Introduction
Breast cancer is one of the most serious threats to
women's health (1). In Western
countries, breast cancer cases account for about 1/3 of all new
cancer cases in women, ranking the first in women with malignant
tumors (2). The incidence of breast
cancer is increasing at an annual rate of 2% in coastal cities of
China (3). The emergence of
multidisciplinary treatment methods such as targeted therapy and
the overall survival of patients with breast cancer has been
prolonged, but the survival rate of patients with advanced breast
cancer is still not significantly improved (4). Understanding the mechanism of breast
cancer is helpful for further improving the level of clinical
treatments of breast cancer.
The occurrence of tumors is caused by the activation
of oncogenes and inactivation of tumor-suppressor genes. The
changes of genes include two aspects, genetics and epigenetics.
Epigenetics can affect gene transcription and translation, but DNA
sequences are not changed. Epigenetics can affect signal
transduction pathway of single cells for several times at different
sites, leading to disorders of cell structure and internal
environment (5). Epigenetic
alterations include DNA methylation, histone modification,
chromatin remodeling and RNA interference (6,7).
Hypermethylation of promoter region of tumor suppressor genes is
the most important research area, because it occurs at the early
stage of malignant tumor formation. DNA methylation is the best
understood and important epigenetic modification, and it converts
cytosine to 5-methylcytocine under the action of DNA methyl
transferases (DNMTs). Aberrant methylation of DNA can lead to the
activation of oncogenes and inactivation of tumor-suppressor genes,
being closely related to the occurrence of multiple tumors
(7,8).
Opioid binding protein/cell adhesion molecule
(OPCML) is a novel tumor-suppressor gene located in the 11q
chromosome segment. The main function of OPCML is to regulate cell
adhesion and recognition, and it plays a role in inhibiting tumor
growth through the activation of relevant ion channels and
adenylate cyclase (9). The
expression and deficiency of OPCML gene are closely related to the
occurrence and development of many tumors (10,11). The
silencing of OPCML gene in human breast cancer cells is related to
its methylation status (12).
Luteolin is a common natural flavonoid compound,
that is rich in celery, green pepper and dandelion leaves. It has a
variety of biological activities, such as anti-oxidation,
anti-inflammation, anti-depression, anti-convulsion, anti-anxiety,
anti-allergy, and immunity improvement (13). Studies show that luteolin can inhibit
the growth of tumor cells through a variety of mechanisms in
gastric cancer and ovarian cancer (14,15), and
effectively improves the methylation status of various tumor cells
(16). These suggest that luteolin
is a potential antitumor adjuvant drug. In the present study, we
evaluate whether luteolin is able to restore OPCML activity in
breast cancer cells, and investigate the molecular mechanism of its
low methylation activity.
Materials and methods
Cells
Human breast cancer BT474, MCF-7 and MDA-MB-231
cells were purchased from American Type Culture Collection
(Manassas, VA, USA), and cultured in RPMI-1640 medium (Mediatech,
Manassas, VA, USA) supplemented with 10% fetal bovine serum in a
humidified incubator with 5% CO2. The number of passages
of all cells was strictly controlled and mycoplasma contamination
was regularly monitored.
MTT assay
MTT assay was used to determine the effect of
luteolin on cell activity. Briefly, 100 µl medium containing
3×103 cells were inoculated in a 96-well plate in
triplicate wells overnight. Then, the medium was replaced with
RPMI-1640 supplemented with 5% fetal bovine serum, and the cells
were incubated with different concentrations of luteolin for 72
h.
Western blotting of protein expression
and phosphorylation
Cells were washed with phosphate-buffered saline and
used for protein extraction from nucleus and cytoplasm. After
determination of protein concentrations, protein samples (50 µg)
were mixed with equal volume of 2X sodium dodecyl sulfate loading
buffer before denaturation in boiling water bath for 5 min.
