Introduction
Mesenchymal stem cells (MSCs) have self-renewal
capabilities and multi-directional differentiation potential, and
are induced by different conditions to differentiate into various
different tissues such as bone, cartilage, adipose, muscle,
endothelial, epithelial and nerve tissue (1,2). MSCs
may be used in transplantation for the treatment of systemic
diseases, including leukemia, autoimmune disease and β-cell defects
in diabetes (3). In vivo
imaging technology may be used to dynamically observe the
short-term distribution, homing and long-term survival of
transplanted MSCs (4,5). The intravital tracer technique means
that in stem cell transplantation, cells are able to be dynamically
monitored in vivo to assess their migration and survival
non-invasively (6–8). To obtain high definition images with
high sensitivity, tracer technology for stem cells should be
combined with multi-mode imaging (9). It is therefore of interest to establish
a stem cell line with multi-mode imaging function. Generally, stem
cell engineering studies require genetic modification of stem
cells. At present, the instruments typically used to modify genes
in mammalian cells include plasmid vectors, adenovirus vectors,
retroviral vectors and lentiviral vectors (10). Lentiviral vectors are vectors for
gene modification developed based on human immunodeficiency virus
(HIV) (11). They are able to infect
dividing and non-dividing cells, and integrate the target gene into
the chromosomes of primary cells, stem cells and practically all
cell types. Furthermore, the use of lentiviral vectors has few
safety concerns and they are able to be expressed in vivo
for a long time (12). These
characteristics make the lentivirus vector an ideal instrument for
gene modification (13,14). In the present study, multiple
labeling was performed for human umbilical cord mesenchymal stem
cells (hUCMSCs) to enable them to be displayed using isotopic
imaging, magnetic resonance imaging (MRI) and in vivo
fluorescence imaging in order to establish effective in vivo
tracer technologies, provide the basis for in-depth studies on
hUCMSCs and enhance their homing ability.
Materials and methods
Extraction of hUCMSCs
hUCMSCs were obtained from umbilical cords harvested
from 3 patients recruited to the Department of Obstetrics and
Gynecology, the First Affiliated Hospital of Jinan University
(Guangzhou, China) April to May 2013. The present study was
approved by the Ethics Committee of the First Affiliated Hospital
of Jinan University, and informed written consent was obtained from
all patients from whom tissue was collected. Blood was removed from
the blood vessels of the umbilical cord, and blood on the surface
was washed off. The outer membrane of the umbilical cord was opened
to excise veins and arteries, and hUCMSCs were harvested under
aseptic conditions within 4–6 h of sample collection. Umbilical
cord amniotic epithelium were removed to obtain Wharton's jelly,
which were further dissected into ~1 mm3 sections. The
sections were incubated with 0.1% collagenase IV and digested at
37°C for 24 h. The mixture was centrifuged at room temperature at
1,000 × g for 5 min. The pellet was resuspended with 0.1% trypsin
and further digested for 30 min at 37°C, and then was filtered
through a 74 µm cell strainer. The filtrate was centrifuged at
1,500 rpm for 10 min. The cells were then completely resuspended
and cultured in a 6-well plate at a density of 5×104
cells/well in DMEM/F12 containing 2 ng/ml bFGF and 10% fetal bovine
serum (all Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
at 37°C under 5% CO2 and relative humidity (100%). The
culture solution was replaced 2 days later and the red blood cells
and parenchyma cells that did not adhere to wall in the culture
solution were removed. The culture solution was added again and
became clear. Cellular morphology of hUCMSCs were observed every
day under an inverted light microscope (Olympus CKX41; Olympus
Corporation, Tokyo, Japan) at a magnification of ×100 under normal
culturing conditions.
hUCMSC identification and Induction of
adipogenic and osteogenic differentiation
To confirm that the isolated and purified hUCMSCs
were differentiated successfully, the following surface markers
were selected: Cluster of differentiation (CD)73, CD90, CD105,
hematopoietic stem/progenitor cell marker CD34, hematopoietic cell
marker CD45. The expression of these markers was examined by flow
cytometry. Briefly, a 50 µl cell suspension was seeded at a
concentration of 2×107 cells/ml in L-15 medium
(Sigma-Aldrich; Merck MgaA). A total of 50 µl CD73 (cat. no.
ab175396), CD105 (cat. no. ab49228), CD90 (cat. no. ab11153), CD34
(cat. no. ab185732), CD45 (cat. no. ab38436; all 1:500; all Abcam,
Cambridge, UK) antibodies was added to the cells and incubated at
4°C in dark for 30 min. The cells were then incubated with goat
anti-rabbit immunoglobulin G (cat. no. ab6795; Abcam) as secondary
antibodies at room temperature for 1 h. Wash cells by adding 2 ml
L-15 and performing centrifugation for 5 min at 300 × g and room
temperature. The stained cell pellet was resuspended in 400 ul L-15
at 4°C and blocked by 5% skimmed milk at room temperature for 15
min. Flow cytometry analysis was conducted by CellQuest Software
Pro 5.1 and a BD FACSVerse™ flow cytometer (both BD Biosciences,
Franklin Lakes, NJ, USA). CD73, CD90, CD105 were highly expressed
on the differentiated hUCMSCs, while CD34 and CD45 were barely
detected on these cells.
The hUCMSC cells (5×103 cells) were
seeded on 3 cm dishes in DMEM/F12 supplemented with 2 ng/ml bFGF
and 10% FBS for 2 days. The cells were incubated with osteogenic or
adipogenic differentiation media for 4 weeks. The adipogenic
differentiation medium was composed of DMEM/F12 supplemented with
10% FBS, 100 nM dexamethasone, 50 µg/ml ascorbic acid and 50 µg/ml
indomethacin. The osteogenic differentiation medium was composed of
DMEM/F12 supplemented with 10% FBS, 10 nM dexamethasone, 10 mM
β-glycerophosphate and 50 µg/ml ascorbic acid. As a negative
control, cells were cultured in DMEM/F12 supplemented with 2 ng/ml
bFGF and 10% FBS. Adipogenic differentiation was detected by Oil
Red ‘O’ staining while osteogenic differentiation was detected by
Alizarin Red-S staining as previously described (15) and digitalized for analysis using a
Leica light microscope DMI3009B (Leica Microsystems GmbH, Wetzlar,
Germany).
Construction of lentiviral vector
Obtainment of target gene human
sodium/iodide symporter (hNIS)
Expression plasmid pCMV-Tag2-hNIS preserved by the
Central Laboratory at the First Affiliated Hospital of Jinan
University was used. Primers were designed using Oligo 5.0 software
(Molecular Biology Insights, Inc., Cascade, CO, USA) with reference
to the hNIS-cDNA sequence registered in GeneBank (NM_000453.2).
Upstream primer for NIS forward,
5′-GAATTCGCCACCATGGAGGCCGTGGAGAC-3′ (EcoR I) and reverse,
5′-GCGGATCCTCAGAGGTTTGTCTCCTGCTGGTCTC-3′ (BamH I), were synthesized
by Sangon Biotech Co., Ltd. (Shanghai, China).
Purification of plasmid
A total of 4 plasmids pSico-enhanced green
fluorescent protein (EGFP), pMDLg-pRRE, pRSV-REV and pMD2G were
transformed. Briefly, 1 µg plasmid and 50 µl E. coli
competent cells (Takara Biotechnology Co., Ltd., Beijing, China)
were mixed in a 1.5 ml tube for 30 min at room temperature, then
the tube was incubated at 42°C for 90 sec. The tube was placed
immediately on ice for 2 min. A total of 500 µl LB medium (Thermo
Fisher Scientific. Inc.) was added to each tube, which was
incubated for 1 h at 37°C. Subsequently, 100 µl of the suspension
was placed on a LB agar plate and incubated at 37°C for 12–16 h. A
fresh toothpick was placed on a colony, which was selected by
adding streptomycin 50 mg/ml, kanamycin 50 mg/ml and ampicillin 60
mg/ml; the toothpick was then dipped into a 15 ml tube containing
3–5 ml super optimal broth (SOB) medium (Spectrum Laboratory
Products, Inc., Shanghai, China). The tube was vigorously shaken at
37°C (speed, 90 × g) for 8 h. The plasmids DNA used for
transfections was prepared with the Endofree Plasmid Maxi kit (cat.
no. 12362, Qiagen, Hilden, Germany) according to manufacturer's
protocol.
Lentiviral titer determination
The 293T cells (Thermo Fisher Scientific. Inc.) were
used for virus titer determination. Cells were inoculated in
96-well plates at a density of ~5×104 cells/well within
100 µl medium of DMEM containing 10% fetal bovine serum (FBS) at a
37°C incubator with 5% CO2. A total of 10 sterile
eppendorf (EP) tubes containing 90 µl fresh medium of DMEM
containing 10% FBS were used; 10 µl virus stock was added into the
first EP tube and mixed well, and 10 µl solution was transferred
from the first EP tube into the second. This process was repeated
for the remaining tubes until the virus stock was diluted 10 times.
