Introduction
Oral squamous cell carcinoma (OSCC) is the most
common malignancy of the oral cavity, accounting for >90% of all
oral cancer cases (1–3). Although substantial advances have been
made in the treatment of OSCC, including surgical resection
combined with chemotherapy and radiotherapy, the prognosis of
patients with advanced OSCC is unsatisfactory (4,5).
Therefore, it is necessary to explore underlying mechanisms of OSCC
tumourigenesis, which may be helpful in the development of novel
therapeutic targets.
Long non-coding RNAs (lncRNAs) are a group of small
and single-stranded non-coding RNAs that are >200 nt in size and
evolutionarily conserved (6,7). Although lncRNAs exhibit no
protein-coding capacity, they are key regulators of gene expression
and serve important roles in various biological processes,
including cell growth, migration, invasion and tumourigenesis
(8,9). In recent years, evidence has suggested
that lncRNAs serve promoting or tumour suppressive roles in the
development and malignant progression of OSCC (10,11). For
instance, lncRNA taurine upregulated gene 1 promotes progression of
OSCC through upregulation of formin-like 2 by acting as molecular
sponges for microRNA (miR)-219 (12). LncRNA maternally expressed 3 inhibits
proliferation and metastasis of OSCC via regulation of the
WNT/β-catenin signalling pathway (13). Recently, lncRNA nuclear enriched
abundant transcript 1 (NEAT1) has been observed to be upregulated
in several human cancer types and studies have revealed its
oncogenic role (14–16). For example, NEAT1 is regulated by the
epidermal growth factor receptor signalling pathway and contributes
to glioblastoma progression through the WNT/β-catenin pathway by
serving as a scaffold for enhancer of zeste homolog 2 (15). NEAT1 contributes to paclitaxel
resistance in ovarian cancer cells through regulation of zinc
finger E-box-binding homeobox 1 expression via miR-194 (16). However, to the best of our knowledge,
the function of NEAT1 in OSCC has not yet been reported.
miRs are a class of small non-coding RNAs that are
composed of ~22–25 nt and have been demonstrated to regulate gene
expression via binding to the 3′-untranslated region (3′-UTR) of
their target mRNAs, which results in mRNA degradation or
translation repression (17,18). Similar to lncRNAs, miRs are also
involved in development and progression of tumours, including OSCC
(2,3,17–19).
Recently, miR-365 has been reported to serve a general
tumour-suppressive role in multiple human cancer types; for
instance, inhibiting ovarian cancer progression by targeting Wnt5a
(20). Additionally, miR-365
inhibits proliferation, migration and invasiveness of glioma cells
by targeting phosphoinositide-3-kinase regulatory subunit 3
(21). However, to the best of our
knowledge, the exact role of miR-365 in OSCC has not been
reported.
In addition, matrix metalloproteinase (MMP)2 and
MMP9 are two key enzymes involved in tumour cell invasiveness
(22). Whether these enzymes are
involved in the function of NEAT1 and miR-365 in OSCC still remains
unclear.
The present study aimed to investigate NEAT1 and
miR-365 expression levels and functions in OSCC, as well as their
underlying molecular mechanism in vitro.
Materials and methods
Clinical tissues
The current study was approved by the Research
Ethics Committee of the Stomatological Hospital of Jinan City
(Jinan, China). A total of 58 OSCC and adjacent non-tumour tissues
were collected from patients with primary OSCC at the
Stomatological Hospital of Jinan City between March 2010 and April
2012. No patient received chemotherapy or radiotherapy before
surgical resection. The clinicopathological characteristics of the
patients are summarized in Table I.
Written informed consent was obtained from all patients. Tissues
were immediately snap-frozen in liquid nitrogen and stored at −80°C
until use.
 | Table I.Association between NEAT1 expression
and clinicopathological characteristics in patients with oral
squamous cell carcinoma. |
Table I.
Association between NEAT1 expression
and clinicopathological characteristics in patients with oral
squamous cell carcinoma.
|
|
| NEAT1 expression |
|
|---|
|
|
|
|
|
|---|
| Variable | Number (n=58) | Low (n=32) | High (n=26) | P-value |
|---|
| Age (years) |
|
|
| 0.791 |
| ≤55 | 24 | 14 | 10 |
|
|
>55 | 34 | 18 | 16 |
|
| Sex |
|
|
| 0.183 |
| Male | 35 | 22 | 13 |
|
|
Female | 23 | 10 | 13 |
|
| Differentiation
grade |
|
|
| 0.213 |
| Well and
moderately | 45 | 27 | 18 |
|
| Poor | 13 | 5 | 8 |
|
| Lymph node
metastasis |
|
|
| 0.009 |
|
Present | 18 | 5 | 13 |
|
|
Absent | 40 | 27 | 13 |
|
| Tumor, node and
metastasis stage |
|
|
| 0.007 |
| I–II | 36 | 25 | 11 |
|
|
III–IV | 22 | 7 | 15 |
|
Cell culture and transfection
Human OSCC cell lines HN4, Tca-8113, UM-SCC-1,
Cal-27, SCC-25 and SCCKN, and the normal human oral keratinocyte
cell line hNOK were purchased from the Cell Bank of the Chinese
Academy of Sciences (Shanghai, China). All cell lines were cultured
in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Thermo Fisher Scientific, Inc.) at 37°C in
humidified atmosphere with 5% CO2. Lipofectamine 2000
(Thermo Fisher Scientific, Inc.) was used to transfect HN4 and
Tca-8113 cells with 100 nM of non-specific short interfering RNA
(NC siRNA; Am10301; Amspring, Changsha, China), NEAT1-specific
siRNA (NEAT1 siRNA; Am10542; Amspring), pcDNA3.1 vector (Am00013;
Amspring), pcDNA-NEAT1 expression plasmid (Am02051; Amspring),
miR-365 inhibitor (HmiR-AN0451-SN-10; Guangzhou Fulengen Co., Ltd.,
Guangzhou, China) and negative control (NC) inhibitor
(CmiR-AN0001-SN; Guangzhou Fulengen Co., Ltd.) according to the
manufacturer's instructions. Subsequent experiments were performed
48 h following transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cells using
TRIzol reagent (Thermo Fisher Scientific, Inc.), while genomic DNA
was removed by treatment with DNase (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Total RNA (1
µg) was reverse transcribed using a RevertAid First Stand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Reverse transcription was performed at
16°C for 30 min, followed by incubation at 42°C for 30 min and
enzyme inactivation at 85°C for 5 min. The expression of miR-365
was detected using miScript SYBR Green PCR kit (Qiagen, Inc.,
Valencia, CA, USA) on an ABI 7500 PCR machine (Thermo Fisher
Scientific, Inc.). U6 was used as the internal reference. To detect
NEAT1 expression, qPCR was performed using Power SYBR Green PCR
Master mix (Thermo Fisher Scientific, Inc.). GAPDH was used as the
internal reference. The reaction conditions for all qPCR
experiments were as follows: 95°C for 5 min, followed by 40 cycles
of 95°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec. The
relative expression was determined using the 2−∆∆Cq
method (23). The following primers
were used: U6, forward 5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse
5′-CGCTTCACGAATTTGCGTGT-3′; GAPDH, forward
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′. miR-365 primers were purchased from
Guangzhou Fulengen Co., Ltd.