Afterwards, 20 µl samples were subjected to sodium dodecyl
sulfate-polyacrylamide gel electrophoresis at 100 V. The resolved
proteins were transferred to polyvinylidene difluoride membranes on
ice (300 mA, 1.5 h) and blocked with 5% skimmed milk at room
temperature for 2 h. Then, the membranes were incubated with rabbit
anti-human polyclonal OPCML primary antibody (ab100923; Abcam,
Cambridge, UK), anti-phospho-Histone H2A.X (Ser139) antibody
(2577), total H2A.X antibody (2595), cleaved caspase-8 antibody
(9496) and cleaved PARP antibody (9541; all from Cell Signaling
Technology, Inc., Beverly, MA, USA) and anti-GAPDH (G9545;
Sigma-Aldrich, St. Louis, MO, USA) at 4°C overnight. After
extensive washing with phosphate-buffered saline with Tween-20 for
3 times of 15 min, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibody for 1 h at room
temperature before washing with phosphate-buffered saline with
Tween-20 for 3 times of 15 min. Then, the membrane was developed
with enhanced chemiluminescence detection kit (EMD Millipore,
Billerica, MA, USA) for imaging (VL Chemi-Smart 3000;
Viogene-Biotek Corp., Sunnyvale, CA, USA). The relative content of
target protein was expressed against GAPDH.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent (Thermo
Fisher Scientific, Inc., Waltham, MA, USA). cDNA was obtained by
reverse transcription using Promega's reverse transcription system
(Promega Corporation, Madison, WI, USA). Then, 10 mmol/l forward
and reverse primers and SYBR were added for RT-qPCR amplification
on ABI 7500 (both from Thermo Fisher Scientific, Inc.). The primers
for OPCML were 5′-GGGTCTGTGGGTACCTGTTC-3′ (forward) and
5′-TATGGACCACTTGTCATTCC-3′ (reverse); and for β-actin were
5′-GTCTTCCCCTCCATCGTG-3′ (forward) and 5′-AGGGTGAGGATGCCTCTCTT-3′
(reverse). qPCR protocol was: Initial denaturation at 95°C for 2
min; 32 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30
sec; final elongation at 72°C for 5 min. The 2−ΔΔCq
method was used to calculate the relative expression of target mRNA
against β-actin. All samples were measured in triplicate, and mean
values were considered for comparative analysis.
Determination of methylation
activity
Cells were washed with 1 ml precooled
phosphate-buffered saline and collected. Then, the cells were
centrifuged at 800 rpm and 4°C for 5 min before removing
supernatants. Total nuclear proteins were extracted from BT474 or
MCF-7 cells using protein extraction kit (Pierce, Rockford, IL,
USA). According to the manufacturer's manual, DNMT analysis kit
(Epigentek Group Inc., Brooklyn, NY, USA) was used to determine
methylation activity. Micropores in the kit were coated with
cytosine-rich DNA. DNMT can catalyze the binding of methyl group of
S-adenosylmethionine onto cytosine of DNA. Methylated DNA can be
recognized by anti-5-methylated cytosine antibody, and
Enzyme-linked immunosorbent assay was performed to determine the
amount of methylated DNA, which indirectly reflected methylation
activity.
Analysis of Sp1 and NF-κB
activities
After treatment with luteolin, nuclear proteins were
extracted from the cells using protein extraction kit (Pierce).
Bradford method was used to determine the concentrations of
proteins. Activities of transcription factors Sp1 and NF-κB were
determined according to the manufacturer's manual (EK1090 and
EK1121; Affymetrix, Santa Clara, CA, USA). Primary antibodies of
Sp1 and p65 were provided by the kits. Absorbance was determined at
450 nm to indirectly reflect the activities of Sp1 and NF-κB.
DNA extraction and hydrolysis
After treatment with luteolin, 1×107 BT474 or MCF-7
cells were used for the extraction of genomic DNA according to a
previous report (17). Briefly, 200
ng genomic DNA was denaturated at 100°C for 3 min, before being
cooled on ice. Then, 1/10 volumes of ammonium acetate (0.1 mol/l,
pH 5.3) and 2 U of nuclease P1 (US Biological, Swampscott, MA, USA)
were added. After incubation at 45°C for 2 h, 1/10 volumes of
NH4HCO3 (1 mol/l) and 0.002 U of snake venom phosphodiesterase I
(Sigma-Aldrich) were added before incubation at 37°C for 1 h. After
addition of 0.5 U alkaline phosphatase, the samples were incubated
at 37°C for 1 h.
High performance liquid
chromatography-electrosprary ionization-mass spectrometry
(HPLC-ESI-MS/MS)
HPLC-ESI-MS/MS was performed using HPLC system
(model 1100; Agilent, Santa Clara, CA, USA) and chromatographic
column (Atlantis dC18 column), and pre-column (both from
Waters, Milford, MA, USA). The mobile phase was 0.1% formic
acid-methanol. Flow velocity was 0.2 ml/min. Electrospray
ionization mode was positive ion. Scanning range was m/z 100–2,000.
Ion source temperature was 450°C. Spray voltage was 415 kV, cluster
voltage was 55 V, and entrance voltage was 6 V. Impact energy was
13 V. Curtain gas pressure was 138 kPa, gas 1 pressure was 221 kPa,
gas 1 pressure was 379 kPa, and collision gas pressure was 41 kPa
(17). Sciex Analyst software
version 1.3.1 was used to analyze the data.