A total 9 µl medium was removed from 10-well in the 96-well plate
and 9 µl virus solutions from each of the prepared 10 tubes were
added to each well, thus there were 5,000 cells in each well, and
the concentration of the virus/well were 1, 10−1 and
10−2 µl, respectively. The cell culture plate was
incubated at 37°C in an atmosphere containing 5% CO2 for
48 h. A total of 100 µl fresh medium was added into each well and
incubation was resumed for 96 h. GFP fluorescence expression was
subsequently observed under a fluorescence microscope and the
number of fluorescing cells in the last two wells were counted.
Superparamagnetic iron oxide
(SPIO)-labeling of hNIS-EGFP-hUCMSCs
hNIS-EGFP-hUCMSCs were incubated with SPIOs (50 µg;
Biopal, Inc., Worcester, MA, USA). SPIO solutions were prepared at
a 2× concentration in DMEM/F12 for 24 h. A total of 5 ml of 2× SPIO
solutions were added to 5 ml of a hUCMSCs suspension in a 10 cm
dish (5×104). The cells were plated on a 96-well plate,
100 µl for each, then 10 µl MTT was added to per well and incubated
for 4 h, finally 100 µl DMSO solution was added per well until the
formazan crystals dissolved, which was detected with 490 nm. Growth
medium without SPIOs was added to sister cultures of hUCMSCs, these
cells were used as the controls. Following incubation with SPIO,
the cells were collected and washed twice in PBS; labeling
efficiency was determined by Prussian Blue staining at room
temperature for 30 min. The cellular morphology of hUCMSCs labeled
with SPIO was observed under the inverted light microscope (Olympus
CKX41, ×400) for 8 days.
In vitro study of iodine uptake in
transfected hUCMSCs
The experimental groups (SPIO-hNIS-EGFP-hUCMSCs)
transfected with the lentivirus-hNIS-EGFP using the same kit and
protocol as the 293T transfection. The control group (hUCMSCs) were
untransfected cells. All groups were conventionally cultured and
incubated in a 24-well plate at a density of 5×104
cells/well at 37°C in an atmosphere containing 5% CO2
overnight. When cells reached 80% confluence, the medium (DMEM/F12
containing 2 ng/ml bFGF and 10% FBS) was discarded and the cells
were washed twice with Hank's Balanced Salt Solution (HBSS;
Invitrogen; Thermo Fisher Scientific, Inc.). The 125I
uptake method was as previously described (16). Briefly, 1 ml HBSS (containing 0.1 µl
of 100 uM sodium citrate 125I) was added to each well
and incubated at 37°C for 30 min. The NaClO4 inhibition
test was conducted by incubation the cells with 300 µmol/l
NaClO4 for 60 min. This medium was discarded, cells were
washed twice with cold HBSS, then cold 95% ethanol was then added
to the cells and they were incubated at room temperature for 20
min. The concentrated iodide in the cytolysate was determined with
a γ-counter (2480 WIZARD2™ gamma counter; PerkinElmer,
Inc., Waltham, MA, USA). The experimental and control groups each
consisted of 6 independent wells in the 24-well plate.
Amplification of the hNIS gene
mRNA was extracted from the hUCMSCs using TRIzol and
hNIS mRNA was amplified with one-step reverse transcription
polymerase chain reaction (RT-PCR). The RT-PCR system solution was
as follows: 20 µl of RNase Free dH2O, 25 µl of 2×1 Step
Buffer, 2 µl of PrimeScript 1 Step Enzyme Mix (Takara Biotechnology
Co., Ltd.), 2 µl of upstream primers, 3 µl of downstream primers
and 1 µl RNA samples. The primers were as follows: hNIS forward,
5′-CACCATGGAGGCCGTGGAG-3′ and reverse,
5′-GAGGTTTGTCTCCTGCTGGTCTC-3′; β-actin forward,
5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse, 5′-GCTGTCACCTTCACCGTTCC-3′.
The thermocycling conditions were as follows: 50°C for 30 min, 94°C
for 2 min, 30 cycles of 94°C for 30 sec, 50–65°C for 30 sec and
72°C for 1 min, and 72°C for 10 min. The enzyme digestion system
was: 1 µl 10X NEB-buffer, 0.5 µl EcorI, 0.5 µl xhol1, 5 µl PCR
products/P3.1 vector (Hangzhou Xiaoyong Biotechnology Co., Ltd.,
Hangzhou, China) and 3 µl ddH2O. The cells were then
incubated with the digestion in 37°C for 2 h prior to the PCR
product was identified by 1% agarose gel electrophoresis, which was
determined to have the same size as the target gene.
Colony PCR for recombinant
plasmid
NIS gene segment and P3.1 vector were linked using
the following protocol: 1 ul 10X buffer, 2 ul NIS gene segment, 4
ul P3.1 vector, 1 ul T4 ligase (Thermo Fisher Scientific, Inc.) and
2 ul ddH2O were mixed overnight at 4°C. The NIS gene
segment and P3.1 vector was then transformed in complete medium
(DMEM and 10% FBS) for 48 h at room temperature and coated on a LB
medium plate without any antibiotics. Two monoclones were selected
from the resistant plate with clear colony growth and amplified by
PCR with Pn1 and Pn2 as the primers. The PCR product was identified
by electrophoresis using a 1% agarose gel with ethidium bromide.
The two monoclones revealed 1,900 bp gene segments, which was
consistent with the theoretical value. It was preliminarily proven
that these two colonies contained the correctly linked recombinant
plasmids.
Screening of cell line stably
expressing hNIS by puromycin
Determination of puromycin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) screening concentration revealed that
hUCMSCs were all killed following 7 days of exposure to 8 µl/ml
puromycin. At concentrations <8 µl/ml, some hUCMSCs were not
apoptosed; therefore 8 µl/ml was selected as the optimal puromycin
screening concentration. hUCMSCs transfected with lentivirus were
subsequently sub-cultured at a dilution of 1:10. When cell
attachment occurred, 8 µl/ml puromycin was added to the medium. At
7 days following screening, positive cell colonies of hUCMSCs
stably expressing hNIS and EGFP were obtained by screening with 8
µg/ml puromycin, and these were named as hNIS-EGFP-hUCMSCs and
transferred to a 6-well plate for multiplication culture.
Identification of hNIS and EGFP
expression in hUCMSCs using western blotting
Protein was extracted from 2×105 cells
using RIPA buffer (Thermo Fisher Scientific, Inc.) containing
protease inhibitors (Roche Diagnostics, Basel, Switzerland).
Protein concentration in the supernatants was measured using a
bicinchoninic acid assay protein assay kit (cat. no. 23222; Thermo
Fisher Scientific, Inc.). A total of 60 µg protein was transferred
into an EP tube with 4X loading buffer and mixed well. The mixture
was boiled for 10 min and centrifuged at 2,000 × g for 5 min.
Proteins (10 µg/lane) were subsequently separated by 10% SDS-PAGE
and transferred onto polyvinylidene fluoride membranes. Membranes
were washed with Tris-buffered saline with 0.1% Tween-20 and
blocked at room temperature in PBS with 0.1% Tween-20 (PBST)
containing 5% skim milk powder for 1 h. Membranes were subsequently
incubated at room temperature for 1 h with anti-hNIS (cat. no.
ab83816; Abcam, Cambridge, UK), anti-EGFP antibodies (cat. no.
2956; Cell Signaling Technology, Inc., Danvers, MA, USA; both
1:3,000) and β-actin (cat. no. BM3501-01, 1:5,000; Biomiga, Inc.,
San Diego, USA) and washed with PBST. Membranes were incubated at
room temperature for 2 h with the secondary horseradish
peroxidase-labeled anti-Rabbit IgG (cat. no. ab191866; 1:10,000;
Abcam), washed with PBST, then treated with SuperSignal West Pico
Chemiluminescent Substrate (cat. no. 34077; Thermo Fisher
Scientific, Inc.) for 1 h; the exposure time of X-ray film was 30
min. The membrane was washed with 0.5 mol/l NaOH for 15 min.
