immunosuppressive interleukins IL-10, IL-35, and transforming
growth factor β (36,37). Supportively, Zhu et al
(38) found that TLR5 polymorphism
might be associated with rheumatic heart disease risk in a Chinese
Han population. Platelet TLR5 was found associated with ratio of
total cholesterol to high-density lipoprotein, thus contributing to
cardiovascular risk (39).
Therefore, it could not rule out the link between TLR5-involved
vasculitic and doxorubicin-induced cardiotoxicity although the
underlying mechanism remains uninvestigated.
For the solute carrier family 25 member 20 (SLC25A20
or CACT), a key carnitine-acylcarnitine translocase exchanging for
free carnitine across the mitochondrial membrane in mitochondrial
beta-oxidation, little information has been acknowledged on its
pathophysiologic mechanisms in CVD except for several case reports
about the CACT-deficiency in cardiomyopathy, arrythmias and
cardiogenic shock (40–42). However, the current study found for
the first time a markedly up-regulation of SLC25A20 transcriptional
level in doxorubicin-induced HF, which was further validated with a
1.74-fold change in a HF cohort compared to the healthy control.
Nevertheless, the underlying mechanism remains unknown and that
whether this is a compensatory upregulation to prevent injury needs
to be studied.
The present study also suggested a potential role of
upregulated alanyl aminopeptidase (ANPEP, APN or CD13) in
doxorubicin-induced heart failure. ANPEP is a conserved type II
integral membrane zinc-dependent metalloprotease in the M1 family
of ectoenzymes (43), widely
expressed on all myeloid cells, activated endothelial cells and
epithelium of the kidney and intestine (44). Evolving evidence supported a role for
ANPEP in controlling arterial blood pressure and the pathogenesis
of hypertension by inhibiting renal tubule Na flux and modulating
salt-adaptation (43,45). Interestingly, circulating ANPEP was
found to be up-regulated in patients with essential hypertension
and in the angiogenic vessels in the infarct area and border zone
after myocardial infarction (44,46).
Further exploration revealed that ANPEP is essential for promoting
optimal post-infarction healing for the ischaemic heart via
compensatory mechanisms including the preparation and sustainment
of the reparative response to relieve potential angiogenic defects
(44). Thus, it is biologically
reasonable to speculate that the increase of ANPEP expression may
be compensatory, which might be predictive for doxorubicin-induced
cardiotoxicity.
Several limitations for the study should be
acknowledged. The underlying molecular mechanism of
doxorubicin-induced HF is not completely consistent with
non-pharmaceutical HF. Some potential more important indicators
specified for doxorubicin-induced HF may be concealed by the
validation with the study GSE9128 designed for the identification
of non-pharmaceutical HF biomarkers. For instance, the present
study showed that DEGs in GSE40447 were mainly related to B cell
receptor signaling pathway in KEGG enrichment analysis, whereas the
focused indicators were not implicated, which may suggest the
specificity of some DEGs that needed to be experimentally
confirmed. Moreover, the potential blood indicators were initially
proposed by the intersection between GSE40447 and CardioGenBase,
which may exclude other important predictors of doxorubicin-induced
heart injury that has not been documented before in CardioGenBase,
and therefore may result in limited ability for diagnosis of
doxorubicin-induced HF. Nevertheless, the highlighted indicators
are biologically reasonable and reliable because of the identical
molecular basis of heart failure and a systematic and scientific
study approach in the study. Moreover, in light of applying
predicament of the current proposed biomarkers in the clinic, these
indicators may contribute to the diagnosis and prevention of
doxorubicin-induced HF and the decision-making for clinicians when
combined with several routine markers for cardiac injury such as
troponins and pro-BNP. Definitely, the aforementioned results were
based on microarray data with limited sample size and a lack of
experimental verification, which was also a limitation of the study
and required to be addressed by future studies.
In conclusion, the present study aimed to identify
potential candidate blood biomarkers to predict doxorubicin-induced
heart failure with comprehensive bioinformatics analysis. A set of
significantly altered genes were identified and five (CD163, CD28,
SLC25A20, ANPEP, TLR5) were highlighted by bioinformatics
validation. Our results suggest that data mining and integration
could be a useful tool to predict doxorubicin-induced heart
failure. Despite the absence of experimental validation, this study
still has significance for further study, providing potential new
key genes for further experimental validation, and shedding light
on the molecular mechanisms of doxorubicin-induced
cardiotoxicity.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
GXW conceived and designed the study, and wrote the
manuscript. LHJ, WBX and LC performed the statistical analysis, and
data collation and interpretation. YGZ designed the study, and
reviewed and edited the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Curigliano G, Cardinale D, Dent S,
Criscitiello C, Aseyev O, Lenihan D and Cipolla CM: Cardiotoxicity
of anticancer treatments: Epidemiology, detection, and management.