Lentiviral infection
About 105 cells were inoculated in 6-well
plates overnight. Before lentiviral infection, 293T cells were used
to pack lentiviral vectors, and then BT474 or MCF-7 cells were
inoculated into 24-well plates at a density of 105/well.
After 24 h, Cells were transformed with lentivirus. On the first
day of transformation, the medium was replaced with fresh medium
containing 5 µg/ml polybrene, and then lentiviral particles were
added until multiplicity of infection reached 10. After incubation
overnight, the medium was replaced with fresh complete medium,
followed by addition of luteolin.
Flow cytometry
Cell cycle and DNA content of apoptotic cells were
detected by flow cytometry after 24 h of drug exposure. Cells were
collected and analyzed using flow cytometry. The number of cells
was quantified by ModFit LT software (BD Biosciences, Franklin
Lakes, NJ, USA).
Statistical analysis
The results were analyzed using SPSS 17.0
statistical software (SPSS, Inc., Chicago, IL, USA). The data were
expressed as the mean ± standard deviation. Comparison between
groups was performed using analysis of variance with Dunnett's test
as the post hoc test. Differences with P<0.05 were considered
statistically significant.
Results
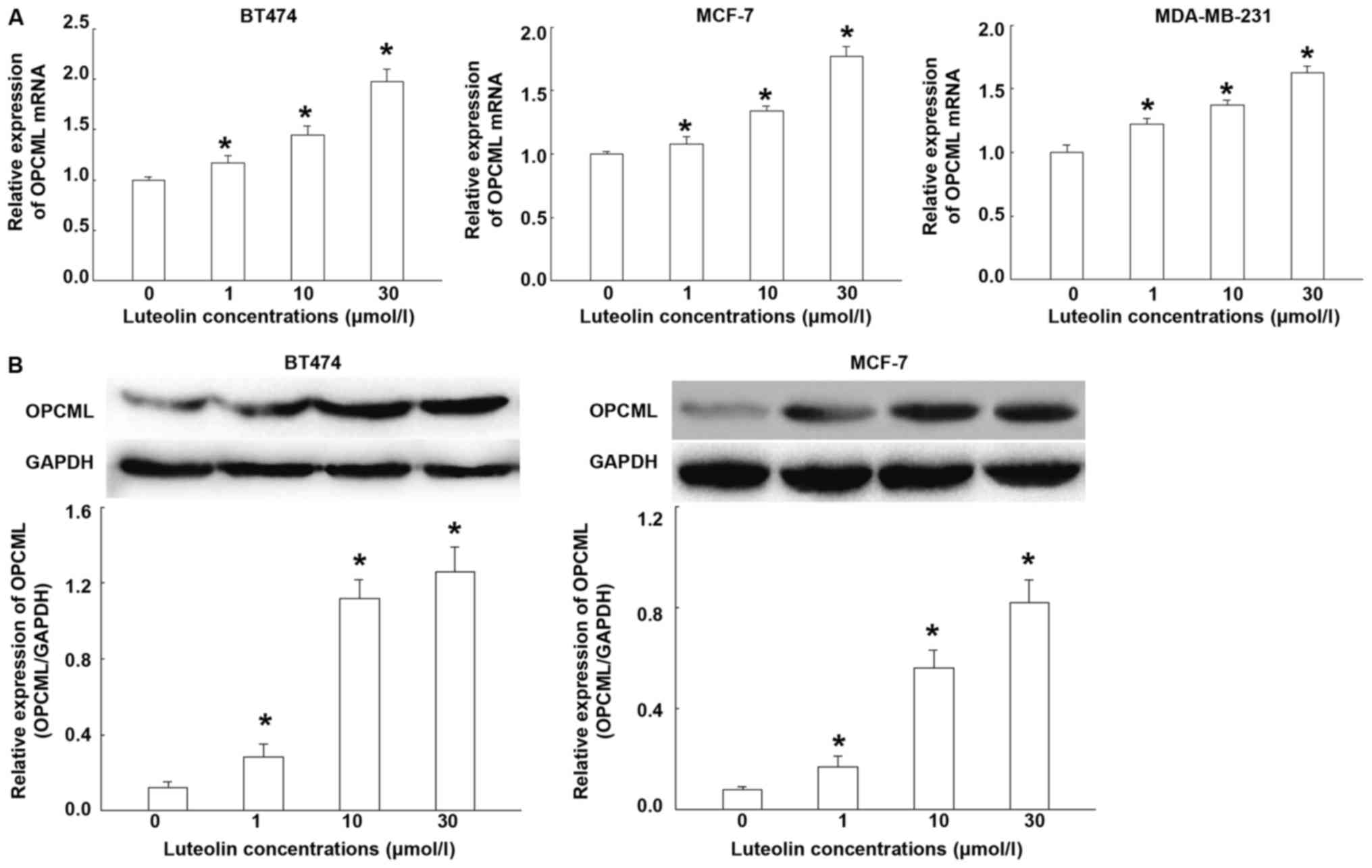
Luteolin effectively upregulates the
expression of OPCML in breast cancer cells
To test the effect of luteolin on the expression of
OPCML mRNA and protein, RT-qPCR and Western blotting were
performed. RT-qPCR showed that the levels of OPCML mRNA were
increased with the increase of luteolin concentrations (Fig. 1A). Western blotting showed that
luteolin treatment effectively increased protein expression of
OPCML, being consistent with mRNA trend (Fig. 1B). In addition, the effect of
luteolin on other breast cancer cells (MCF-7 and MDA-MB-231) were
also tested and similar results were obtained. The results suggest
that luteolin effectively upregulates the expression of OPCML in
breast cancer cells.