Statistical analysis
SPSS 13.0 (SPSS, Inc., Chicago, IL, USA) was used to
perform statistic analysis. Experimental data are expressed as the
mean ± standard deviation. Independent-samples t-tests were used to
compare differences between groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
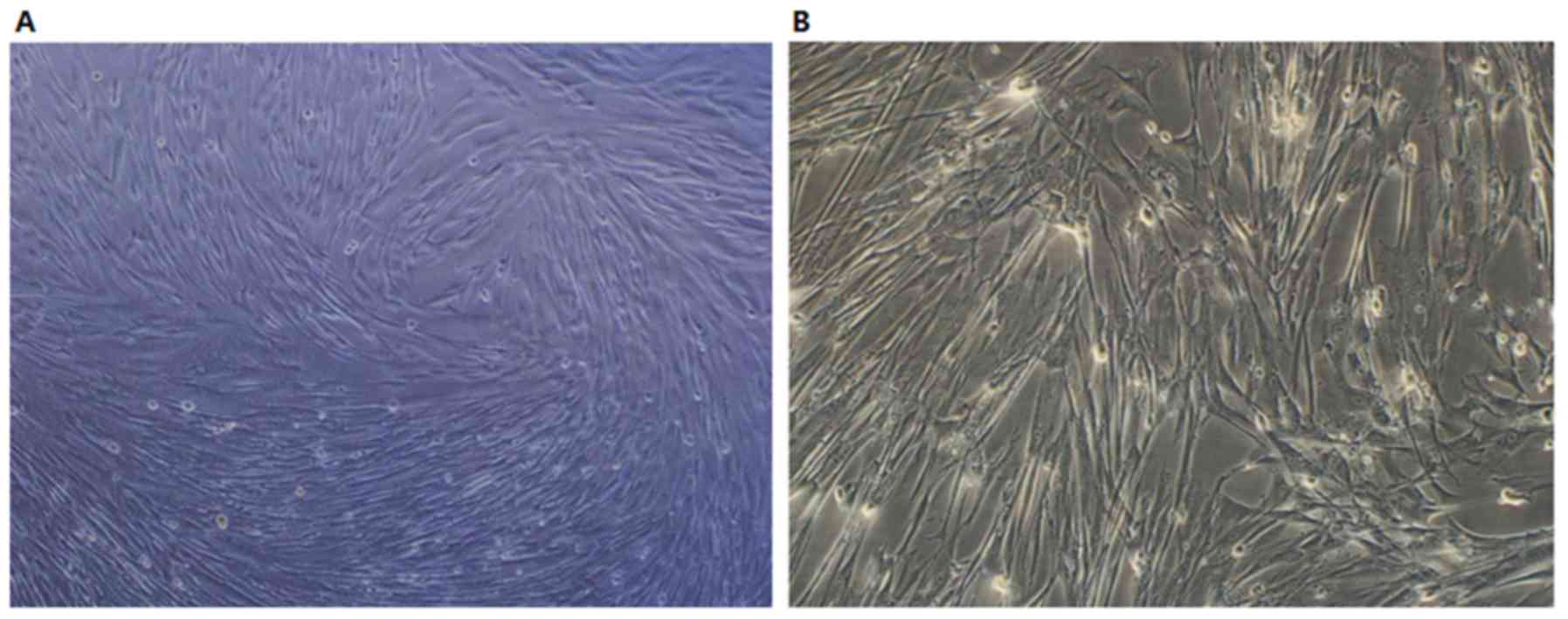
Morphological observation of hUCMSCs
in vitro
Following 12 h of culture, the majority of hUCMSCs
had adhered to the bottom of the plate and spread out gradually,
with flat and spindle-like shapes and fibroblast-like growth. After
five days of cell proliferation, confluence reached 80–90%, with
cells distributed in clusters growing in a whirl or
fingerprint-like manner (Fig. 1A).
The sub-cultured hUCMSCs exhibited increased proliferative
abilities and spindle-like growth (Fig.
1B). The cellular morphology of hUCMSCs after the third
generation was uniform and cell growth entered plateau phase.
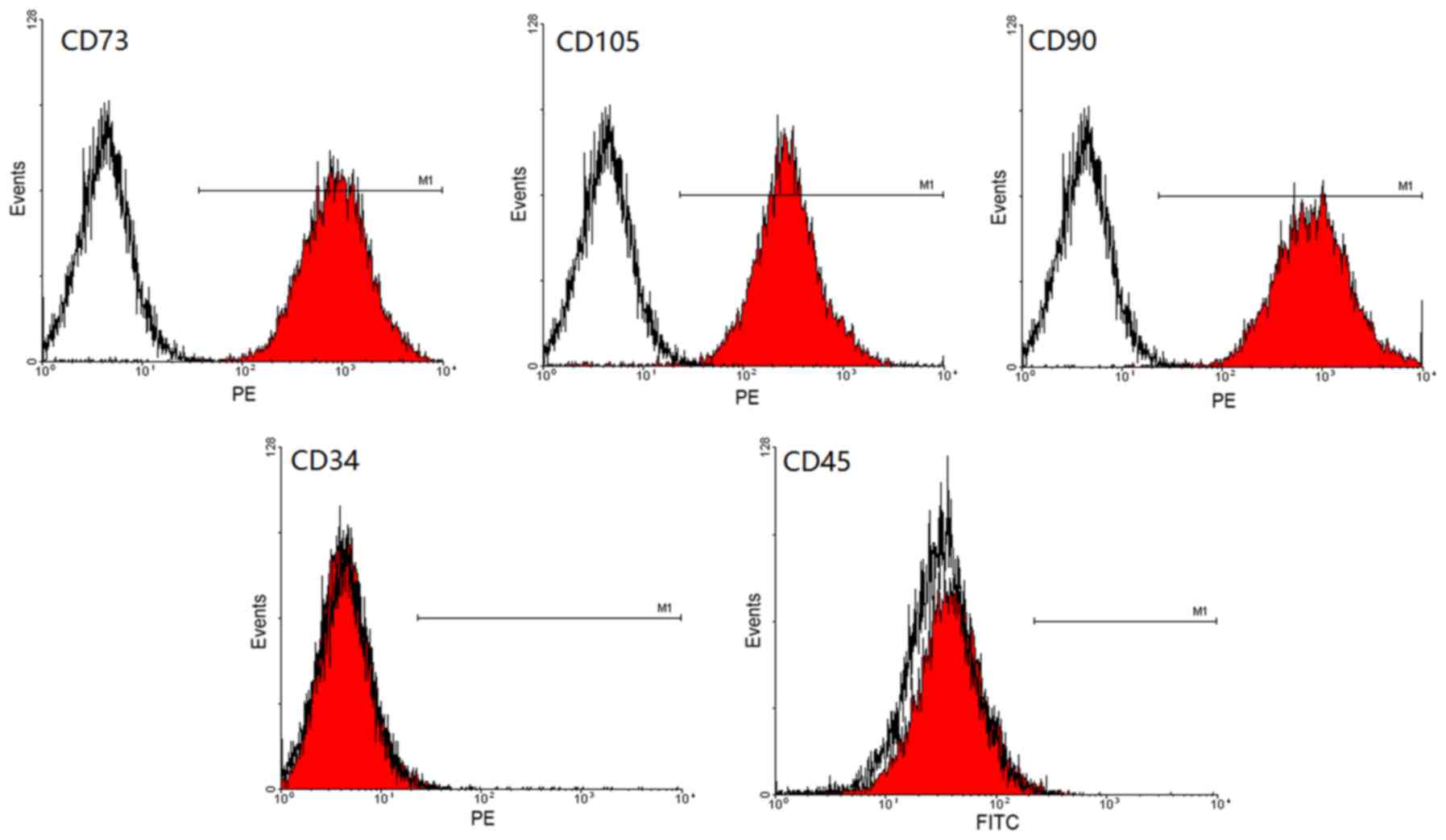
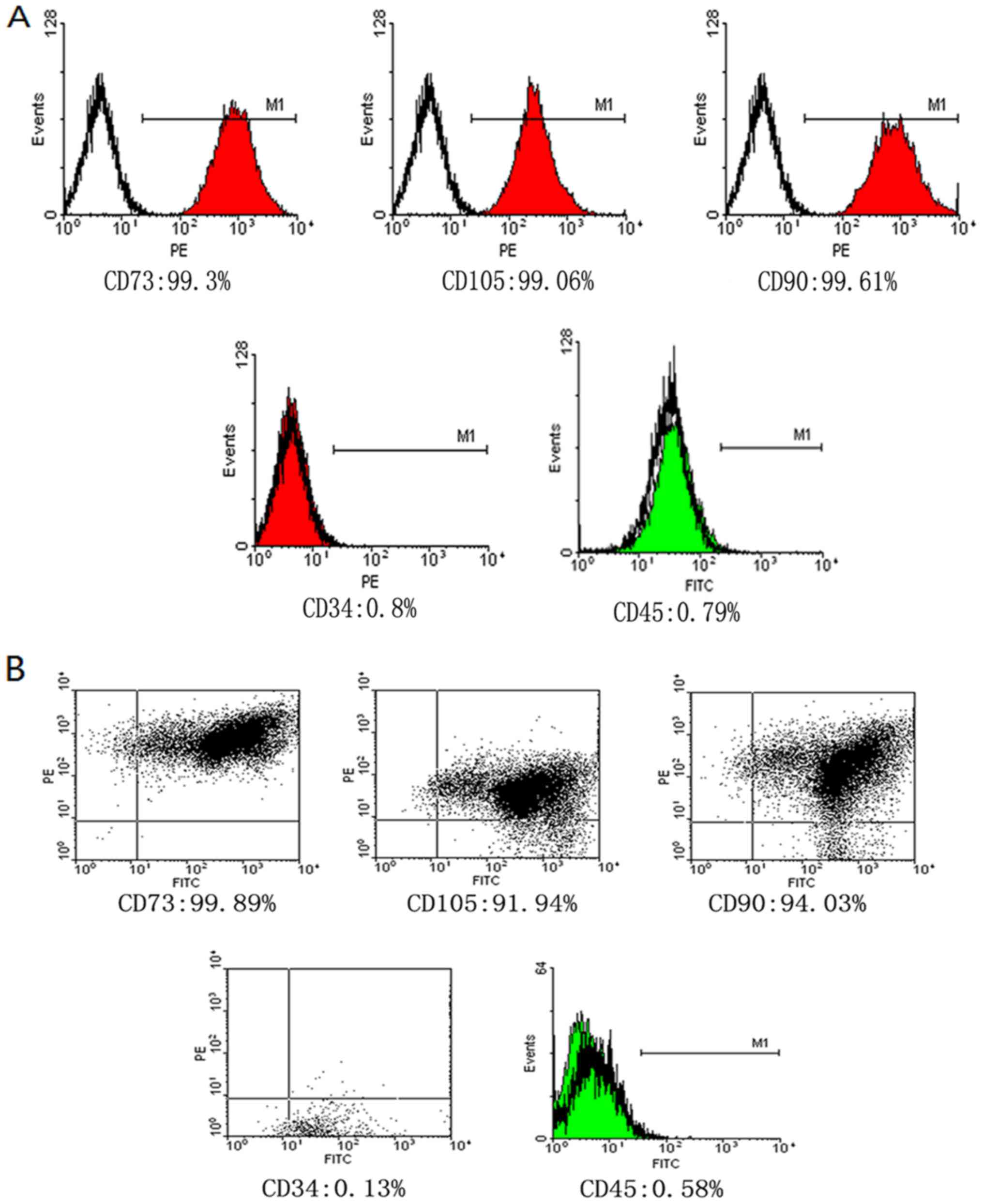
Expression of surface antigen of
hUCMSCs by flow cytometry
Third generation hUCMSCs were identified by flow
cytometry and they were found to be positive for CD73 (93.30%),
CD90 (99.61%) and CD105 (99.06%); however, cells were negative for
CD34 (0.80%) and CD45 (0.79%; Fig.
2), which was consistent with the phenotypic features of
hUCMSCs.
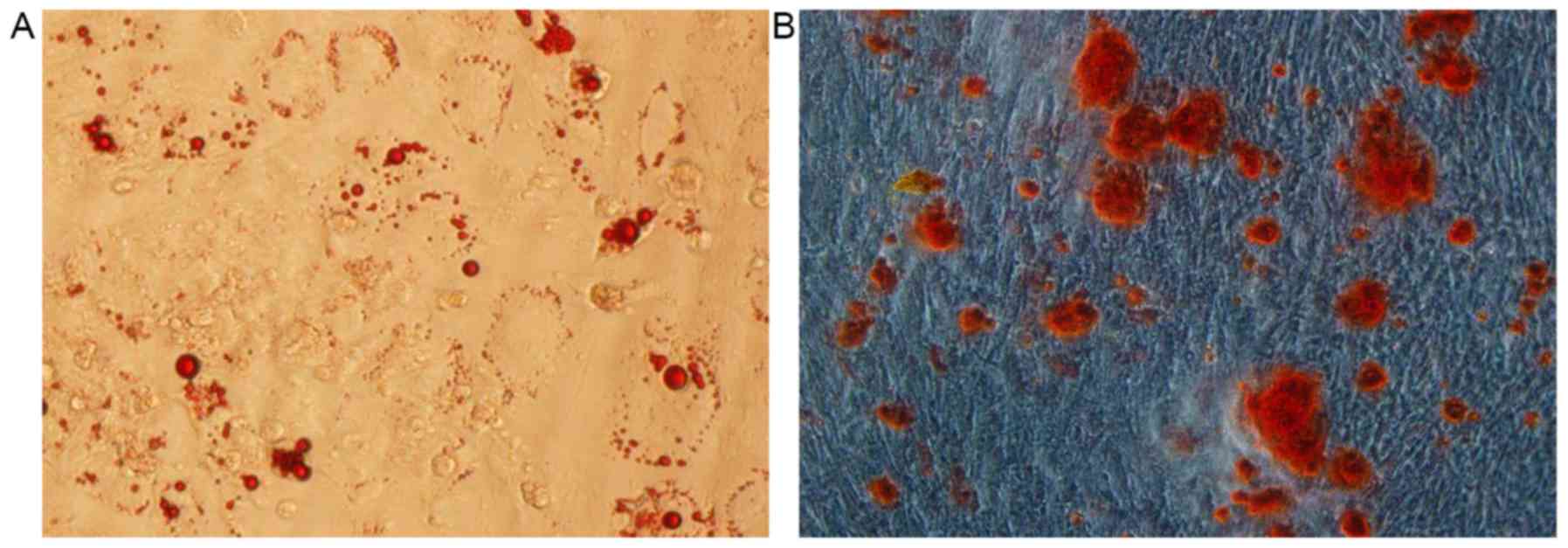