Bioinformatics analysis
Target genes for NEAT1 and miR-365 were predicted
using RNAhybrid 2.12 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/).
Luciferase reporter gene assay
Fragments of NEAT1 containing wild-type (WT) or
mutant type (MT) miR-365 binding sites were cloned into pmirGLO
Dual-luciferase Target Expression Vector (Promega Corporation,
Madison, WI, USA), which generated WT or MT NEAT1 plasmids. HN4 and
Tca-8113 cells were co-transfected with miR-365 mimic, scramble miR
mimic (miR-NC), WT or MT NEAT1 plasmid using Lipofectamine 2000.
Following 48 h of transfection, luciferase reporter gene assays
were performed using the Dual-Luciferase Reporter Assay System
(Promega Corporation). The firefly luciferase activity was
normalized against Renilla luciferase activity.
Wound-healing assay
Wound-healing assays were conducted to determine the
migratory capacity of cells. HN4 and Tca-8113 cells were cultured
to full confluence, wounds of ~1 mm in width were generated with a
plastic scriber and cells were washed with PBS. Cells were then
cultured at 37°C with 5% CO2 for 48 h and assessed with
an inverted microscope (magnification, ×40).
Transwell assay
HN4 and Tca-8113 cells (10,000/well) in DMEM were
added to the upper chamber of Transwell inserts, pre-coated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) and DMEM
supplemented with 10% FBS was added to the lower chamber. HN4 and
Tca-8113 cells were then incubated at 37°C for 24 h. HN4 and
Tca-8113 cells that had not migrated through the membrane of the
insert were removed using a cotton-tipped swab, while the cells on
the lower surface of the membrane were stained with gentian violet
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at room temperature
for 10 min and counted under an inverted microscope (magnification,
×400).
Western blotting
Tissues and cells were lysed in cold
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) and the protein concentration was determined using a
Bicinchoninic Acid Protein Assay kit (Thermo Fisher Scientific,
Inc.). Proteins (50 µg) were separated on 10% SDS-PAGE gels and
transferred to a polyvinylidene difluoride membrane (Thermo Fisher
Scientific, Inc.). The membrane was blocked in 5% non-fat milk in
PBS containing 0.1% Tween-20 (Sigma-Aldrich; Merck KGaA) at room
temperature for 3 h. It was then incubated with rabbit polyclonal
anti-human MMP2 (1:200; ab37150; Abcam, Cambridge, MA, USA), rabbit
polyclonal anti-human MMP9 (1:200; ab38898; Abcam) or rabbit
polyclonal anti-human GAPDH (1:100; ab9485; Abcam) at room
temperature for 3 h. This was followed by incubation with the
horseradish peroxidase-conjugated goat anti-rabbit secondary IgG
antibody (1:5,000; ab6721; Abcam) at room temperature for 1 h.
Enhanced chemiluminescence (Thermo Fisher Scientific, Inc.) was
used to examine protein expression, which was analysed using
Image-Pro Plus software 6.0 (Media Cybernetics, Inc., Rockville,
MD, USA) according to the manufacturer's protocol.
Statistical analysis
All data are presented as the mean ± standard
deviation. SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) was used for
statistical analysis. Student's t-test was used for comparisons
between two groups, while one-way analysis of variance followed by
Tukey's post hoc test was used for comparisons of >2 groups. The
associations between NEAT1 expression and the clinicopathological
characteristics of OSCC were examined using the χ2 test.
The Kaplan-Meier method was applied to analyse overall survival of
patients with OSCC. Pearson's correlation was used to analyze the
correlation between NEAT1 and miR-365 expression in OSCC tissues.
P<0.05 was considered to indicate a statistically significant
difference.