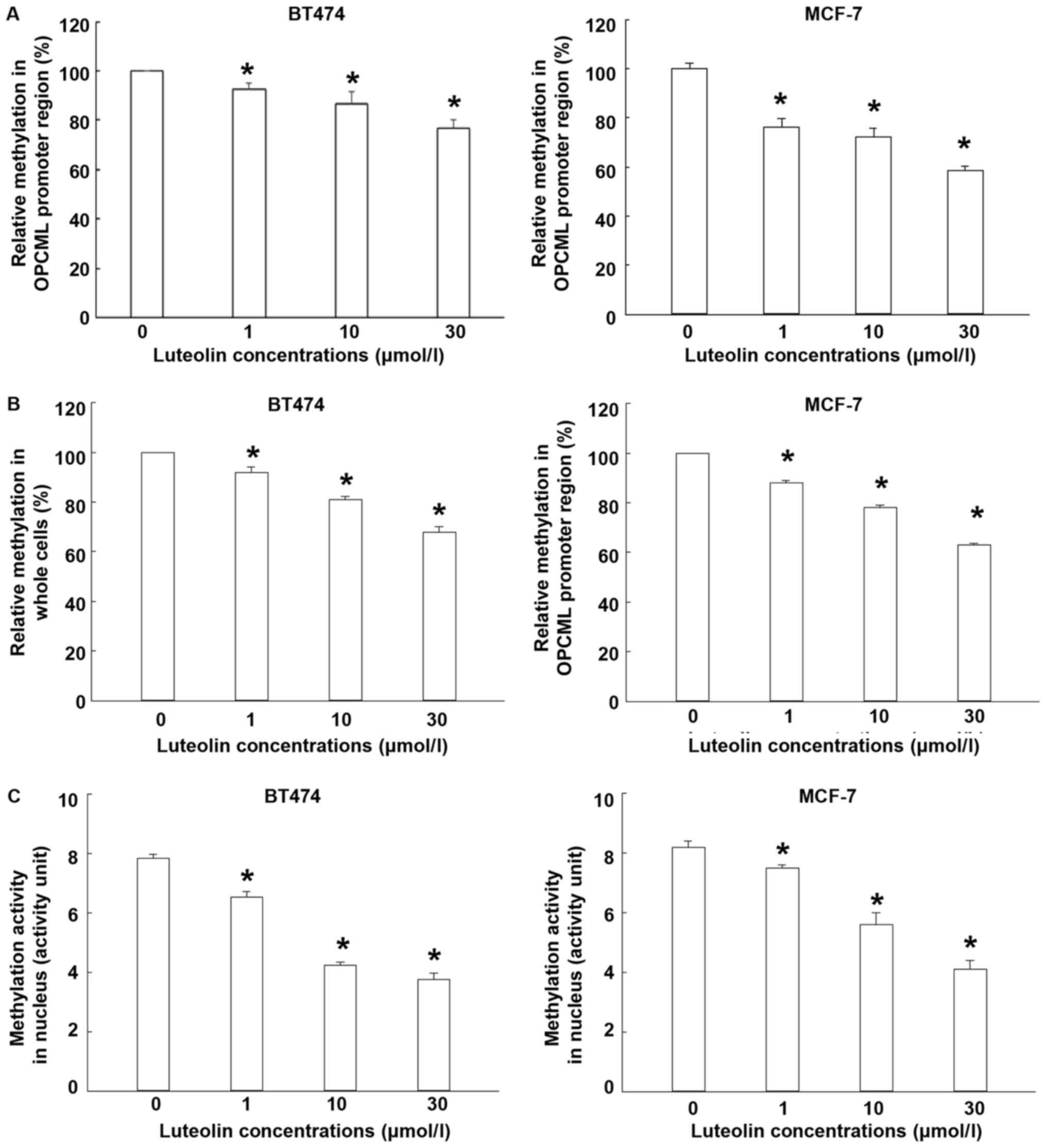
Luteolin activates OPCML by reducing
intracellular methylation levels
To determine the methylation level and activity,
LC-MS/MS and EpiQuik TM DNMT1 detection kit were used. The data
showed that treatment with different concentrations of luteolin
significantly decreased the methylation level of OPCML promoter
region (P<0.05; Fig. 2A). In the
meantime, luteolin treatment reduced the global DNA methylation
level compared with control (P<0.05; Fig. 2B). Moreover, methylation activity of
nucleoprotein in BT474 and MCF-7 cells were significantly reduced
by luteolin treatment (Fig. 2C). The
results indicate that luteolin activates OPCML by reducing
intracellular methylation levels.