Adipogenic induction for hUCMSCs
Cellular morphology
hUCMSCs were cultured with the adipogenic inducer
and their appearance changed from spindle shaped to irregularly
circular or polygonal, indicating clustering growth. At 4 days
following the induction, intracellular circular lipid droplets were
observed under the inverted microscope. Following continuous
differentiation, the number of lipid droplets markedly increased
and some fused to form large liquid droplets.
Oil red ‘O’ staining
At 28 days following adipogenic differentiation,
third generation hUCMSCs were stained with oil red ‘O’. Lipid
droplets were observed to possess a red color in >80% of the
cell with sharp contrast (Fig.
3A).
Osteogenic induction for hUCMSCs
Cellular morphology
Following osteogenic induction, the cellular
morphology changed. On the fifth day of osteogenic induction, cell
volume was observed to have increased and hUCMSCs exhibited short
spindle-like or irregular polygonal shapes. Following continuous
differentiation, hUCMSCs clustered and cascaded to form colonies.
Calcified nodules were observed in the central regions and
increased gradually.
Alizarin red staining
Following 4 weeks of osteogenic induction, third
generation hUCMSCs were stained with alizarin red. Multiple black
calcium nodules were observed at the central region of the cell
colonies, which were stained black by reduced metallic silver
(Fig. 3B).