Results
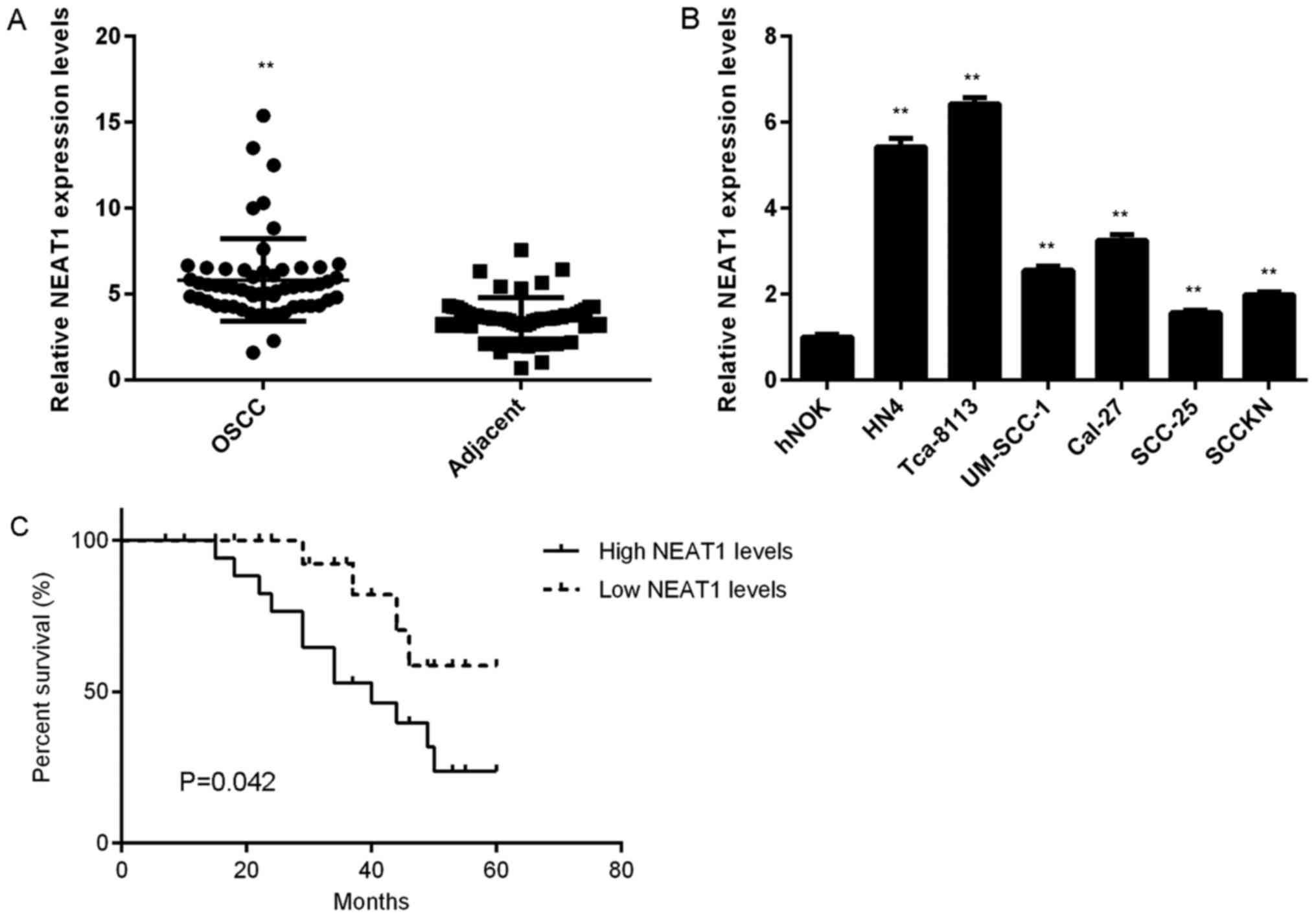
NEAT1 is upregulated in OSCC
In the present study, NEAT1 expression in OSCC and
adjacent non-tumour tissue was evaluated. RT-qPCR assay data
indicated that NEAT1 was significantly upregulated in OSCC tissue
compared with adjacent non-tumour tissue (Fig. 1A). Based on the mean expression value
of NEAT1 as cut-off value (5.54), these patients with OSCC were
divided into high and low expression groups. Further investigation
indicated that increased NEAT1 expression was significantly
associated with positive lymph node metastasis and advanced
clinical stage (Table I). NEAT1 was
significantly upregulated in OSCC cell lines compared with hNOK
cells (Fig. 1B). Additionally,
patients with OSCC and high NEAT1 expression exhibited a shorter
survival time compared with patients with low NEAT1 expression
(Fig. 1C).
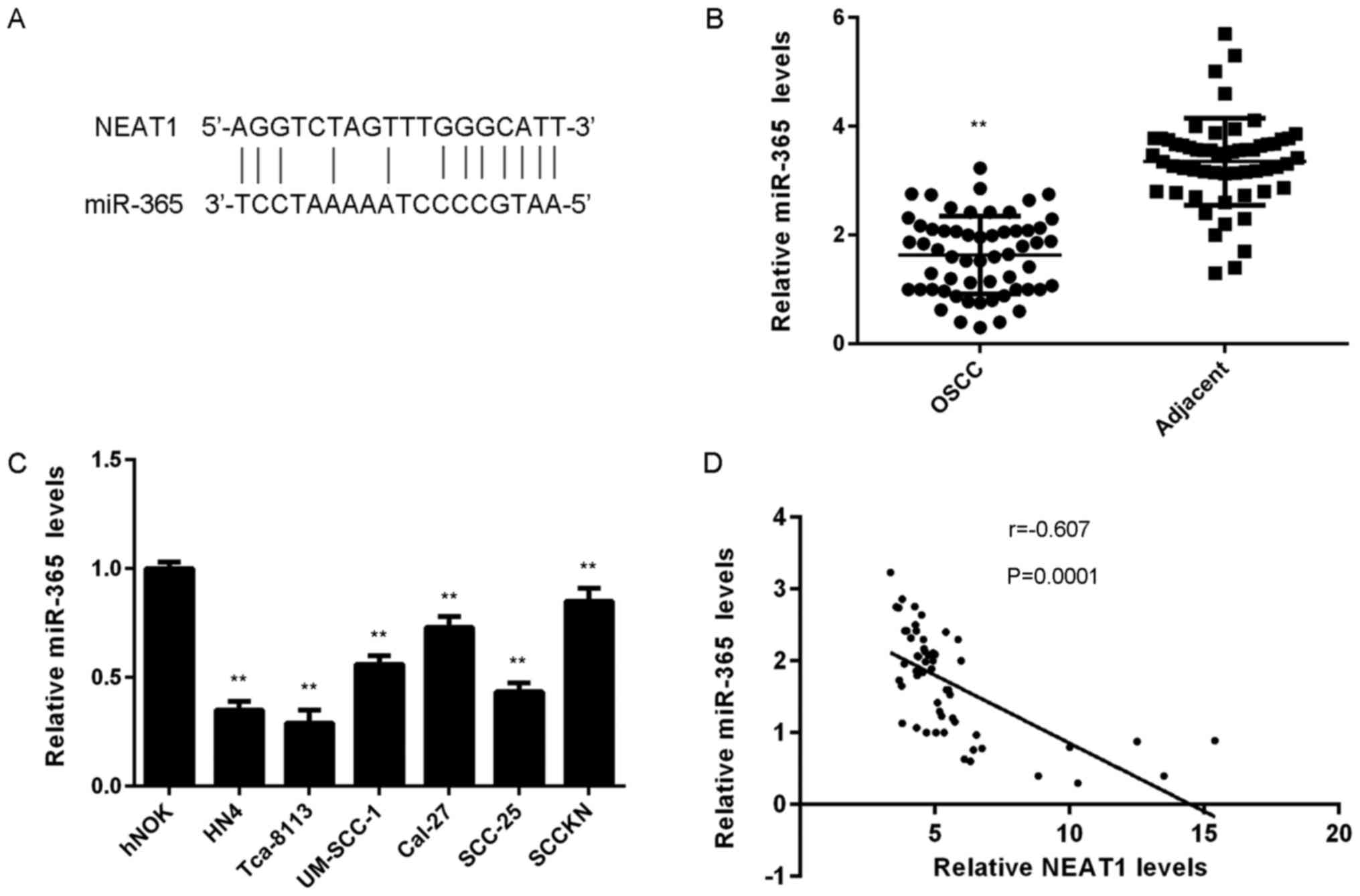
MiR-365 is downregulated in OSCC
Potential target miRs of NEAT1 in OSCC cells were
evaluated. A bioinformatics analysis revealed that miR-365 was
predicted to bind to NEAT1 (Fig.
2A). To examine the association between NEAT1 and miR-365 in
OSCC, miR-365 levels in OSCC tissue and cell lines were determined.
As indicated in Fig. 2B, RT-qPCR
assay data demonstrated that miR-365 expression was significantly
lower in OSCC tissue compared with adjacent non-tumour tissue. In
addition, miR-365 expression was significantly reduced in OSCC cell