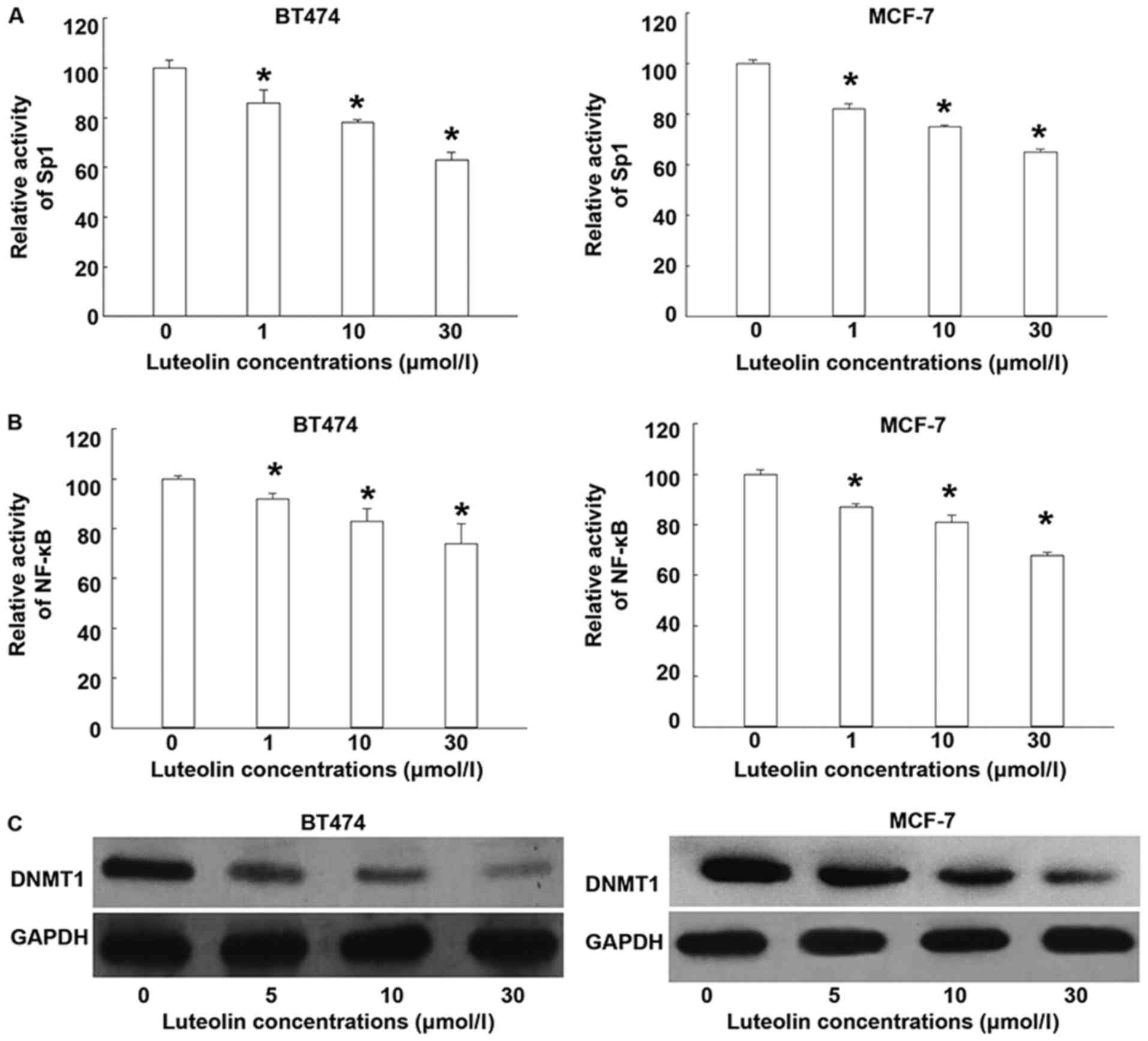
Luteolin downregulates intracellular
methylation levels by decreasing Sp1 and NF-κB activities
To test whether luteolin affects the activities of
Sp1 and NF-κB that regulate DNMT1 activity, ELISA and western
blotting were used. ELISA showed that luteolin treatments
significantly reduced the activities of Sp1 and NF-κB (P<0.05;
Fig. 3A and B). Western blotting
showed that luteolin inhibited the expression of DNMT1 protein
(Fig. 3C). The results suggest that
luteolin downregulates intracellular methylation levels by
decreasing Sp1 and NF-κB activities.