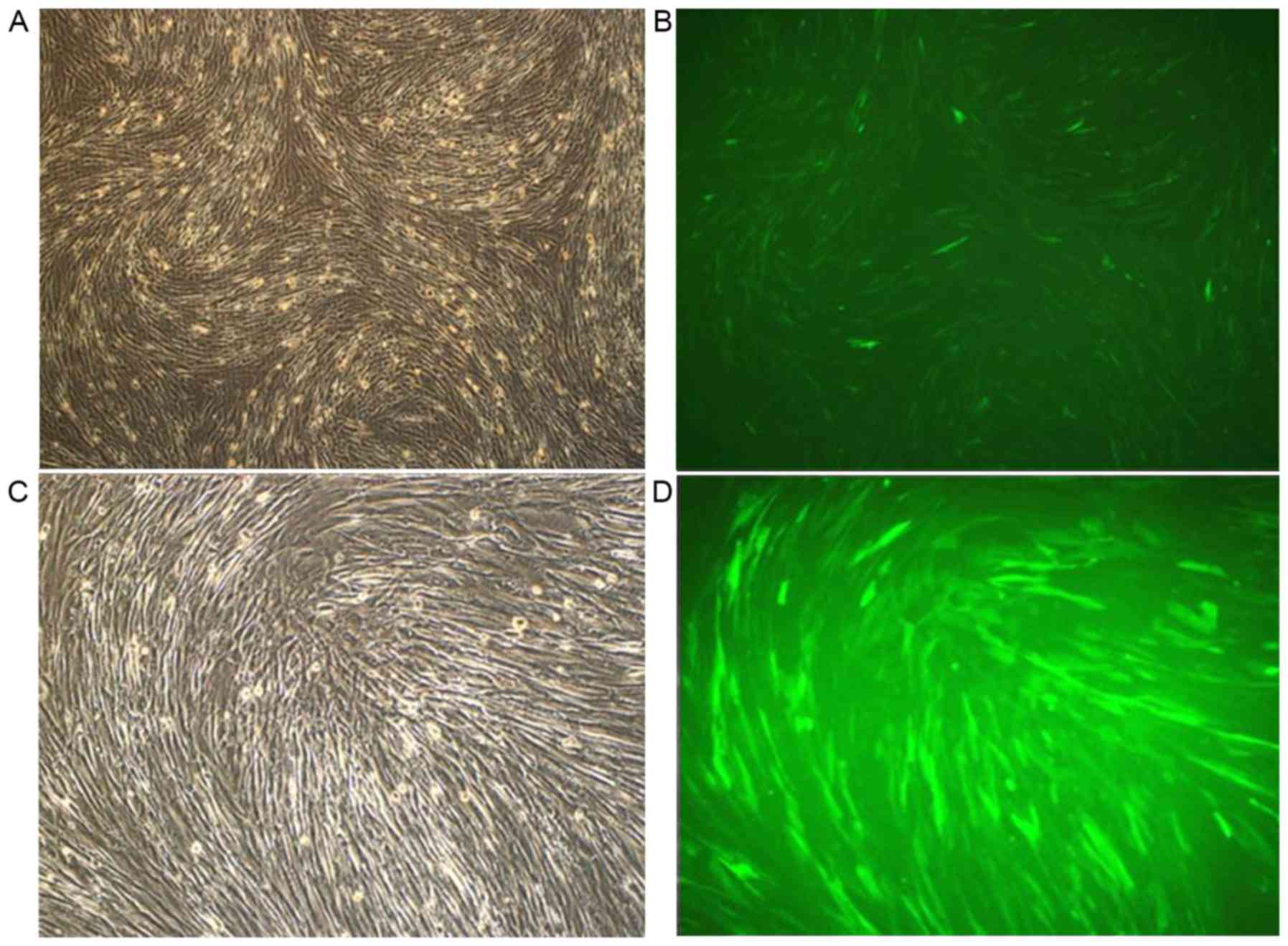
Positive expression of hNIS-EGFP
observed using fluorescence microscopy
Following transfection with EGFP and hNIS, the
expression of hNIS in cells was assessed via fluorescence
microscopy. At 12 h following transient transfection, a small
number of hUCMSCs were observed to express green fluorescence with
weak intensity. Following another 24 h, the number of fluorescing
cells and the intensity of fluorescence increased, with 4–10/field
in each field (magnification, ×10). Over the subsequent 48 h, the
number of human amniotic epithelial cells with green fluorescence
increased continuously, with >10 in each field. No obvious
differences were observed in the number of hUCMSCs with green
fluorescence at 72 and 48 h. The majority of hNIS-EGFP-hUCMSCs
fluoresced green under a fluorescence microscope (Fig. 4). It was observed that the virus
infection efficiency for the NIS gene was >95%.
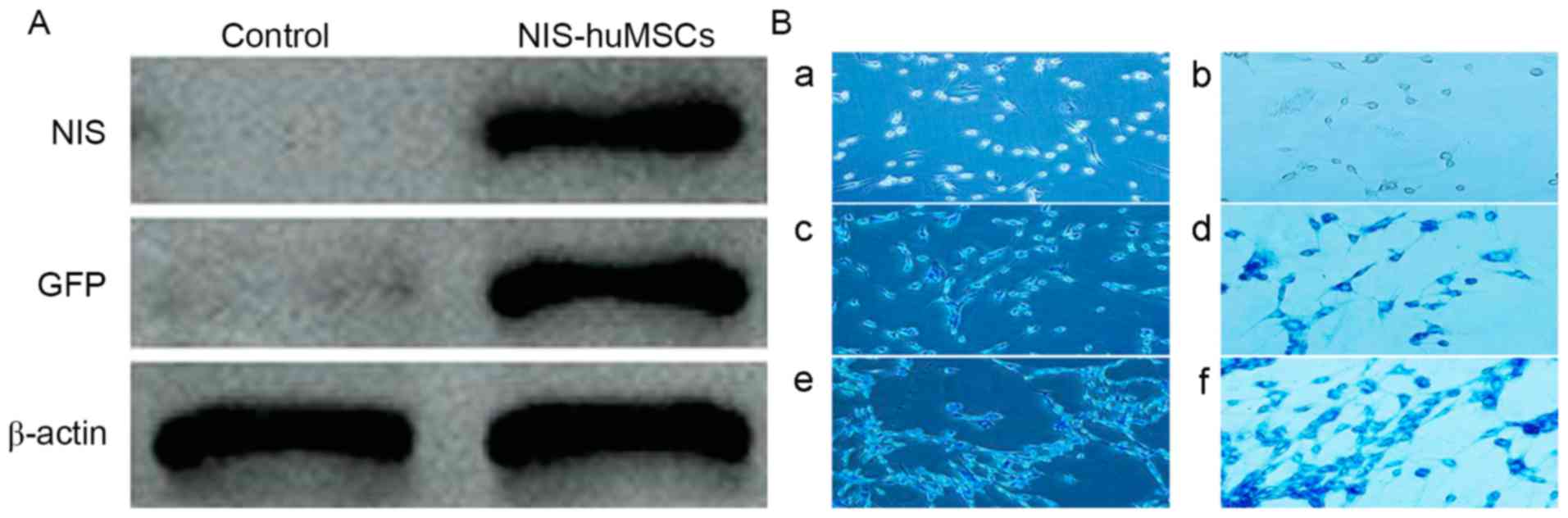
Identification of hNIS and EGFP
expression in hUCMSCs by western blot
The cell lysate of the experimental
(hNIS-EGFP-hUCMSCs) and control groups (hUCMSCs) underwent western
blot analysis. Western blotting revealed positive bands at 90 kD
(NIS) and 57 kD (eGFP) in the experimental group. Western blotting
results for the control group did not reveal any specific bands
(Fig. 5A).
SPIO-labeled hNIS-EGFP-hUCMSCs
hNIS-EGFP-hUCMSCs were co-incubated with SPIO again.
It was confirmed by Prussian blue staining that SPIO transfection
efficiency was >98%. The results are presented in Fig. 5B.
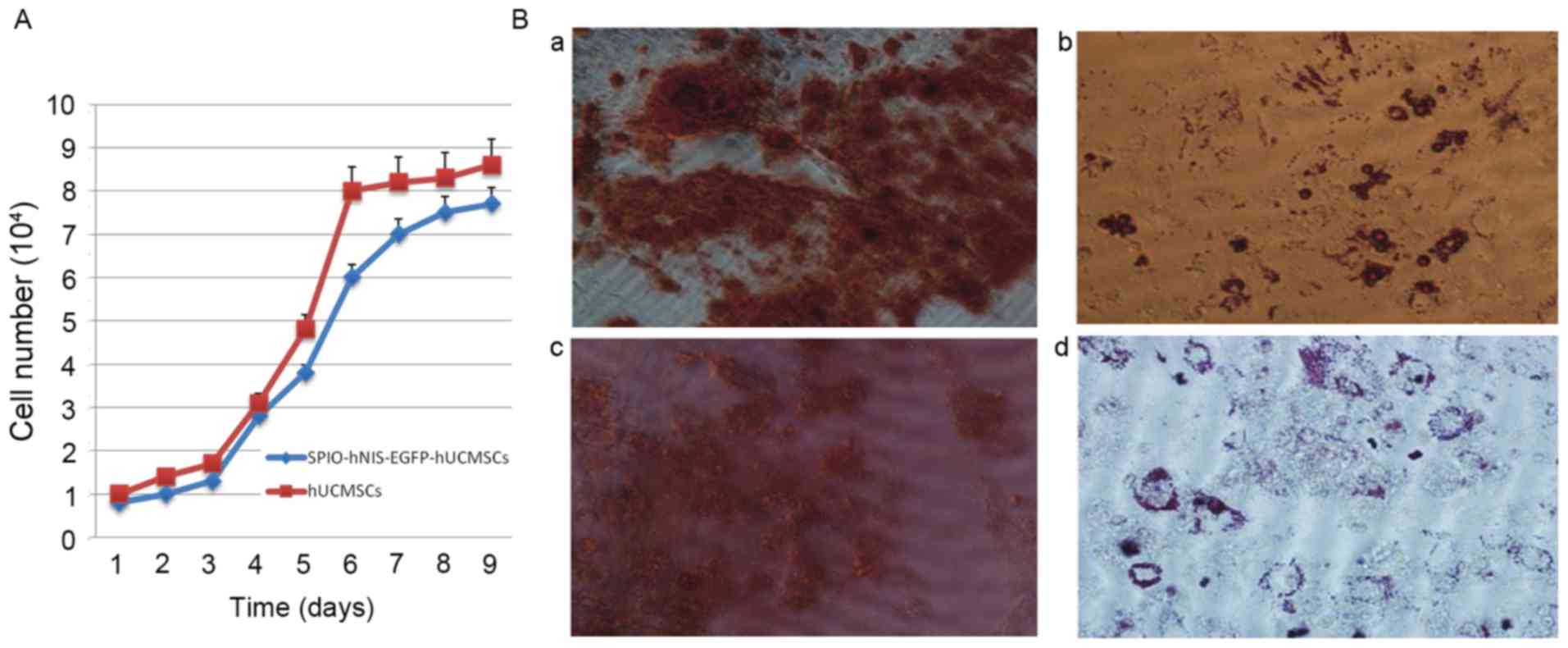
Study on cell biology of SPIO-labeled
hNIS-EGFP-hUCMSCs Cell growth curve of hNIS-EGFP-hUCMSCs following
labeling with SPIO
SPIO, hNIS and EGFP co-labeled hUCMSCs were
sub-cultured. On days 1 and 2, hUCMSCs were latent and exhibited no
marked proliferation. On days 3–7, hUCMSCs entered the logarithmic
phase, and on days 7–8, hUCMSCs entered the plateau phase. Two
double periods of cell growth were observed, the first on days 3–4
and the second on days 4–7. Compared with the control group
(hUCMSCs), there was no significant difference in cell growth at
any time point (Fig. 6A).