lines compared with hNOK cells (Fig.
2C). It was demonstrated that miR-365 was downregulated in
OSCC. In addition, the present study revealed an inverse
correlation between NEAT1 and miR-365 expression in OSCC tissue
(Fig. 2D), which suggests that
reduced miR-365 expression may be involved in OSCC development.
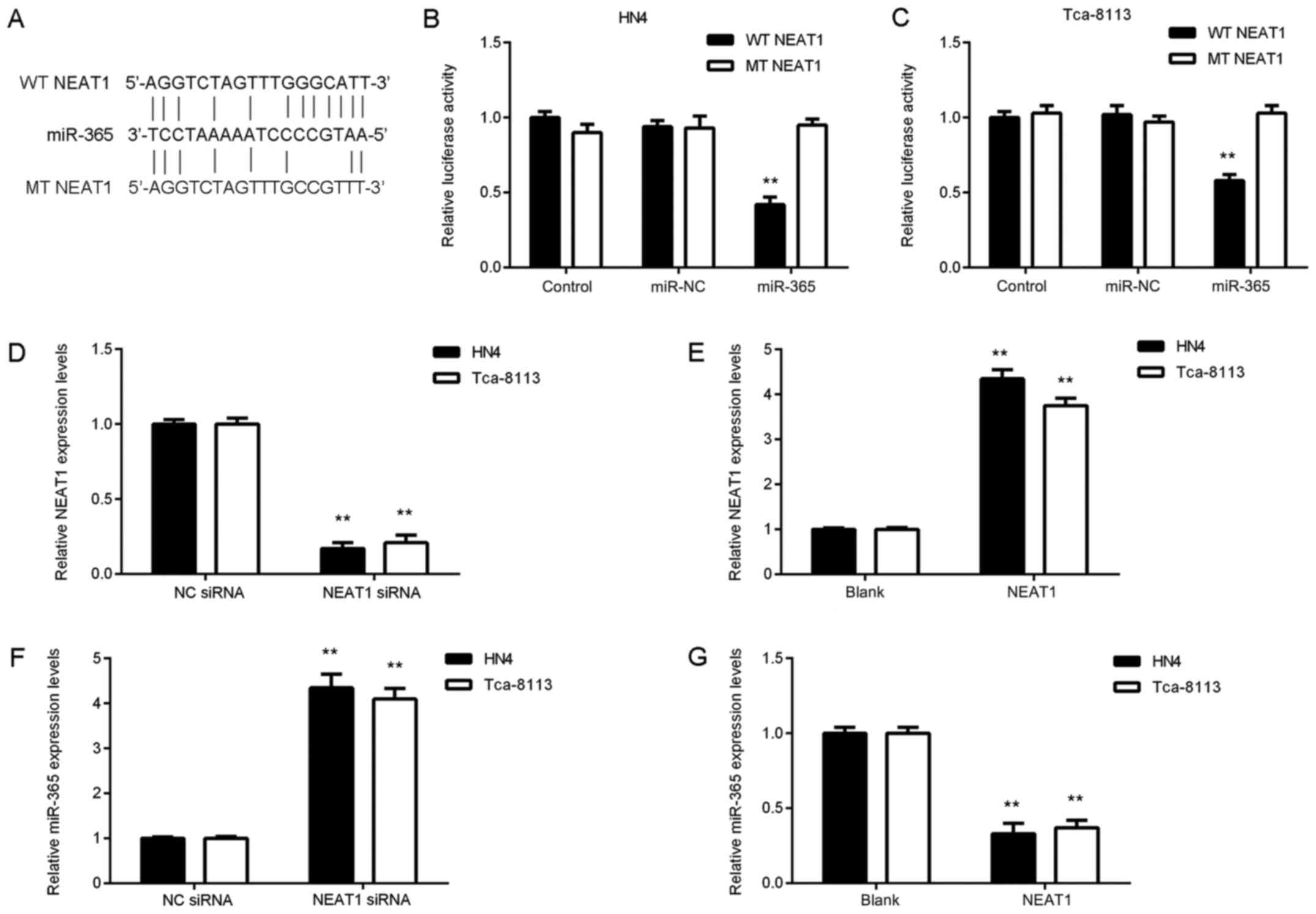
NEAT1 downregulates miR-365 expression
via sponging in OSCC cells
As HN4 and Tca-8113 cells exhibited the highest
expression levels of NEAT1 among the tested OSCC cell lines, these
cell lines were selected for in vitro experiments. To
further clarify the association between miR-365 and NEAT1 in HN4
and Tca-8113 cells, a luciferase reporter plasmid containing WT or
MT miR-365 binding sites of NEAT1 was constructed in the present
study (Fig. 3A). A luciferase
reporter gene assay was then performed. The data revealed that
transfection with an miR-365 mimic significantly inhibited
luciferase activity of WT NEAT1 in OSCC cells but did not affect
luciferase activity of MT NEAT1 (Fig. 3B
and C). These findings suggest that NEAT1 may be able to sponge
miR-365 in OSCC cells.
Effects of NEAT1 on miR-365 expression
of in OSCC cells were evaluated
HN4 and Tca-8113 cells were transfected with NEAT1
siRNA to knock down NEAT1 levels or cells were transfected with
NEAT1 plasmid to upregulate its expression. Following transfection,