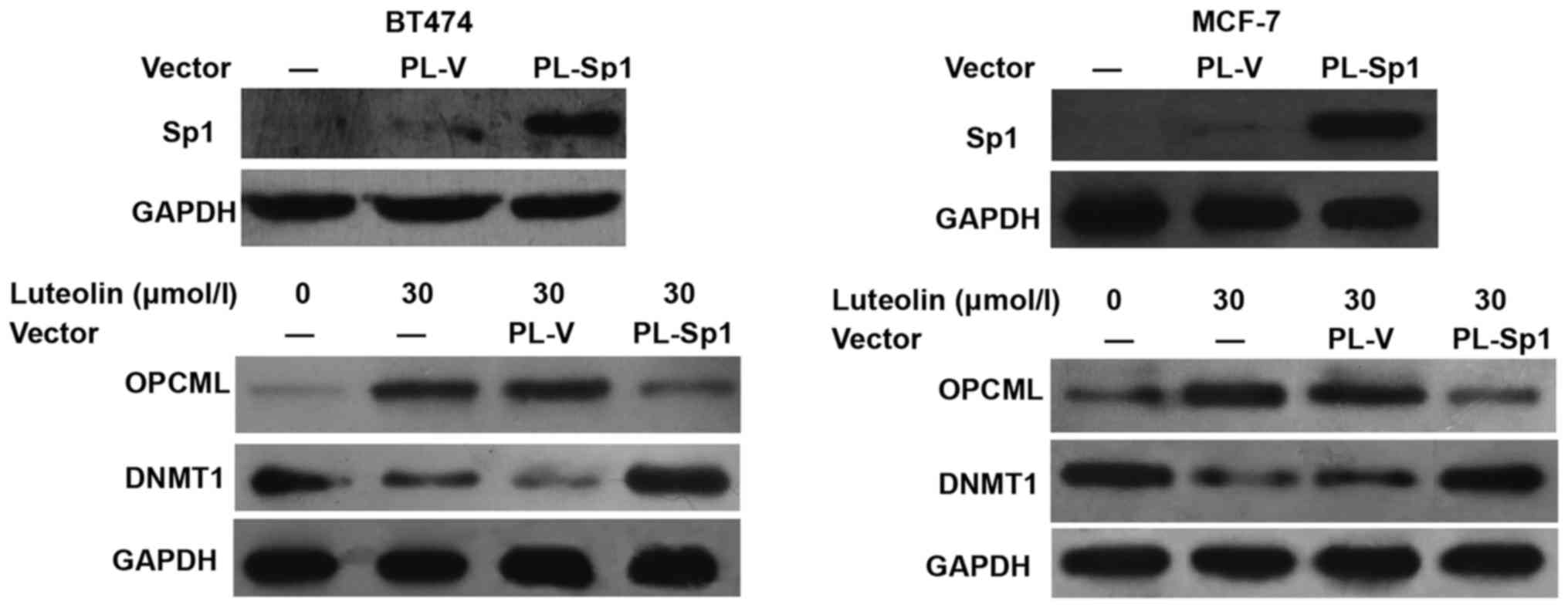
Luteolin affects the expression of
DNMT1 and OPCML by downregulating Sp1 activity
To examine whether luteolin affects the expression
of DNMT1 and OPCML via Sp1, we overexpressed Sp1 in BT474 and MCF-7
cells. The data showed that elevated expression of Sp1 attenuated
the reduction of DNMT1 expression induced by luteolin. In the
meantime, OPCML expression was inhibited by Sp1 overexpression
(Fig. 4). The result indicates that
luteolin affects the expression of DNMT1 and OPCML by
downregulating Sp1 activity.
