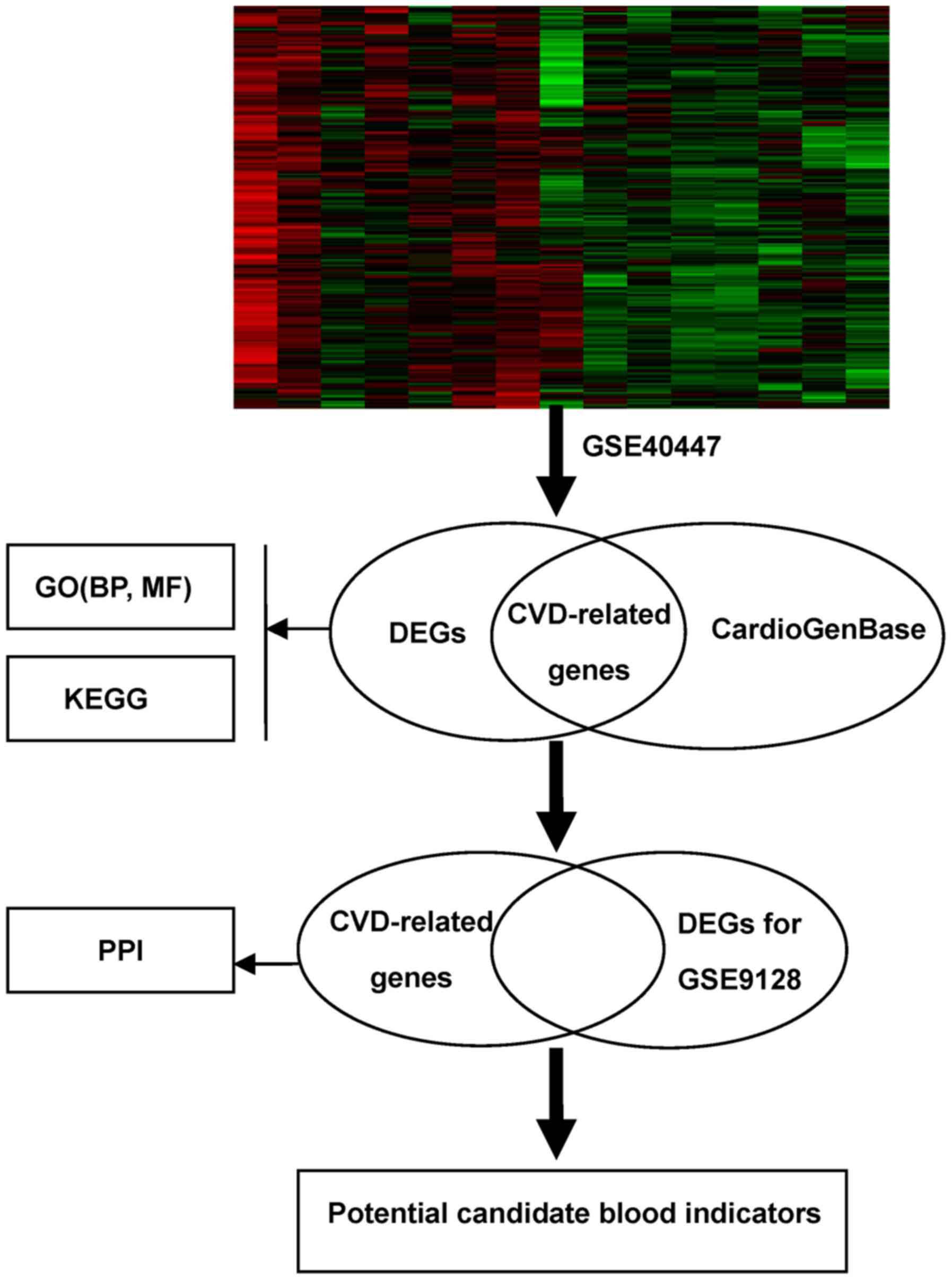
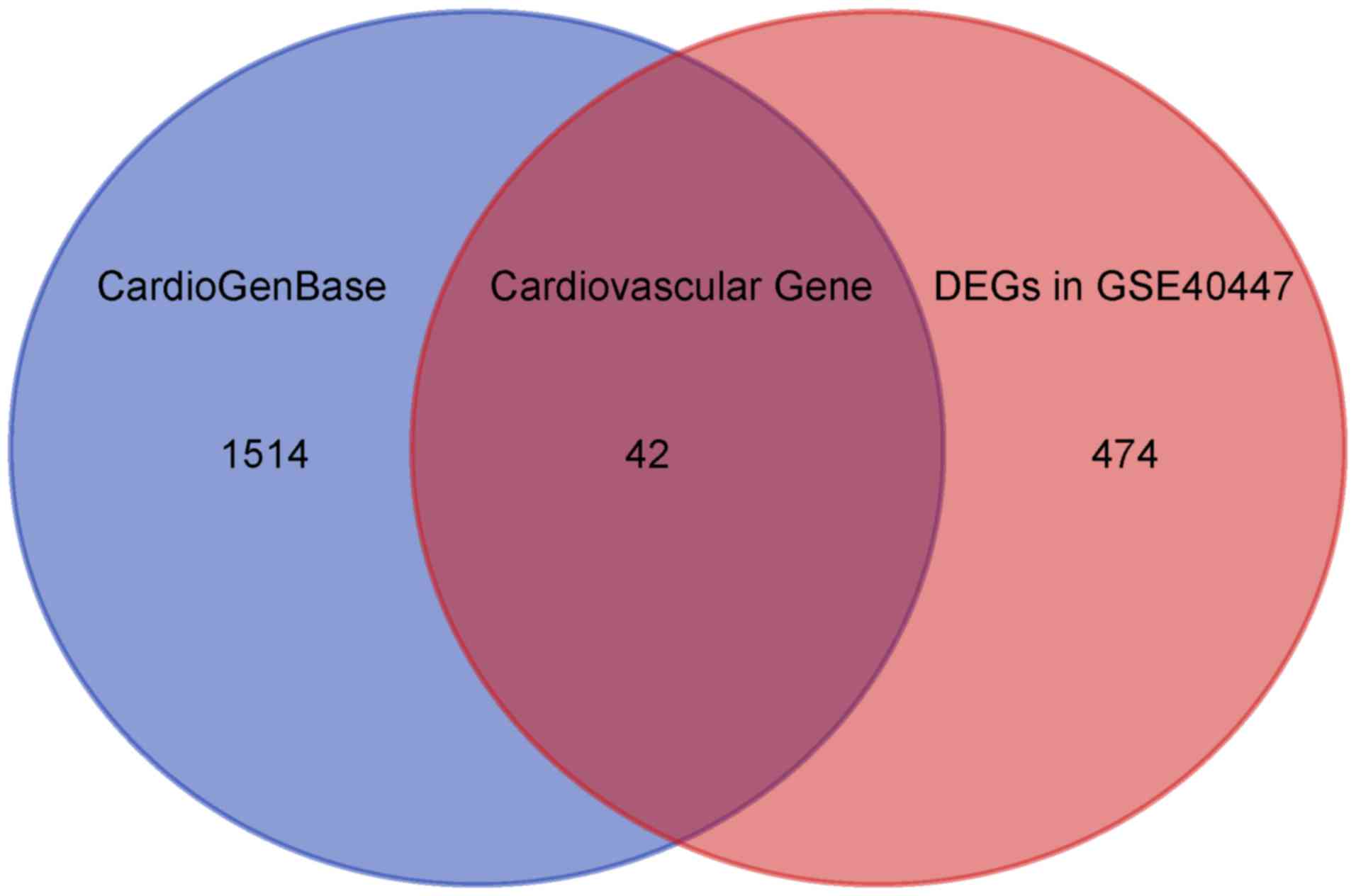
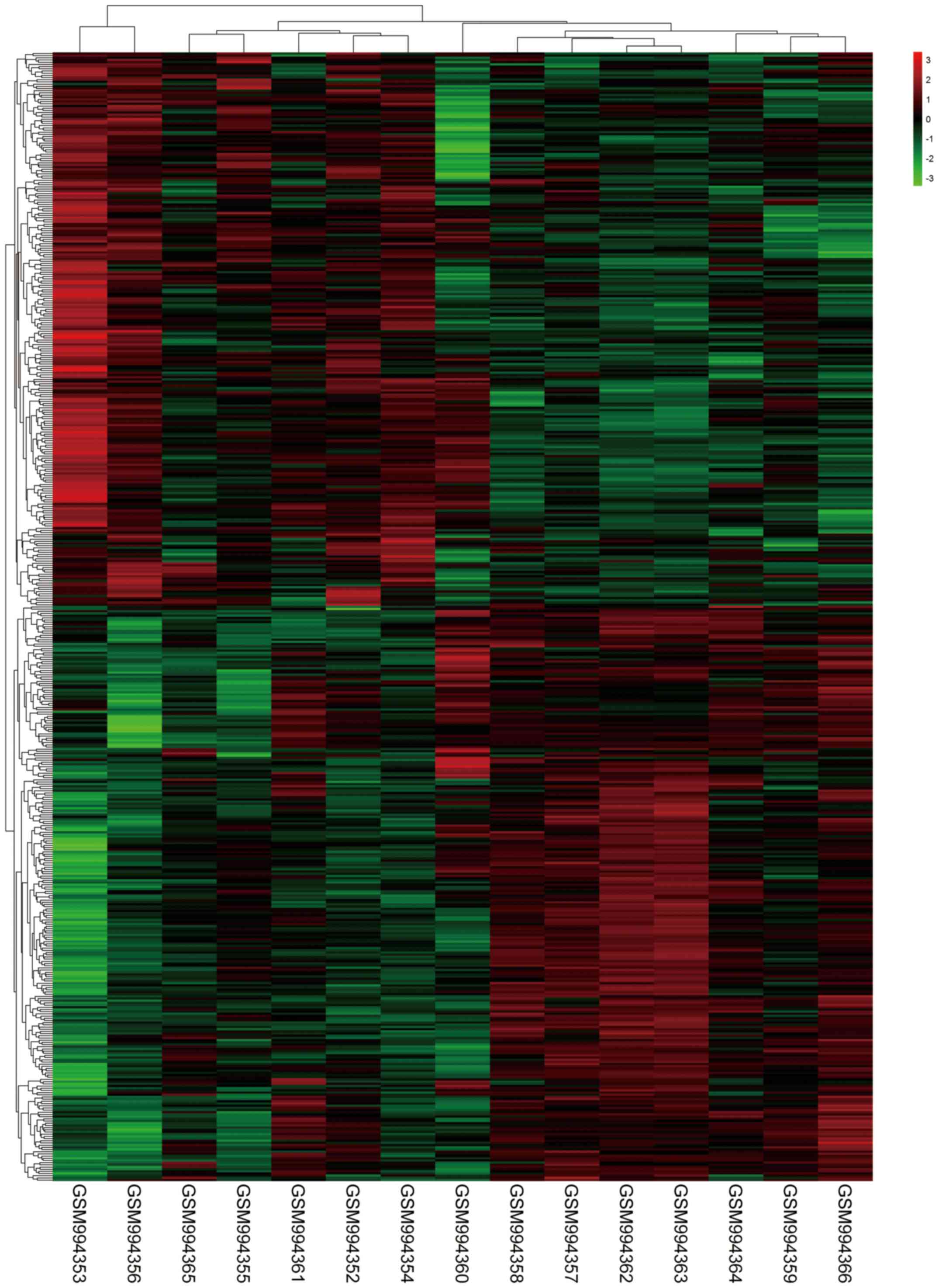