NEAT1 expression was significantly decreased in cells transfected
with NEAT1 siRNA compared with the NC siRNA group and was
significantly increased in the NEAT1 group compared with the blank
control group (Fig. 3D and E). The
current study demonstrated that knockdown of NEAT1 enhanced miR-365
expression in OSCC cells and NEAT1 overexpression significantly
inhibited miR-365 expression (Fig. 3F
and G). These findings indicated that NEAT1 downregulates
miR-365 expression via sponging in OSCC cells.
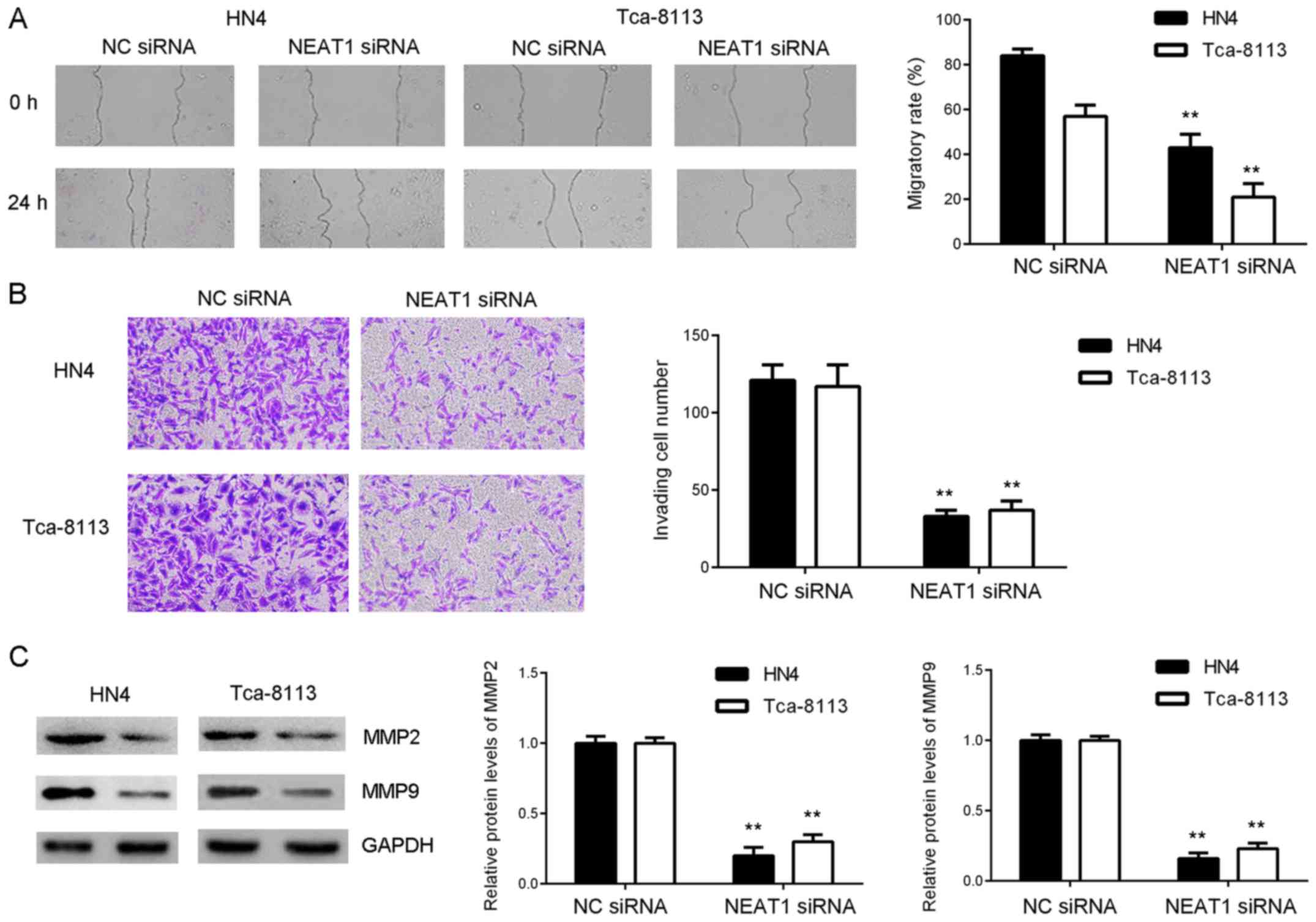
Knockdown of NEAT1 inhibits OSCC cell
invasion
To further study the function of NEAT1 in OSCC
cells, wound-healing assays were performed to examine effects of
NEAT1 downregulation on OSCC cell migration. As presented in
Fig. 4A, migratory capacity of HN4
and Tca-8113 cells was significantly reduced in the NEAT1 siRNA
group compared with the NC siRNA group. Transwell assays were
performed to evaluate cell invasion. As presented in Fig. 4B, invasiveness of HN4 and Tca-8113
cells in the NEAT1 siRNA group was significantly reduced following
inhibition of NEAT1 expression. Consistently, western blot data
indicated that protein expression of MMP2 and MMP9, two key enzymes
involved in tumour cell invasiveness (22), was significantly reduced in the NEAT1
siRNA group compared with the NC siRNA group (Fig. 4C). These findings indicate that
knockdown of NEAT1 expression inhibits OSCC cell migration and
invasion.