Adipogenic and osteogenic
differentiation of hNIS-EGFP-hUCMSCs following labeling with
SPIO
Experimental results demonstrated that modification
with hNIS and EGFP and labeling with SPIO had no effect on
adipogenic and osteogenic differentiation (Fig. 6B).
Phenotype identification of
hNIS-EGFP-hUCMSCs following labeling with SPIO determined by flow
cytometry
The adipogenic and osteogenic differentiation of
normal MSCs and MSCs transfected with EGFP and SPIO were
identified. The results revealed that the stemness of MSCs was not
significantly decreased. Following transfection with EGFP and SPIO,
and they were determined to be positive for CD73 (99.89%), CD90
(94.03%) and CD105 (91.94%) and negative for CD34 (0.13%) and CD45
(0.58%). There was no significant difference between the control
and experimental groups (Fig.
7).
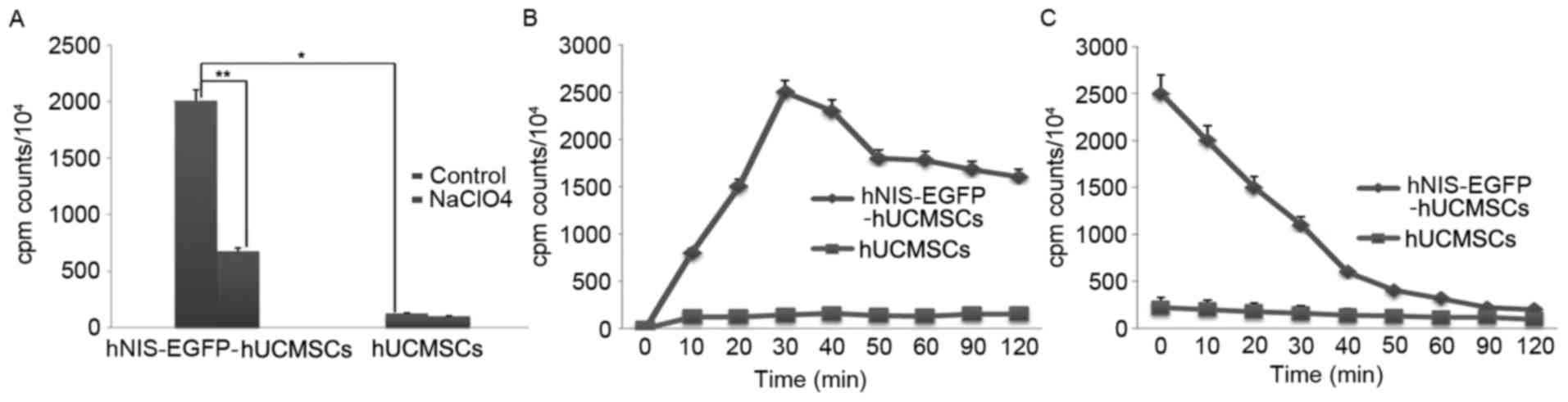
In vitro study on 125I
uptake of SPIO-labeled hNIS-EGFP-hUCMSCs
Uptake of 125I was determined as
previously described (17,18). 125I uptake activity
significantly increased by 16.43±2.30 times in the experimental
group compared with the control group (P<0.05; Table I; Fig.
8A), accumulation was faster in SPIO-hNIS-EGFP-hUCMSCs than in
hUCMSCs, reaching maximal levels within 30 min (Fig. 8B). The efflux of 125I from
SPIO-hNIS-EGFP-hUCMSCs was rapid, with half-maximal activity levels
reached after 7.95 min (Fig.
8C).
 | Table I.125I uptake activity of
superparamagnetic iron oxide-labeled human sodium/iodide
symporter-enhanced green fluorescent protein-human umbilical cord
mesenchymal stem cells. |
Table I.
125I uptake activity of
superparamagnetic iron oxide-labeled human sodium/iodide
symporter-enhanced green fluorescent protein-human umbilical cord
mesenchymal stem cells.
| Group | Without
NaClO4 (cpm) | With
NaClO4 (cpm) |
|---|
| Experimental |
2005.2±52.0a,b | 673.8.2±31.8 |
| Control | 122.7±20.9 | 98.3±10.5 |
Discussion
Compared with bone marrow mesenchymal stem cells
(BMSCs), dental pulp stem cells (DPSCs), adipose derived stem cells
(ADSCs) and embryonic stem cells (ESCs), hUCMSCs possess more
primitive properties, strong plasticity and differentiation
potential (19). Furthermore,
hUCMSCs express the specific markers (CD73, CD90 and CD105) of stem
cells (20). Compared with BMSCs,
UMSCs have a greater proliferative ability, making them more
suitable for clinical applications (21). Compared with ADSCs, hUCMSCs exhibit
lower immunogenicity and no oncogenicity (22). hUCMSCs do not express human leukocyte
antigen-antigen D related, which is a major causative factor of the
immune response, suggesting that hUCMSCs may be suitable for
transplantation between different individuals (23). Compared with other stem cells, the
proliferative activity and differentiation ability of hUCMSCs does
not decrease with sub-culture. Due to the aforementioned features,
hUCMSCs were selected for use in the present study. Isotopic
imaging, MRI and in vivo fluorescence imaging were compared
or combined in order to identify effective in vivo tracer
technologies, provide the basis for in-depth studies on hUCMSCs and
enhance their homing ability.
hNIS is an endogenous gene derived from human
thyrocytes, and its encoding product is a physiological protein
that has no immunogenicity (24,25). The
product of the hNIS reporter gene is expressed on the cell
membrane, making it easier to capture the reporter probe compared
with herpes simplex virus type 1 thymidine kinase and other
reporter genes of intracellular enzymes (26). Endogenous hNIS is only expressed in
certain tissues and organs, and commonly used radionuclides, such
as 123I and 99mTc may be used as reporter
probes of hNIS; as such, there is no labeling requirement, which
reduces the cost and increases accessibility (27). hNIS may be used for single-photon
emission computed tomography (SPECT) or positron emission
tomography and, as a reporter gene, hNIS is more stable than
stomatostatin receptor 2 and human dopamine 2 receptor (28,29).
However, the hNIS reporter gene lso has some deficiencies. As it is
an endogenous gene, it is expressed in different degrees by several
organs and tissues, such as the thyroid, gastric mucosa, mammary
glands and salivary glands, which affects the monitoring of hNIS
gene expression (30). In addition,
in non-thyroid tissues and cells, iodine is released quickly
because of a lack of thyroid specific proteins that hold iodine,
therefore radioactive iodine does not accumulate (31–33).
Although this reduces radiation damages to tissues, the signal
detection is also affected.
For molecular biological techniques, the
transfection of reporter genes into quiescent cells, in particular
hUCMSCs, is difficult. The methods for transfecting target genes
into target cells primarily include the transfection of eukaryotic
expression plasmids and the transfection of viral vector-mediated
genes (34). Studies on lentiviral
vector (35) demonstrate that these
vectors are able to effectively integrate exogenous genes into host
chromosome to establish persistent expression (36). Lentiviral vectors are also able to
effectively transfect neuronal, hepatic, myocardial, tumor,
endothelial and stem cells (37).