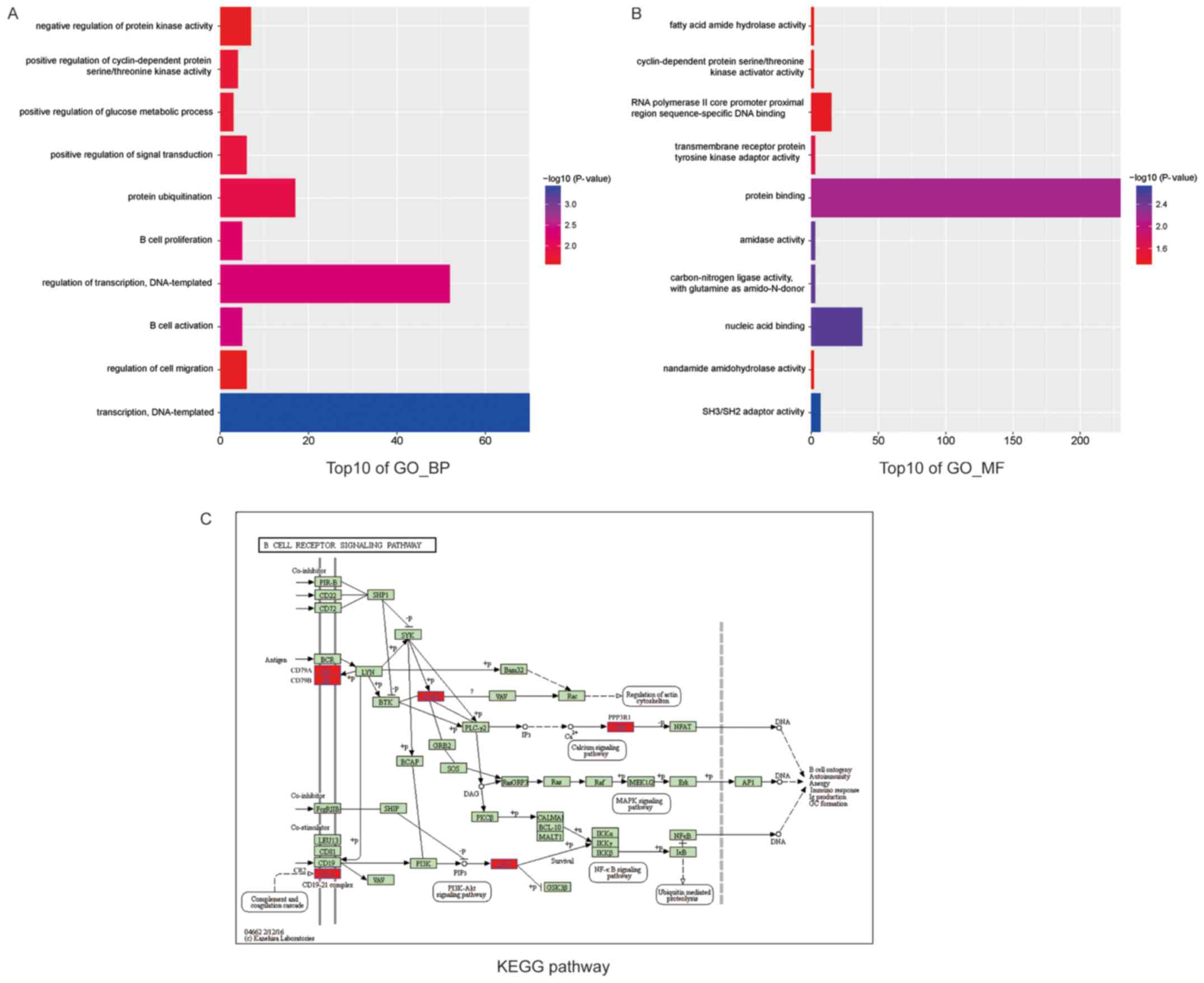
|
1
|
Curigliano G, Cardinale D, Dent S,
Criscitiello C, Aseyev O, Lenihan D and Cipolla CM: Cardiotoxicity
of anticancer treatments: Epidemiology, detection, and management.