Inhibition of NEAT1 suppresses OSCC
cell migration and invasion by sponging miR-365
Based on the aforementioned findings, it was
speculated that NEAT1 may regulate migration and invasiveness of
OSCC cells by sponging miR-365. To clarify this, NEAT1
siRNA-transfected OSCC cells were co-transfected with NC inhibitor
or miR-365 inhibitor. Following co-transfection, miR-365 levels
were significantly reduced in the NEAT1 siRNA+miR-365 inhibitor
group compared with the NEAT1 siRNA+NC inhibitor group (Fig. 5A). Further investigation indicated
that migration ability and invasiveness of OSCC cells was increased
in the NEAT1 siRNA+miR-365 inhibitor group compared with the NEAT1
siRNA+NC inhibitor group (Fig. 5B and
C). Consistently, suppressive effects of NEAT1 inhibition on
MMP2 and MMP9 protein expression were reversed following
co-transfection with miR-365 inhibitor (Fig. 5D). These findings demonstrated that
inhibition of NEAT1 suppressed OSCC migration and invasiveness via
inhibition of miR-365 sponging.
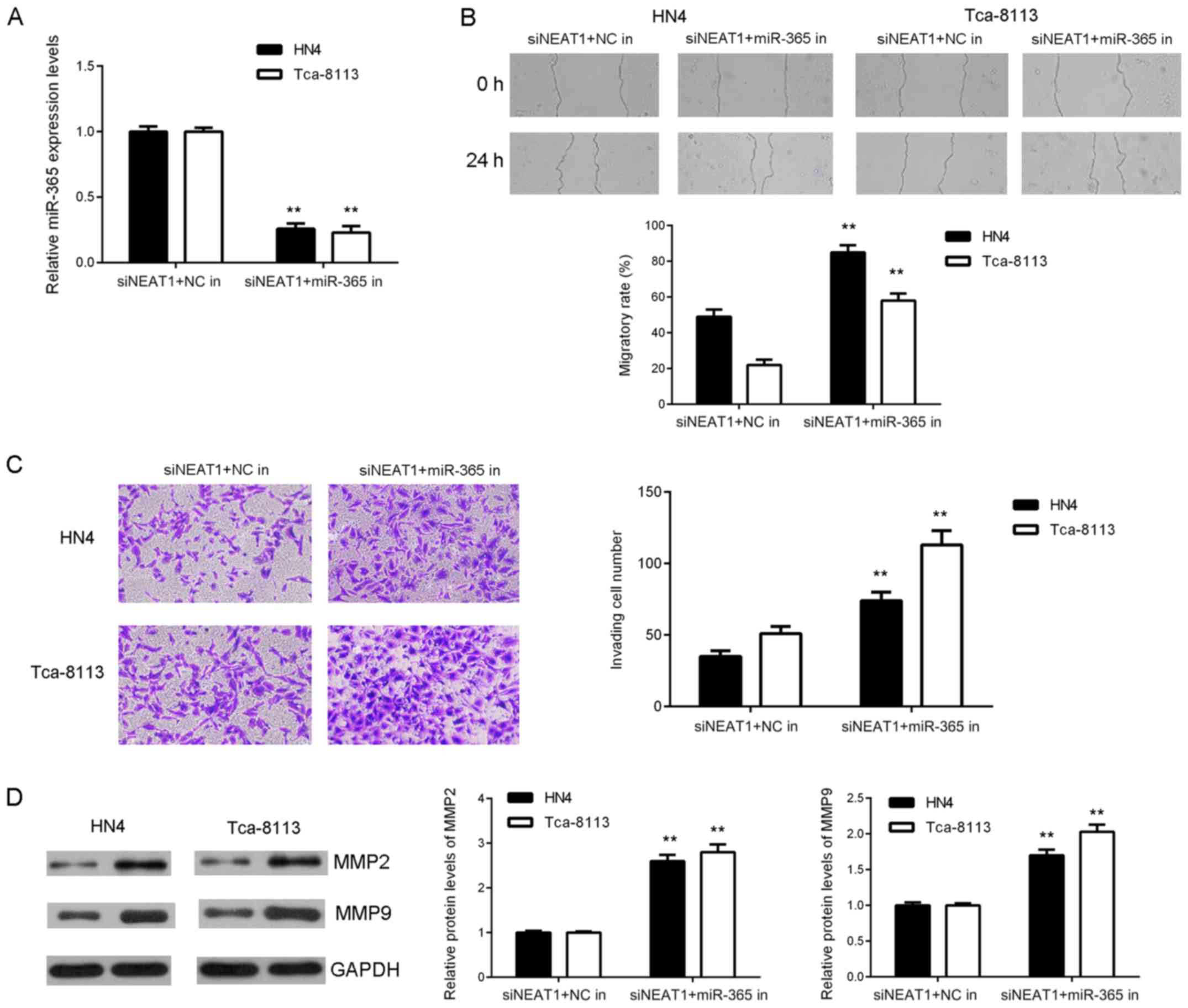 | Figure 5.Inhibition of NEAT1 suppresses OSCC
cell migration and invasion by sponging miR-365. NEAT1 siRNA
transfected OSCC cells co-transfected with NC or miR-365 inhibitor.