For cells that are difficult to transfect, such as primary, stem
and non-dividing cells, the lentiviral vector increases the
transduction efficiency of target genes greatly, allowing for the
incorporation of RNAi, cDNA and study reporter genes (38). The lentiviral packaging system used
in the present study is a four-plasmid system, consisting of
pRsc-REV, pMD1g-pRRE, pMD2G and interference plasmid. The
interference plasmid is able to express EGFP, pRsv-REV, pMDlg-pRRE
and pMD2G, and contains the necessary elements for virus packaging
(39). Eukaryotic plasmids
containing the target gene, hNIS, were established and transfected
with the aforementioned four-plasmids system to obtain
hNIS-EGFP-hUCMSCs. Western blot analysis revealed that hNIS
expression was normal. 125I uptake activity of the
experimental group (hNIS-EGFP-hUCMSCs) increased by 16.43±2.30
times in comparison with that of the control group (hUCMSCs).
125I influx and efflux experiments indicated that the
function of hUCMSCs was normal following the expression of hNIS,
which indicates that it may be an effective reporter for SPECT
in vivo cell tracing.
SPIO is a novel MR contrast, with a 20–200 nm
diameter core of Fe2O3 coated with glucan
(40). SPIO has very small crystal
structures; under an applied magnetic field, SPIO exhibits single
magnetic moment along the magnetic field (41). Even in a weak magnetic field, SPIO is
highly magnetic (42). When the
applied magnetic field is removed, this magnetism quickly subsides,
and this is called superparamagnetism (43). Uneven distribution of SPIO in tissues
results in an uneven local magnetic field (44), which shortens the transverse
relaxation time (T2) and longitudinal relaxation time (T1) of
tissues. The shortening of T2 is more marked, manifesting as the
decrease in T2 signals (45). SPIO
is negatively charged, and so in the present study, SPIO was coated
with positively charged polylysine to reduce electrostatic
interaction (29,32,33,46,47).
Thus, SPIO was able to bind with cell surface receptors via
electrostatic interaction and enter into cells via endocytosis.
hNIS-EGFP-hUCMSCs are labeled with SPIO using the aforementioned
technique, allowing for the successful establishment of SPIO, hNIS
and EGFP co-labeled hUCMSCs. Staining with Prussian blue confirmed
that SPIO entered cells, with a labeling rate of 98%.
The purpose of establishing SPIO, hNIS and EGFP
co-labeled hUCMCs was to horizontally compare the advantages and
disadvantages of several imaging technologies in in vivo
stem cell transplantation. A series of tests were performed to
determine whether the biological activity, stemness, proliferative
activity or differentiation ability of hUCMSCs were affected
following experimental interventions. On days 1 and 2 of
sub-culture, hUCMSCs were at a latent phase with no obvious
proliferation; on days 3–7, hUCMSCs entered the logarithmic phase,
and on days 7–8, hUCMSCs entered the plateau phase. Cell growth had
two double periods, the first on days 3–4 and the second on days
4–7. Compared with the control group (hUCMSCs), there were no
significant differences at each time point. Adipogenic and
osteogenic differentiation were further assessed prior to and
following experimental treatments, and the experimental results
suggest that the stemness of hNIS-EGFP-hUCMSCs was slightly
reduced; however, the overall characteristics of stem cells
remained unchanged. The results for adipogenic and osteogenic
differentiation indicate that there are no significant differences
between hNIS-EGFP-hUCMSCs and normal primary hUCMSCs. Western
blotting also demonstrated that hNIS expression was good, and
125I influx and 125I efflux experiments
indicated that the function of hUCMSCs was good following
transfection with hNIS. These results suggest that
hNIS-EGFP-hUCMSCs may be effective reporters for SPECT in
vivo cell tracing. hNIS-EGFP-hUCMSCs were labeled with SPIO
under the mediation of poly-L-lysine, and SPIO, hNIS and EGFP
co-labeled hUCMSCs were successfully established. Staining with
Prussian blue confirmed that SPIO successfully entered the cells,
with a labeling rate of 98%. There were no significant differences
in biological activity, stemness, proliferative activity or
differentiation ability between SPIO, hNIS and EGFP co-labeled
hUCMSCs and primary hUCMSCs. Therefore, the results of the present
study suggest that, SPIO, hNIS and EGFP co-labeled hUCMSCs may be
an effective treatment cell for animal models as it may be
appropriate for stem cell tracing by SPECT, MRI and in vivo
fluorescence imaging of animals.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Science and Technology Planning Project of Guangdong Province,
China (grant nos. 2016A040403054 and 2013B021400002), Important
Guangdong Province Science and Technology Specific Projects (grant
nos. 2003A3080501 and 2015B010106008) and Important
Industry-Academia-Research, Collaboration and Innovation Specific
Projects of Guangzhou City (grant no. 201508020101).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YM, HL and NB designed the experiment, established
the lentivirus infection system, performed the statistical analysis
and wrote the manuscript. JG processed and analyzed the
single-photon emission computed tomography imaging. XZ provided
plasmid of sodium/iodide co-rotor, designed the plasmid experiment
and in vitro transfection. CH and WL performed magnetic
resonance imaging scans and pathological research. HX guided the
experiment, assisted with experiments where issues arose and
reviewed the whole manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Crisan M, Yap S, Casteilla L, Chen CW,
Corselli M, Park TS, Andriolo G, Sun B, Zheng B, Zhang L, et al: A
perivascular origin for mesenchymal stem cells in multiple human
organs. Cell Stem Cell. 3:301–313. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tyndall A and Uccelli A: Multipotent
mesenchymal stromal cells for autoimmune diseases: Teaching new
dogs old tricks. Bone Marrow Transplant. 43:821–828. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang H, Zhang X, Hu X, Shao Z, Zhu J, Dai
L, Man Z, Yuan L, Chen H, Zhou C and Ao Y: A functional biphasic
biomaterial homing mesenchymal stem cells for in vivo cartilage
regeneration. Biomaterials. 35:9608–9619. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao C, Tian M and Zhang H: In vivo stem
cell imaging. Open Nucl Med J. 2:171–177. 2010. View Article : Google Scholar
|
|
6
|
Ezzat T, Dhar DK, Malago M and Damink Olde
SW: Dynamic tracking of stem cells in an acute liver failure model.
World J Gastroenterol. 18:507–516. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xia T, Jiang H, Li C, Tian M and Zhang H:
Molecular imaging in tracking tumor stem-like cells. J Biomed
Biotechnol. 2012:4203642012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schwarz M, Dörfler A, Engelhorn T,
Struffert T, Tietze T, Janko C, Tripal P, Cicha I, Dürr S, Alexiou
C and Lyer S: Imaging modalities using magnetic
nanoparticles-overview of the developments in recent years.
Nanotechnology Reviews. 2013:142013.
|
|
9
|
Wolfs E, Holvoet B, Gijsbers R, Casteels
C, Roberts SJ, Struys T, Maris M, Ibrahimi A, Debyser Z, Van Laere
K, et al: Optimization of multimodal imaging of mesenchymal stem
cells using the human sodium iodide symporter for PET and Cerenkov
luminescence imaging. PLoS One. 9:e948332014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shearer RF and Saunders DN: Experimental
design for stable genetic manipulation in mammalian cell lines:
Lentivirus and alternatives. Genes Cells. 20:1–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pavlovic M, Koehler N, Anton M,
Dinkelmeier A, Haase M, Stellberger T, Busch U and Baiker AE:
Reverse transcription quantitative polymerase chain reaction for
detection of and differentiation between RNA and DNA of HIV-1-based
lentiviral vectors. Hum Gene Ther Methods. 28:215–221. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kantor B, McCown T, Leone P and Gray SJ:
Clinical applications involving CNS gene transfer. Adv Genet.