CA Cancer J Clin. 66:309–325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shabalala S, Muller CJF, Louw J and
Johnson R: Polyphenols, autophagy and doxorubicin-induced
cardiotoxicity. Life Sci. 180:160–170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Argun M, Üzüm K, Sönmez MF, Özyurt A,
Derya K, Çilenk KT, Unalmış S, Pamukcu Ö, Baykan A, Narin F, et al:
Cardioprotective effect of metformin against doxorubicin
cardiotoxicity in rats. Anatol J Cardiol. 16:234–241.
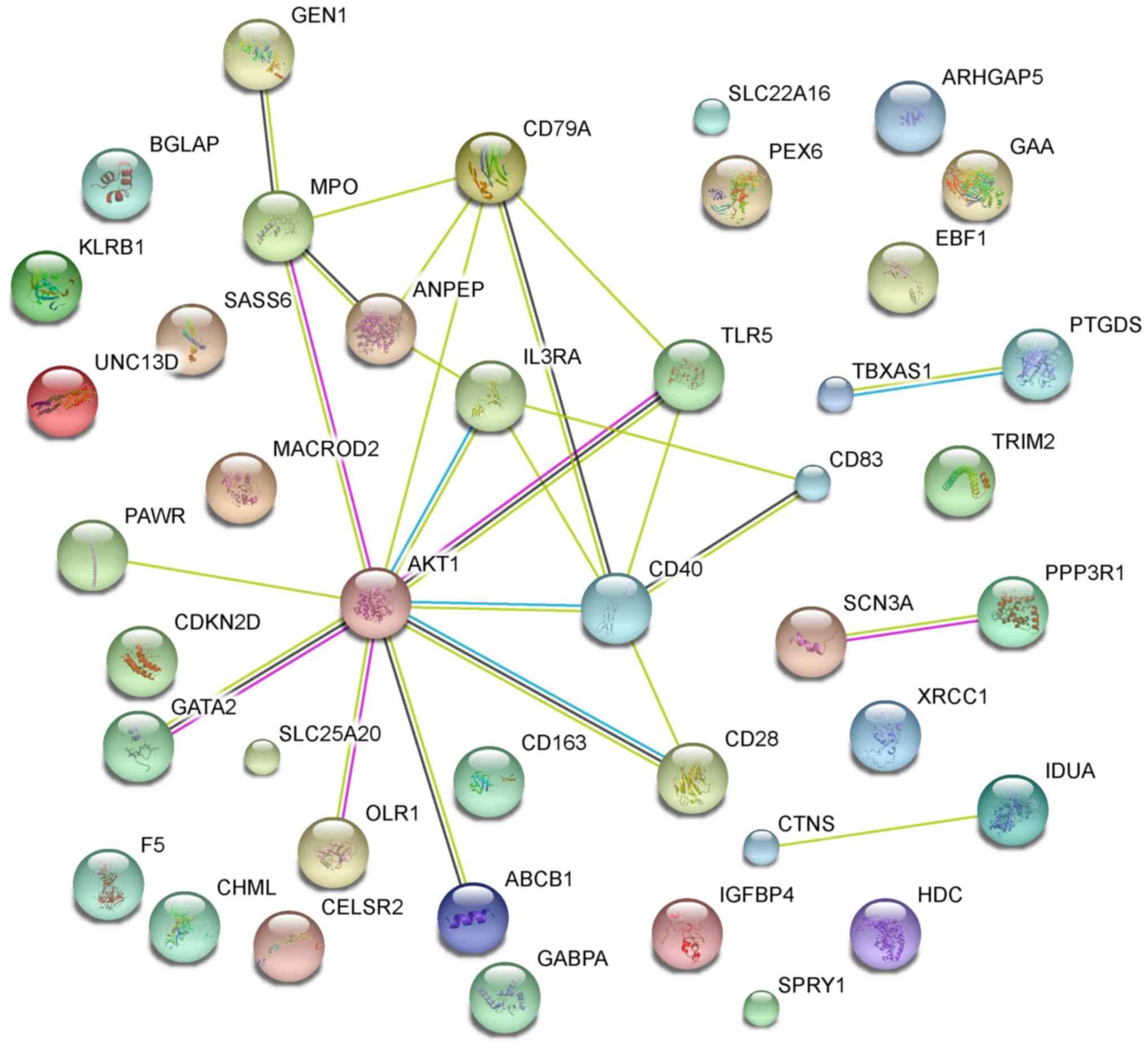
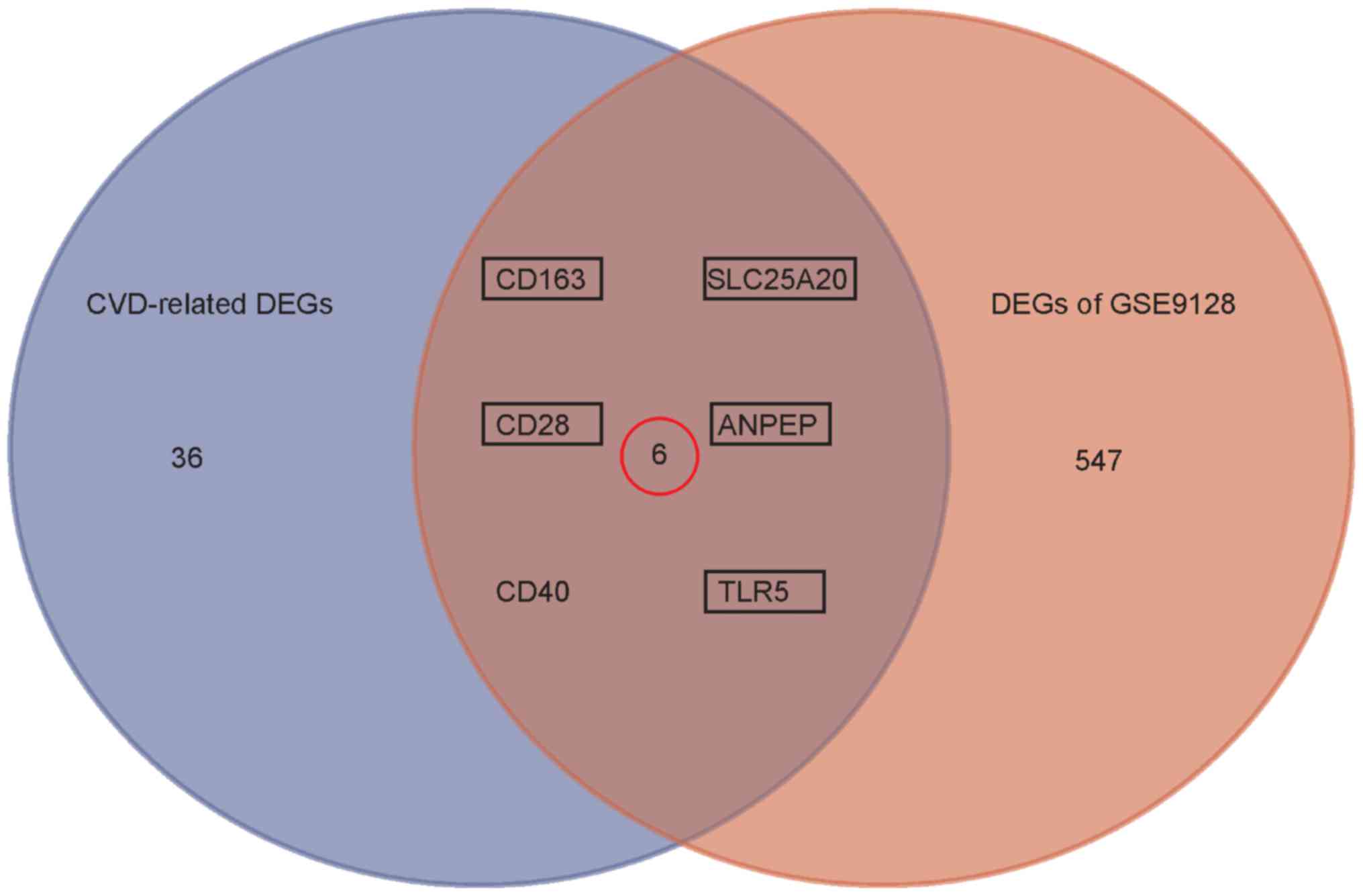
2016.PubMed/NCBI
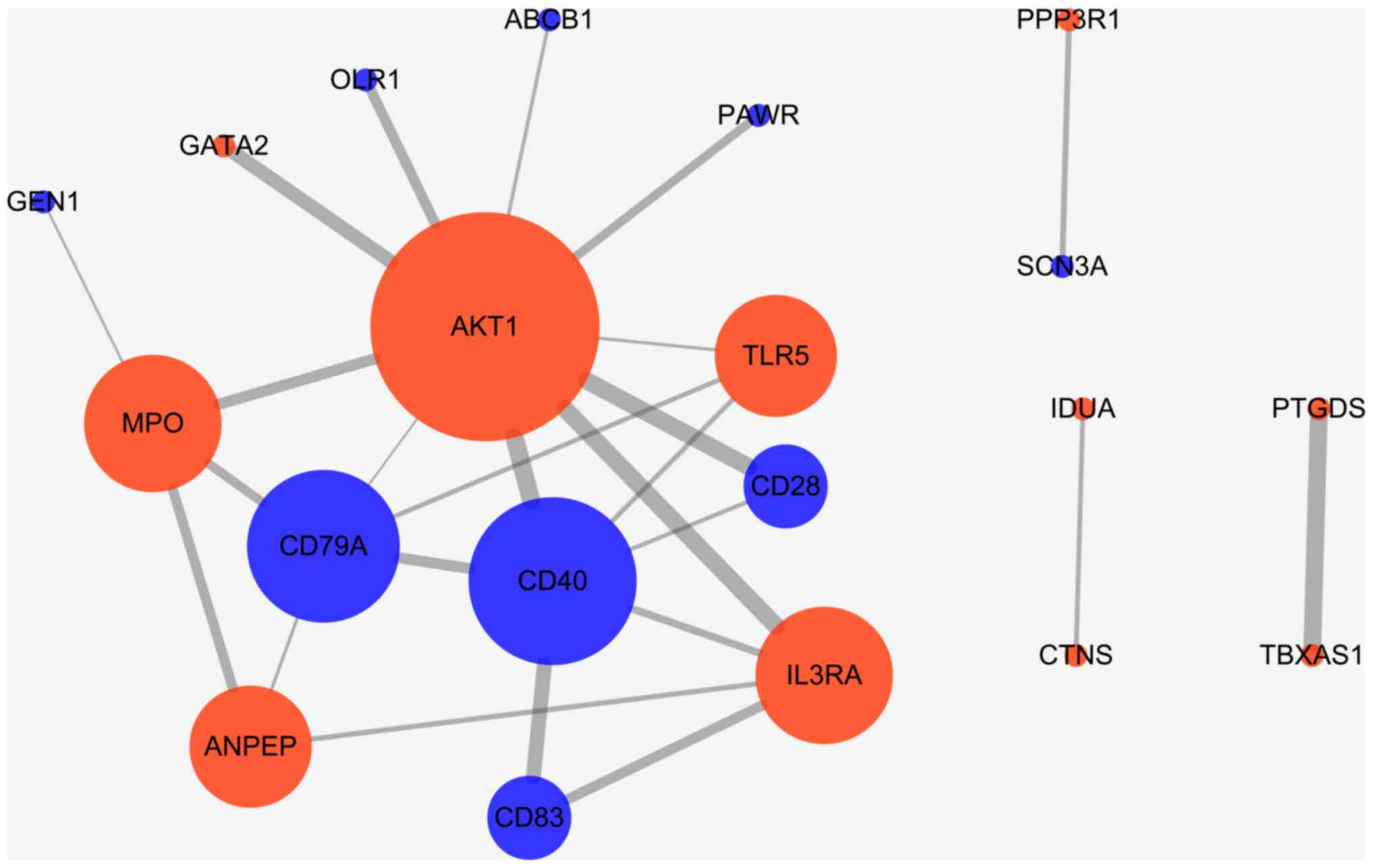
|
|
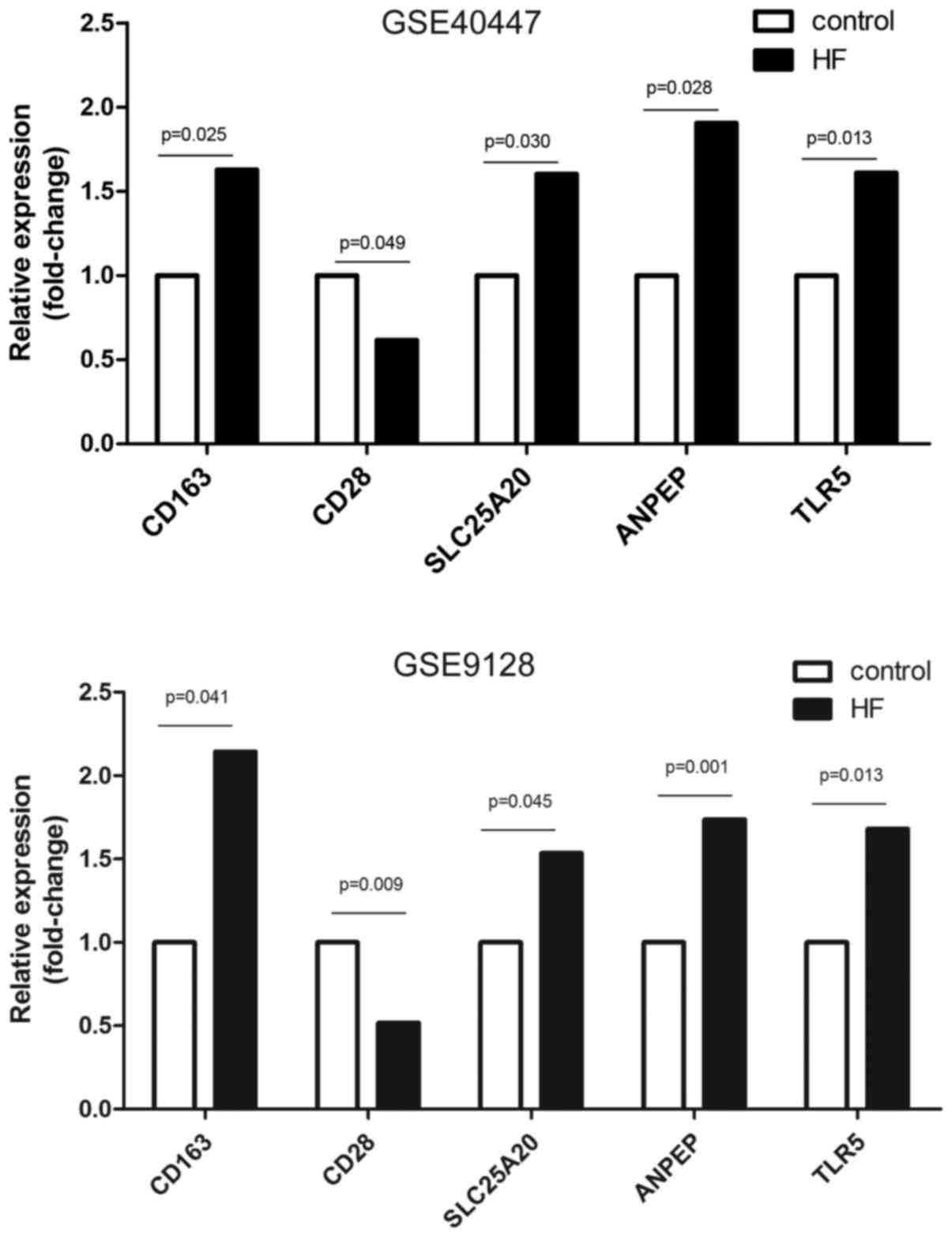
4
|
McGowan JV, Chung R, Maulik A, Piotrowska
I, Walker JM and Yellon DM: Anthracycline chemotherapy and
cardiotoxicity. Cardiovasc Drugs Ther. 31:63–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bahadir A, Kurucu N, Kadıoğlu M and
Yenilme E: The role of nitric oxide in Doxorubicin-induced
cardiotoxicity: Experimental study. Turk J Haematol. 31:68–74.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bryant J, Picot J, Baxter L, Levitt G,
Sullivan I and Clegg A: Use of cardiac markers to assess the toxic