(A) Reverse transcription-quantitative polymerase chain reaction
analysis of miR-365 levels. (B) Wound healing (magnification, ×40)
and (C) Transwell assay performed to examine cell migration and
invasion (magnification, ×400). (D) Western blot analysis of MMP2
and MMP9 protein expression. **P<0.01 vs. siNEAT1+NCin. NEAT1,
nuclear enriched abundant transcript 1; siRNA, short interfering
RNA; OSCC, oral squamous cell carcinoma; NC, negative control;
miR-365, microRNA-365; MMP, matrix metalloproteinase; NC, negative
control; in, inhibitor. |
Discussion
To the best of our knowledge, the function and
regulatory mechanism of NEAT1 in OSCC has not been reported
previously. The present study revealed that NEAT1 was significantly
upregulated in OSCC tissue and cell lines and that its upregulation
was significantly associated with advanced clinical stage and
shorter survival time of patients with OSCC. Data from luciferase
reporter gene assays confirmed an interaction between miR-365 and
NEAT1 and demonstrated that NEAT1 negatively regulated miR-365
expression in two OSCC cell lines. Furthermore, inhibition of NEAT1
expression led to a significant decrease in OSCC cell migration and
invasion, whereas inhibition of miR-365 eliminated NEAT1
knockdown-induced suppressive effects on OSCC cell migration and
invasion. In addition, miR-365 was significantly downregulated in
OSCC tissue and cell lines and an inverse correlation was observed
between miR-365 and NEAT1 expression in OSCC tissue.
Previous studies have demonstrated that NEAT1
functions as an oncogene in multiple types of human cancer
(24,25). For instance, NEAT1 promotes laryngeal
squamous cell cancer through regulation of the miR-107/CDK6
signalling pathway (24). NEAT1 is
an unfavourable prognostic factor in gastric cancer, as it promotes
tumour cell migration and invasion (25). However, the role of NEAT1 in OSCC
remains unclear. In the present study, it was demonstrated that
NEAT1 expression was significantly higher in OSCC tissue and cell
lines compared with adjacent non-tumour tissue and normal oral
keratinocytes, respectively. Additionally, upregulation of NEAT1
may be associated with lymph node metastasis, advanced clinical
stage and poor prognosis of patients with OSCC. Similarly,
increased NEAT1 expression was further associated with unfavourable
clinical characteristics and poor prognosis in ovarian cancer
(26), oesophageal squamous cell
carcinoma (27), gastric cancer
(28) and liver cancer (29).
The present study investigated underlying regulatory
mechanisms of NEAT1 in OSCC metastasis. A bioinformatics analysis
predicted that miR-365 is a potential target of NEAT1. Luciferase
reporter gene assays revealed that transfection with a miR-365
mimic significantly inhibited luciferase activity of WT NEAT1 in
OSCC cells but did not affect luciferase activity of MT NEAT1,
which indicates that NEAT1 may sponge miR-365 in OSCC cells. To
further study the relationship between miR-365 and NEAT1 in OSCC,
experiments that demonstrated that miR-365 was significantly
downregulated in OSCC tissue and cell lines were performed. The
current study observed an inverse correlation between miR-365 and
NEAT1 in OSCC tissue.
The function of NEAT1 in OSCC metastasis was
examined in vitro and it was revealed that knockdown of
NEAT1 reduced OSCC cell migration and invasion. As miR-365
expression was downregulated by NEAT1 in OSCC cells, it was
speculated that miR-365 may be involved in NEAT1-mediated OSCC cell
migration and invasion. Experimental data confirmed suppressive
effects of NEAT1 inhibition on OSCC cell migration and invasion, as
well as downregulation of MMP2 and MMP9 protein expression.
To the best of our knowledge, this is the first
study to report that NEAT1 promotes migration and invasiveness of
OSCC cells by sponging miR-365. Therefore, the current study
suggests that NEAT1 may be used as a novel therapeutic target when
treating patients with OSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XL wrote the manuscript and performed the
experiments. WS collected clinical samples and performed the
statistical analysis. FZ designed the present study and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Stomatological Hospital of Jinan City (Jinan, China).
All patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shan Z, Yang G, Xiang W, Pei-Jun W and Bin
Z: Effects of resveratrol on oral squamous cell carcinoma (OSCC)
cells in vitro. J Cancer Res Clin Oncol. 140:371–374. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lu L, Xue X, Lan J, Gao Y, Xiong Z, Zhang
H, Jiang W, Song W and Zhi Q: MicroRNA-29a upregulates MMP2 in oral
squamous cell carcinoma to promote cancer invasion and
anti-apoptosis. Biomed Pharmacother. 68:13–19. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu R, Zeng G, Gao J, Ren Y, Zhang Z, Zhang
Q, Zhao J, Tao H and Li D: MiR-138 suppresses the proliferation of
oral squamous cell carcinoma cells by targeting Yes-associated
protein 1. Oncol Rep. 34:2171–2178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tan DS, Wang W, Leong HS, Sew PH, Lau DP,
Chong FT, Krisna SS, Lim TK and Iyer NG: Tongue carcinoma
infrequently harbor common actionable genetic alterations. BMC
Cancer. 14:6792014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shi Z and Stack MS: Molecules of cell
adhesion and extracellular matrix proteolysis in oral squamous cell
carcinoma. Histol Histopathol. 25:917–932. 2010.PubMed/NCBI
|
|
6
|
Zhu J, Shi H, Liu H, Wang X and Li F: Long
non-coding RNA TUG1 promotes cervical cancer progression by
regulating the miR-138-5p-SIRT1 axis. Oncotarget. 8:65253–65264.