87:71–124. 2014.PubMed/NCBI
|
|
13
|
Hu YL, Fu YH, Tabata Y and Gao JQ:
Mesenchymal stem cells: A promising targeted-delivery vehicle in
cancer gene therapy. J Control Release. 147:154–162. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao Z, Zhang L, Hu J and Sun Y:
Mesenchymal stem cells: A potential targeted-delivery vehicle for
anti-cancer drug, loaded nanoparticles. Nanomedicine. 9:174–184.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Salehinejad P, Alitheen NB,
Nematollahi-Mahani SN, Ali AM, Omar AR, Janzamin E and Hajghani M:
Effect of culture media on expansion properties of human umbilical
cord matrix-derived mesenchymal cells. Cytotherapy. 14:948–953.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dwyer RM, Ryan J, Havelin RJ, Morris JC,
Miller BW, Liu Z, Flavin R, O'Flatharta C, Foley MJ, Barrett HH, et
al: Mesenchymal stem cell-mediated delivery of the sodium iodide
symporter supports radionuclide imaging and treatment of breast
cancer. Stem Cells. 29:1149–1157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhong X, Shi C, Gong J, Guo B, Li M and Xu
H: Experimental study of nasopharyngeal carcinoma radionuclide
imaging and therapy using transferred human sodium/iodide symporter
gene. PLoS One. 10:e01170532015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Visser FW, Muntinga JH, Dierckx RA and
Navis G: Feasibility and impact of the measurement of extracellular
fluid volume simultaneous with GFR by 125I-iothalamate. Clin J Am
Soc Nephrol. 3:1308–1315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Riekstina U, Cakstina I, Parfejevs V,
Hoogduijn M, Jankovskis G, Muiznieks I, Muceniece R and Ancans J:
Embryonic stem cell marker expression pattern in human mesenchymal
stem cells derived from bone marrow, adipose tissue, heart and
dermis. Stem Cell Rev. 5:378–386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nan C, Shi Y, Zhao Z, Ma S, Liu J, Yan D,
Song G and Liu H: Monosialoteterahexosyl ganglioside induces the
differentiation of human umbilical cord-derived mesenchymal stem
cells into neuron-like cells. Int J Mol Med. 36:1057–1062. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ding DC, Chang YH, Shyu WC and Lin SZ:
Human umbilical cord mesenchymal stem cells: A new era for stem
cell therapy. Cell Transplant. 24:339–347. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang C, Lei D, Ouyang W, Ren J, Li H, Hu J
and Huang S: Conditioned media from human adipose tissue-derived
mesenchymal stem cells and umbilical cord-derived mesenchymal stem
cells efficiently induced the apoptosis and differentiation in
human glioma cell lines in vitro. Biomed Res Int. 2014:1093892014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dayem M, Basquin C, Navarro V, Carrier P,
Marsault R, Chang P, Huc S, Darrouzet E, Lindenthal S and Pourcher
T: Comparison of expressed human and mouse sodium/iodide symporters
reveals differences in transport properties and subcellular
localization. J Endocrinol. 197:95–109. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou QB, Chen RF, Li ZH, Pan QH, Zhou JJ,
Tang QB and Chen JS: Human mucin 1 promoter drives human
sodium/iodide symporter gene targeting expression in pancreatic
carcinoma cells. Zhonghua Yi Xue Za Zhi. 87:2780–2784. 2007.(In
Chinese). PubMed/NCBI
|
|
25
|
Han H, He W, Wang W and Gao B: Inhibitory
effect of aqueous Dandelion extract on HIV-1 replication and
reverse transcriptase activity. BMC Complement Altern Med.
11:1122011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Che J, Doubrovin M, Serganova I, Ageyeva
L, Beresten T, Finn R and Blasberg R: HSP70-inducible
hNIS-IRES-eGFP reporter imaging: Response to heat shock. Mol
Imaging. 6:404–416. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Merron A, Peerlinck I, Martin-Duque P,
Burnet J, Quintanilla M, Mather S, Hingorani M, Harrington K, Iggo
R and Vassaux G: SPECT//CT imaging of oncolytic adenovirus
propagation in tumours in vivo using the Na//I symporter as a
reporter gene. Gene Ther. 14:1731–1738. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Spitzweg C, Baker CH, Bergert ER, O'Connor
MK and Morris JC: Image-guided radioiodide therapy of medullary
thyroid cancer after carcinoembryonic antigen promoter-targeted
sodium iodide symporter gene expression. Hum Gene Ther. 18:916–924.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eekels JJ, Pasternak AO, Schut AM, Geerts
D, Jeeninga RE and Berkhout B: A competitive cell growth assay for
the detection of subtle effects of gene transduction on cell
proliferation. Gene Ther. 19:1058–1064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Serrano-Nascimento C, da Silva Teixeira S,
Nicola JP, Nachbar RT, Masini-Repiso AM and Nunes MT: The acute
inhibitory effect of iodide excess on sodium/iodide symporter
expression and activity involves the PI3K/Akt signaling pathway.
Endocrinology. 155:1145–1156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Siddiqui F, Barton KN, Stricker HJ, Steyn
PF, Larue SM, Karvelis KC, Sparks RB, Kim JH, Brown SL and Freytag
SO: Design considerations for incorporating sodium iodide symporter
reporter gene imaging into prostate cancer gene therapy trials. Hum
Gene Ther. 18:312–322. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bitsika V, Roubelakis MG, Zagoura D,
Trohatou O, Makridakis M, Pappa KI, Marini FC, Vlahou A and Anagnou
NP: Human amniotic fluid-derived mesenchymal stem cells as
therapeutic vehicles: A novel approach for the treatment of bladder
cancer. Stem Cells Dev. 21:1097–1111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nedeau AE, Gallagher KA, Liu ZJ and
Velazquez OC: Elevation of hemopexin-like fragment of matrix
metalloproteinase-2 tissue levels inhibits ischemic wound healing
and angiogenesis. J Vasc Surg. 54:1430–1438. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu Y, Cheng M, Lu N, Luo S, Xie Y and Liu
D: Construction of NK4 gene lentiviral vector and its expression in
bone mesenchymal stem cells. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi.
28:976–981. 2011.(In Chinese). PubMed/NCBI
|
|
35
|
Notka F and Wagner R: Reprogramming a GFP
reporter gene subjects it to complex lentiviral gene regulation.
Methods Mol Biol. 813:85–106. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nethercott HE, Brick DJ and Schwartz PH:
Derivation of induced pluripotent stem cells by lentiviral
transduction. Methods Mol Biol. 767:67–85. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Buzon MJ, Erkizia I, Pou C, Minuesa G,
Puertas MC, Esteve A, Castello A, Santos JR, Prado JG,
Izquierdo-Useros N, et al: A non-infectious cell-based phenotypic
assay for the assessment of HIV-1 susceptibility to protease
inhibitors. J Antimicrob Chemother. 67:32–38. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim JE, Ahn BC, Hwang MH, Jeon YH, Jeong
SY, Lee SW and Lee J: Combined RNA interference of hexokinase II
and (131)I-sodium iodide symporter gene therapy for anaplastic
thyroid carcinoma. J Nucl Med. 52:1756–1763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Timmons CL, Shao Q, Wang C, Liu L, Liu H,
Dong X and Liu B: GB virus type C E2 protein inhibits human
immunodeficiency virus type 1 assembly through interference with
HIV-1 gag plasma membrane targeting. J Infect Dis. 207:1171–1180.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen L, Altmann A, Mier W, Eskerski H,
Leotta K, Guo L, Zhu R and Haberkorn U: Radioiodine therapy of
hepatoma using targeted transfer of the human sodium/iodide
symporter gene. J Nucl Med. 47:854–862. 2006.PubMed/NCBI
|
|
41
|
Saraswathy A, Nazeer SS, Nimi N, Arumugam
S, Shenoy SJ and Jayasree RS: Synthesis and characterization of
dextran stabilized superparamagnetic iron oxide nanoparticles for
in vivo MR imaging of liver fibrosis. Carbohydr Polym. 101:760–768.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ittrich H, Peldschus K, Raabe N, Kaul M
and Adam G: Superparamagnetic iron oxide nanoparticles in
biomedicine: Applications and developments in diagnostics and
therapy. Rofo. 185:1149–1166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hansen L, Hansen AB, Mathiasen AB, Ng M,
Bhakoo K, Ekblond A, Kastrup J and Friis T: Ultrastructural
characterization of mesenchymal stromal cells labeled with
ultrasmall superparamagnetic iron-oxide nanoparticles for clinical
tracking studies. Scand J Clin Lab Invest. 74:437–446. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chao Y, Karmali PP and Simberg D: Role of
carbohydrate receptors in the macrophage uptake of dextran-coated
iron oxide nanoparticles. Adv Exp Med Biol. 733:115–123. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
van Buul GM, Kotek G, Wielopolski PA,
Farrell E, Bos PK, Weinans H, Grohnert AU, Jahr H, Verhaar JA,
Krestin GP, et al: Clinically translatable cell tracking and
quantification by MRI in cartilage repair using superparamagnetic
iron oxides. PLoS One. 6:e170012011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Levin MC, Lidberg U, Jirholt P, Adiels M,
Wramstedt A, Gustafsson K, Greaves DR, Li S, Fazio S, Linton MF, et
al: Evaluation of macrophage-specific promoters using lentiviral
delivery in mice. Gene Ther. 19:1041–1047. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sykova E and Jendelova P: In vivo tracking
of stem cells in brain and spinal cord injury. Prog Brain Res.
161:367–383. 2007. View Article : Google Scholar : PubMed/NCBI
|