effects of anthracyclines given to children with cancer: A
systematic review. Eur J Cancer. 43:1959–1966. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Conway A, McCarthy AL, Lawrence P and
Clark RA: The prevention, detection and management of cancer
treatment-induced cardiotoxicity: A meta-review. BMC Cancer.
15:3662015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Senkus E and Jassem J: Cardiovascular
effects of systemic cancer treatment. Cancer Treat Rev. 37:300–311.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai AD, Agarwal A, Steinberg M, Showler A,
Burry L, Tomlinson GA, Bell CM and Morris AM: Clinical predictors
and clinical prediction rules to estimate initial patient risk for
infective endocarditis in Staphylococcus aureus bacteraemia: A
systematic review and meta-analysis. Clin Microbiol Infect.
23:900–906. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lotrionte M, Biondi-Zoccai G, Abbate A,
Lanzetta G, D'Ascenzo F, Malavasi V, Peruzzi M, Frati G and
Palazzoni G: Review and meta-analysis of incidence and clinical
predictors of anthracycline cardiotoxicity. Am J Cardiol.
112:1980–1984. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jain D, Russell RR, Schwartz RG, Panjrath
GS and Aronow W: Cardiac complications of cancer therapy:
Pathophysiology, identification, prevention, treatment, and future
directions. Curr Cardiol Rep. 19:362017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cardinale D and Sandri MT: Role of
biomarkers in chemotherapy-induced cardiotoxicity. Prog Cardiovasc
Dis. 53:121–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dolci A, Dominici R, Cardinale D, Sandri
MT and Panteghini M: Biochemical markers for prediction of
chemotherapy-induced cardiotoxicity: Systematic review of the
literature and recommendations for use. Am J Clin Pathol.
130:688–695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fallah-Rad N, Walker JR, Wassef A, Lytwyn
M, Bohonis S, Fang T, Tian G, Kirkpatrick ID, Singal PK, Krahn M,
et al: The utility of cardiac biomarkers, tissue velocity and
strain imaging, and cardiac magnetic resonance imaging in
predicting early left ventricular dysfunction in patients with
human epidermal growth factor receptor II-positive breast cancer
treated with adjuvant trastuzumab therapy. J Am Coll Cardiol.
57:2263–2270. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McCaffrey TA, Tziros C, Lewis J, Katz R,
Siegel R, Weglicki W, Kramer J, Mak IT, Toma I, Chen L, et al:
Genomic profiling reveals the potential role of TCL1A and MDR1
deficiency in chemotherapy-induced cardiotoxicity. Int J Biol Sci.
9:350–360. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cappuzzello C, Napolitano M, Arcelli D,
Melillo G, Melchionna R, Di Vito L, Carlini D, Silvestri L,
Brugaletta S, Liuzzo G, et al: Gene expression profiles in
peripheral blood mononuclear cells of chronic heart failure
patients. Physiol Genomics. 38:233–240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Troyanskaya O, Cantor M, Sherlock G, Brown
P, Hastie T, Tibshirani R, Botstein D and Altman RB: Missing value
estimation methods for DNA microarrays. Bioinformatics. 17:520–525.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nayar PG, Murugesan R, Mary BPD and Ahmed
SS: CardioGenBase: A literature based Multi-Omics database for
major cardiovascular diseases. PLoS One. 10:e1431882015.
|
|
23
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(Database Issue): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Etzerodt A and Moestrup SK: CD163 and
inflammation: Biological, diagnostic, and therapeutic aspects.