Luteolin inhibits the proliferation
and induces the apoptosis of breast cancer cells
To evaluate the effect of luteolin on the
proliferation of breast cancer cells, MTT assay and flow cytometry
were carried out after treating the cells with different
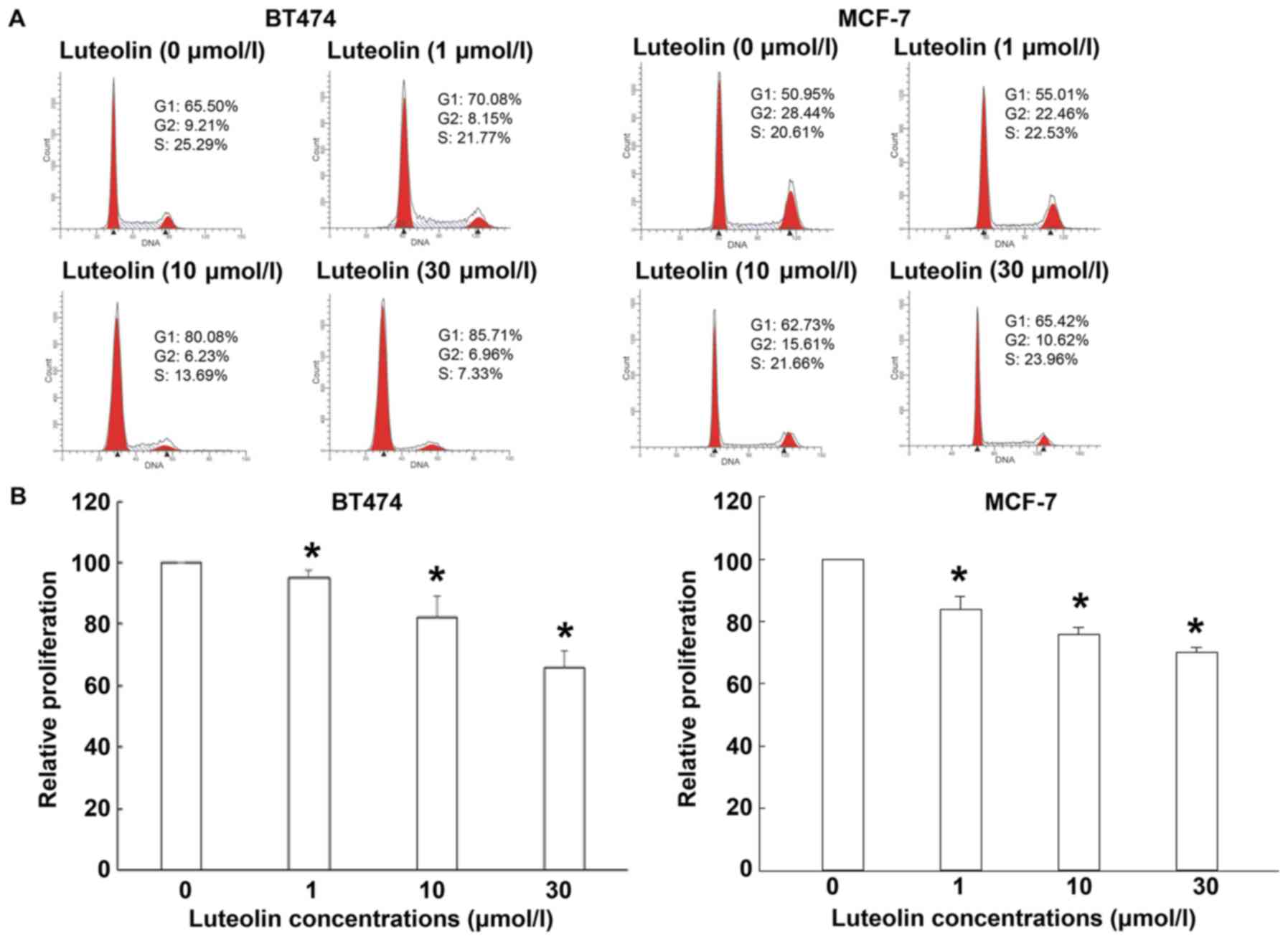
concentrations of luteolin. Cell cycle analysis showed that the
percentages of BT474 and MCF-7 cells in G1 phase after treatment
with luteolin were significantly increased, while the percentages
of BT474 and MCF-7 cells in G2 phase were significantly decreased
(P<0.05). In addition, the percentage of BT474 cells in S phase
was significantly decreased (P<0.05) (Fig. 5A). MTT assay showed that luteolin
inhibited the proliferation of BT474 and MCF-7 cells in a
dose-dependent manner (Fig. 5B).