2017.PubMed/NCBI
|
|
7
|
Zhang Y, Dai Q, Zeng F and Liu H: MALAT1
promotes the proliferation and metastasis of osteosarcoma cells by
activating the Rac1/JNK pathway via targeting MiR-509. Oncol Res.
Apr 27–2018.(Epub ahead of print). View Article : Google Scholar
|
|
8
|
Zhang JJ, Wang DD, Du CX and Wang Y: Long
noncoding RNA ANRIL promotes cervical cancer development by acting
as a sponge of miR-186. Oncol Res. May 22–2017.(Epub ahead of
print). View Article : Google Scholar
|
|
9
|
Wang S, Hui Y, Li X and Jia Q: Silencing
of lncRNA-CCDC26 restrains the growth and migration of glioma cells
in vitro and in vivo via targeting miR-203. Oncol Res. Jun
9–2017.(Epub ahead of print). View Article : Google Scholar
|
|
10
|
Zhu G, Wang S, Chen J, Wang Z, Liang X,
Wang X, Jiang J, Lang J and Li L: Long noncoding RNA HAS2-AS1
mediates hypoxia-induced invasiveness of oral squamous cell
carcinoma. Mol Carcinog. 56:2210–2222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan G, Wang X, Yang M, Lu L and Zhou Q:
Long non-coding RNA TUG1 promotes progression of oral squamous cell
carcinoma through upregulating FMNL2 by sponging miR-219. Am J
Cancer Res. 7:1899–1912. 2017.PubMed/NCBI
|
|
13
|
Liu Z, Wu C, Xie N and Wang P: Long
non-coding RNA MEG3 inhibits the proliferation and metastasis of
oral squamous cell carcinoma by regulating the WNT/β-catenin
signaling pathway. Oncol Lett. 14:4053–4058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li S, Yang J, Xia Y, Fan Q and Yang KP:
Long noncoding RNA NEAT1 promotes proliferation and invasion via
targeting miR-181a-5p in non-small cell lung cancer. Oncol Res.
26:289–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Q, Cai J, Wang Q, Wang Y, Liu M, Yang
J, Zhou J, Kang C, Li M and Jiang C: Long noncoding RNA NEAT1,
regulated by the EGFR pathway, contributes to glioblastoma
progression through the WNT/β-Catenin pathway by scaffolding EZH2.
Clin Cancer Res. 24:684–695. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
An J, Lv W and Zhang Y: LncRNA NEAT1
contributes to paclitaxel resistance of ovarian cancer cells by
regulating ZEB1 expression via miR-194. Onco Targets Ther.
10:5377–5390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Y, Yang C, Wang K, Liu X and Liu Q:
MicroRNA-33b inhibits the proliferation and migration of
osteosarcoma cells via targeting hypoxia-inducible factor-1α. Oncol
Res. 25:397–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Xu C and Zhang X: MicroRNA-365
inhibits ovarian cancer progression by targeting Wnt5a. Am J Cancer
Res. 7:1096–1106. 2017.PubMed/NCBI
|
|
21
|
Zhu Y, Zhao H, Rao M and Xu S:
MicroRNA-365 inhibits proliferation, migration and invasion of
glioma by targeting PIK3R3. Oncol Rep. 37:2185–2192. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pietruszewska W, Bojanowska-Poźniak K and
Kobos J: Matrix metalloproteinases MMP1, MMP2, MMP9 and their
tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: An
immunohistochemical study. Otolaryngol Pol. 70:32–43. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang P, Wu T, Zhou H, Jin Q, He G, Yu H,
Xuan L, Wang X, Tian L, Sun Y, et al: Long noncoding RNA NEAT1
promotes laryngeal squamous cell cancer through regulating
miR-107/CDK6 pathway. J Exp Clin Cancer Res. 35:222016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fu JW, Kong Y and Sun X: Long noncoding
RNA NEAT1 is an unfavorable prognostic factor and regulates
migration and invasion in gastric cancer. J Cancer Res Clin Oncol.
142:1571–1579. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen ZJ, Zhang Z, Xie BB and Zhang HY:
Clinical significance of up-regulated lncRNA NEAT1 in prognosis of
ovarian cancer. Eur Rev Med Pharmacol Sci. 20:3373–3377.
2016.PubMed/NCBI
|
|
27
|
Chen X, Kong J, Ma Z, Gao S and Feng X: Up
regulation of the long non-coding RNA NEAT1 promotes esophageal
squamous cell carcinoma cell progression and correlates with poor
prognosis. Am J Cancer Res. 5:2808–2815. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Liu L, Yan F, Wei W, Deng J and Sun
J: Enhanced expression of long non-coding RNA NEAT1 is associated
with the progression of gastric adenocarcinomas. World J Surg
Oncol. 14:412016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Z, Chang Q, Yang F, Liu B, Yao HW, Bai
ZG, Pu CS, Ma XM, Yang Y, Wang TT, et al: Long non-coding RNA NEAT1
overexpression is associated with unfavorable prognosis in patients
with hepatocellular carcinoma after hepatectomy: A Chinese
population-based study. Eur J Surg Oncol. 43:1697–1703. 2017.
View Article : Google Scholar : PubMed/NCBI
|