Antioxid Redox Signal. 18:2352–2363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Landis RC, Philippidis P, Domin J, Boyle
JJ and Haskard DO: Haptoglobin genotype-dependent anti-inflammatory
signaling in CD163(+) macrophages. Int J Inflam. 2013:9803272013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhong SM, Qin YH, Li ZC and Wei YS:
Clinical value of detecting serum soluble CD163 level in patients
with atrial fibrillation. Nan Fang Yi Ke Da Xue Xue Bao.
36:1406–1409. 2016.(In Chinese). PubMed/NCBI
|
|
28
|
Zou LY, Peng CQ, Li CZ, Zhao CL, Zhu JM,
Liu JL and Zhang CX: Association between hemoglobin scavenger
receptor CD163 expression and coronary atherosclerotic severity in
patients with coronary heart disease. Zhonghua Xin Xue Guan Bing Za
Zhi. 37:605–609. 2009.(In Chinese). PubMed/NCBI
|
|
29
|
Ptaszynska-Kopczynska K,
Marcinkiewicz-Siemion M, Lisowska A, Waszkiewicz E, Witkowski M,
Jasiewicz M, Miklasz P, Jakim P, Galar B, Musial WJ and Kaminski
KA: Alterations of soluble TWEAK and CD163 concentrations in
patients with chronic heart failure. Cytokine. 80:7–12. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aruffo A and Seed B: Molecular cloning of
a CD28 cDNA by a high-efficiency COS cell expression system. Proc
Natl Acad Sci USA. 84:8573–8577. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Porciello N and Tuosto L: CD28
costimulatory signals in T lymphocyte activation: Emerging
functions beyond a qualitative and quantitative support to TCR
signalling. Cytokine Growth Factor Rev. 28:11–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sulzgruber P, Koller L, Winter MP, Richter
B, Blum S, Korpak M, Hulsmann M, Goliasch G, Wojta J and Niessner
A: The impact of CD4+CD28null T-lymphocytes
on atrial fibrillation and mortality in patients with chronic heart
failure. Thromb Haemost. 117:349–356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dumitriu IE, Araguás ET, Baboonian C and
Kaski JC: CD4+ CD28 null T cells in coronary artery disease: When
helpers become killers. Cardiovasc Res. 81:11–19. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kyaw T, Tipping P, Toh BH and Bobik A:
Killer cells in atherosclerosis. Eur J Pharmacol. 816:67–75. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kyaw T, Peter K, Li Y, Tipping P, Toh BH
and Bobik A: Cytotoxic lymphocytes and atherosclerosis:
Significance, mechanisms and therapeutic challenges. Br J
Pharmacol. 174:3956–3972. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Piggott K, Biousse V, Newman NJ, Goronzy
JJ and Weyand CM: Vascular damage in giant cell arteritis.
Autoimmunity. 42:596–604. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Garib FY and Rizopulu AP: T-regulatory
cells as part of strategy of immune evasion by pathogens.
Biochemistry (Mosc). 80:957–971. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu L, Zou LJ, Hua R and Li B: Association
of single-nucleotide polymorphisms in toll-like receptor 5 gene
with rheumatic heart disease in Chinese Han population. Int J
Cardiol. 145:129–130. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Koupenova M, Mick E, Mikhalev E, Benjamin
EJ, Tanriverdi K and Freedman JE: Sex differences in platelet
toll-like receptors and their association with cardiovascular risk
factors. Arterioscler Thromb Vasc Biol. 35:1030–1037. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mahapatra S, Ananth A, Baugh N, Damian M
and Enns GM: Triheptanoin: A rescue therapy for cardiogenic shock
in carnitine-acylcarnitine translocase deficiency. JIMD Rep.
39:19–23. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Choong K, Clarke JT, Cutz E, Pollit RJ and
Olpin SE: Lethal cardiac tachyarrhythmia in a patient with neonatal
carnitine-acylcarnitine translocase deficiency. Pediatr Dev Pathol.
4:573–579. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rubio-Gozalbo ME, Bakker JA, Waterham HR
and Wanders RJ: Carnitine-acylcarnitine translocase deficiency,
clinical, biochemical and genetic aspects. Mol Aspects Med.
25:521–532. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Danziger RS: Aminopeptidase N in arterial
hypertension. Heart Fail Rev. 13:293–298. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pereira FE, Cronin C, Ghosh M, Zhou SY,
Agosto M, Subramani J, Wang R, Shen JB, Schacke W, Liang B, et al:
CD13 is essential for inflammatory trafficking and infarct healing
following permanent coronary artery occlusion in mice. Cardiovasc
Res. 100:74–83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sánchez-Agesta Ortega R, Arias de
Saavedra-Alías JM, Liébana-Cañada A, Sánchez-Muñoz B,
Martínez-Martos JM and Ramírez-Expósito MJ: Circulating
aminopeptidase activities in men and women with essential
hypertension. Curr Med Chem. 20:4935–4945. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Buehler A, van Zandvoort MA, Stelt BJ,
Hackeng TM, Schrans-Stassen BH, Bennaghmouch A, Hofstra L,
Cleutjens JP, Duijvestijn A, Smeets MB, et al: cNGR: A novel homing
sequence for CD13/APN targeted molecular imaging of murine cardiac
angiogenesis in vivo. Arterioscler Thromb Vasc Biol. 26:2681–2687.
2006. View Article : Google Scholar : PubMed/NCBI
|