CA Cancer J Clin. 66:309–325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shabalala S, Muller CJF, Louw J and
Johnson R: Polyphenols, autophagy and doxorubicin-induced
cardiotoxicity. Life Sci. 180:160–170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Argun M, Üzüm K, Sönmez MF, Özyurt A,
Derya K, Çilenk KT, Unalmış S, Pamukcu Ö, Baykan A, Narin F, et al:
Cardioprotective effect of metformin against doxorubicin
cardiotoxicity in rats. Anatol J Cardiol. 16:234–241.
2016.PubMed/NCBI
|
|
4
|
McGowan JV, Chung R, Maulik A, Piotrowska
I, Walker JM and Yellon DM: Anthracycline chemotherapy and
cardiotoxicity. Cardiovasc Drugs Ther. 31:63–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bahadir A, Kurucu N, Kadıoğlu M and
Yenilme E: The role of nitric oxide in Doxorubicin-induced
cardiotoxicity: Experimental study. Turk J Haematol. 31:68–74.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bryant J, Picot J, Baxter L, Levitt G,
Sullivan I and Clegg A: Use of cardiac markers to assess the toxic
effects of anthracyclines given to children with cancer: A
systematic review. Eur J Cancer. 43:1959–1966. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Conway A, McCarthy AL, Lawrence P and
Clark RA: The prevention, detection and management of cancer
treatment-induced cardiotoxicity: A meta-review. BMC Cancer.
15:3662015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Senkus E and Jassem J: Cardiovascular
effects of systemic cancer treatment. Cancer Treat Rev. 37:300–311.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai AD, Agarwal A, Steinberg M, Showler A,
Burry L, Tomlinson GA, Bell CM and Morris AM: Clinical predictors
and clinical prediction rules to estimate initial patient risk for
infective endocarditis in Staphylococcus aureus bacteraemia: A
systematic review and meta-analysis. Clin Microbiol Infect.
23:900–906. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lotrionte M, Biondi-Zoccai G, Abbate A,
Lanzetta G, D'Ascenzo F, Malavasi V, Peruzzi M, Frati G and
Palazzoni G: Review and meta-analysis of incidence and clinical
predictors of anthracycline cardiotoxicity. Am J Cardiol.
112:1980–1984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jain D, Russell RR, Schwartz RG, Panjrath
GS and Aronow W: Cardiac complications of cancer therapy:
Pathophysiology, identification, prevention, treatment, and future
directions. Curr Cardiol Rep. 19:362017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cardinale D and Sandri MT: Role of
biomarkers in chemotherapy-induced cardiotoxicity. Prog Cardiovasc
Dis. 53:121–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dolci A, Dominici R, Cardinale D, Sandri
MT and Panteghini M: Biochemical markers for prediction of
chemotherapy-induced cardiotoxicity: Systematic review of the
literature and recommendations for use. Am J Clin Pathol.
130:688–695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fallah-Rad N, Walker JR, Wassef A, Lytwyn
M, Bohonis S, Fang T, Tian G, Kirkpatrick ID, Singal PK, Krahn M,
et al: The utility of cardiac biomarkers, tissue velocity and
strain imaging, and cardiac magnetic resonance imaging in
predicting early left ventricular dysfunction in patients with
human epidermal growth factor receptor II-positive breast cancer
treated with adjuvant trastuzumab therapy. J Am Coll Cardiol.
57:2263–2270. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McCaffrey TA, Tziros C, Lewis J, Katz R,
Siegel R, Weglicki W, Kramer J, Mak IT, Toma I, Chen L, et al:
Genomic profiling reveals the potential role of TCL1A and MDR1
deficiency in chemotherapy-induced cardiotoxicity. Int J Biol Sci.
9:350–360. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cappuzzello C, Napolitano M, Arcelli D,
Melillo G, Melchionna R, Di Vito L, Carlini D, Silvestri L,
Brugaletta S, Liuzzo G, et al: Gene expression profiles in
peripheral blood mononuclear cells of chronic heart failure
patients. Physiol Genomics. 38:233–240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Troyanskaya O, Cantor M, Sherlock G, Brown
P, Hastie T, Tibshirani R, Botstein D and Altman RB: Missing value
estimation methods for DNA microarrays. Bioinformatics. 17:520–525.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nayar PG, Murugesan R, Mary BPD and Ahmed
SS: CardioGenBase: A literature based Multi-Omics database for
major cardiovascular diseases. PLoS One. 10:e1431882015.
|
|
23
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database Issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Etzerodt A and Moestrup SK: CD163 and
inflammation: Biological, diagnostic, and therapeutic aspects.
Antioxid Redox Signal. 18:2352–2363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Landis RC, Philippidis P, Domin J, Boyle
JJ and Haskard DO: Haptoglobin genotype-dependent anti-inflammatory
signaling in CD163(+) macrophages. Int J Inflam. 2013:9803272013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhong SM, Qin YH, Li ZC and Wei YS:
Clinical value of detecting serum soluble CD163 level in patients
with atrial fibrillation. Nan Fang Yi Ke Da Xue Xue Bao.
36:1406–1409. 2016.(In Chinese). PubMed/NCBI
|
|
28
|
Zou LY, Peng CQ, Li CZ, Zhao CL, Zhu JM,
Liu JL and Zhang CX: Association between hemoglobin scavenger
receptor CD163 expression and coronary atherosclerotic severity in
patients with coronary heart disease. Zhonghua Xin Xue Guan Bing Za
Zhi. 37:605–609. 2009.(In Chinese). PubMed/NCBI
|
|
29
|
Ptaszynska-Kopczynska K,
Marcinkiewicz-Siemion M, Lisowska A, Waszkiewicz E, Witkowski M,
Jasiewicz M, Miklasz P, Jakim P, Galar B, Musial WJ and Kaminski
KA: Alterations of soluble TWEAK and CD163 concentrations in
patients with chronic heart failure. Cytokine. 80:7–12. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aruffo A and Seed B: Molecular cloning of
a CD28 cDNA by a high-efficiency COS cell expression system. Proc
Natl Acad Sci USA. 84:8573–8577. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Porciello N and Tuosto L: CD28
costimulatory signals in T lymphocyte activation: Emerging
functions beyond a qualitative and quantitative support to TCR
signalling. Cytokine Growth Factor Rev. 28:11–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sulzgruber P, Koller L, Winter MP, Richter
B, Blum S, Korpak M, Hulsmann M, Goliasch G, Wojta J and Niessner
A: The impact of CD4+CD28null T-lymphocytes
on atrial fibrillation and mortality in patients with chronic heart
failure. Thromb Haemost. 117:349–356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dumitriu IE, Araguás ET, Baboonian C and
Kaski JC: CD4+ CD28 null T cells in coronary artery disease: When
helpers become killers. Cardiovasc Res. 81:11–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kyaw T, Tipping P, Toh BH and Bobik A:
Killer cells in atherosclerosis. Eur J Pharmacol. 816:67–75. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kyaw T, Peter K, Li Y, Tipping P, Toh BH
and Bobik A: Cytotoxic lymphocytes and atherosclerosis:
Significance, mechanisms and therapeutic challenges. Br J
Pharmacol. 174:3956–3972. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Piggott K, Biousse V, Newman NJ, Goronzy
JJ and Weyand CM: Vascular damage in giant cell arteritis.
Autoimmunity. 42:596–604. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Garib FY and Rizopulu AP: T-regulatory
cells as part of strategy of immune evasion by pathogens.
Biochemistry (Mosc). 80:957–971. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu L, Zou LJ, Hua R and Li B: Association
of single-nucleotide polymorphisms in toll-like receptor 5 gene
with rheumatic heart disease in Chinese Han population. Int J
Cardiol. 145:129–130. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Koupenova M, Mick E, Mikhalev E, Benjamin
EJ, Tanriverdi K and Freedman JE: Sex differences in platelet
toll-like receptors and their association with cardiovascular risk
factors. Arterioscler Thromb Vasc Biol. 35:1030–1037. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mahapatra S, Ananth A, Baugh N, Damian M
and Enns GM: Triheptanoin: A rescue therapy for cardiogenic shock
in carnitine-acylcarnitine translocase deficiency. JIMD Rep.
39:19–23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Choong K, Clarke JT, Cutz E, Pollit RJ and
Olpin SE: Lethal cardiac tachyarrhythmia in a patient with neonatal
carnitine-acylcarnitine translocase deficiency. Pediatr Dev Pathol.
4:573–579. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rubio-Gozalbo ME, Bakker JA, Waterham HR
and Wanders RJ: Carnitine-acylcarnitine translocase deficiency,
clinical, biochemical and genetic aspects. Mol Aspects Med.
25:521–532. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Danziger RS: Aminopeptidase N in arterial
hypertension. Heart Fail Rev. 13:293–298. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pereira FE, Cronin C, Ghosh M, Zhou SY,
Agosto M, Subramani J, Wang R, Shen JB, Schacke W, Liang B, et al:
CD13 is essential for inflammatory trafficking and infarct healing
following permanent coronary artery occlusion in mice. Cardiovasc
Res. 100:74–83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sánchez-Agesta Ortega R, Arias de
Saavedra-Alías JM, Liébana-Cañada A, Sánchez-Muñoz B,
Martínez-Martos JM and Ramírez-Expósito MJ: Circulating
aminopeptidase activities in men and women with essential
hypertension. Curr Med Chem. 20:4935–4945. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Buehler A, van Zandvoort MA, Stelt BJ,
Hackeng TM, Schrans-Stassen BH, Bennaghmouch A, Hofstra L,
Cleutjens JP, Duijvestijn A, Smeets MB, et al: cNGR: A novel homing
sequence for CD13/APN targeted molecular imaging of murine cardiac
angiogenesis in vivo. Arterioscler Thromb Vasc Biol. 26:2681–2687.
2006. View Article : Google Scholar : PubMed/NCBI
|