Flow cytometry showed that luteolin induced apoptosis of BT474 and
MCF-7 cells (Fig. 5C). To test
whether luteolin induces DNA double-strand breaks and cell
apoptosis, Western blotting was performed. The data showed that
luteolin (5–30 µmol/l) significantly increased the expression of
γ-H2AX (Ser139) and slicing of caspase-8 and PARP (Fig. 5D). The results suggest that luteolin
inhibits the proliferation and induces the apoptosis of breast
cancer cells.
Discussion
Over the past half century, the incidence of breast
cancer has steadily increased, but the mortality rate of breast
cancer is relatively stable. This change is mainly due to the
deepening of knowledge of breast cancer. Like other malignant
tumors, breast cancer is the result of the activation of oncogenes
and inactivation of tumor-suppressor genes. Hypermethylation of the
promoter region of tumor-suppressor gene occurs at the early stage
of malignant tumor formation, and is the most studied topic. DNA
methylation is an important epigenetic mechanism of transcriptional
regulation in eukaryotes. Changes in methylation patterns may
promote the occurrence of tumors. OPCML gene is a tumor-suppressor
gene discovered in recent years. It is a member of IgLON family
that belongs to immunoglobulin superfamily. OPCML widely exists in
nervous system cells, and plays important roles in regulating
opioid receptors and signal transduction, promoting cell
differentiation and changing cell membrane properties (18). It is reported that the expression of
OPCML in a variety of malignant tumor tissues is deficient or low,
and the promoter region of OPCML is usually methylated (19). In addition, the expression of OPCML
is closely related to the recurrence, metastasis and prognosis of
tumor patients (19).
Because of the reversibility of epigenetic changes,
a new method of tumor therapy has been developed, and it has
achieved some effect in the treatment of leukemia. Currently, DNA
demethylation agents such as decitabine and azacytidine are already
used in the clinical treatment of myelodysplastic syndrome
(20). Luteolin is a natural
flavonoid that is extracted from mignonette. The two adjacent polar
-OH groups at C3 and C4 of luteolin benzene ring B are essential
for the inhibition of enzyme activity. The conjugated double bond
between C2 and C3 makes B rings and C rings in the same plane,
being beneficial for approaching kinase substrate binding sites.
These two kinds of structures are crucial for the inhibition of
cell proliferation by luteolin (21,22).
Luteolin inhibits tumor cell growth factors by changing cell
signaling pathways or resists cancer cell invasion by changing
kinase activity (23). It can also
inhibit tumor cell growth by blocking cell cycles (24). Our study on OPCML mRNA and protein
expression shows that luteolin increases OPCML mRNA and protein
expression in a dose-dependent manner, suggesting that luteolin
induces the reexpression of OPCML by some unknown mechanism. Our
data also show that the methylation status upstream of OPCML gene
and in the cells is higher, and treatment with luteolin reduces the
methylation level, suggesting that luteolin reverses the
methylation status of OPCML gene in tumor cells, which is
beneficial to its transcription and expression. The intracellular
methylation activity is mainly related to DNMTs, so the decrease of
intracellular methylation level after luteolin treatment may be
caused by the inhibition of DNMTs expression or biological activity
(25). Our data on DNMT1 expression
and intracellular methylation show that luteolin downregulates the
expression of DNMT1, and inhibits intracellular methylation
activity. This may be a direct mechanism by which OPCML expression
is upregulated.
Sp1 transcription factor belongs to the Sp protein
family. Sp1 promotes the expression of a number of molecules that
positively regulate cell cycle, such as cyclin D1, E2F1, c-fos, and
TGF-α, etc. In addition, Sp1 can also affect the methylation of CpG
island of DNA. In a variety of tumor tissues, such as gastric
cancer, pancreatic cancer, breast cancer and thyroid tumor tissues,
Sp1 is often highly expressed, and its activity is positively
correlated with the degree of tumor infiltration, and negatively
correlated with the prognosis of patients (26,27). In
addition, Sp1 can also promote the expression of DNMT1 by forming a
complex with NF-κB. Our data show that the activities of Sp1 and
NF-κB in control group are high, and treatment with different
concentrations of luteolin significantly reduces the activities of
the two transcription factors. Yu et al (28) report that curcumin inhibits the
expression of DNMT1 in human leukemia cells, possibly by inhibiting
the activity of Sp1. Du et al (29) have shown similar results. In
addition, overexpression of Sp1 in cells attenuates the inhibitory
effect of luteolin on DNMT1 expression, and reverses the
upregulating effect of luteolin on OPCML, suggesting that Sp1
affects the methylation and expression of OPCML by regulating
intracellular levels of DNMT1. Sp1 expression is elevated in
various tumor tissues, and correlated with poor prognosis.
Therefore, targeted therapy against Sp1 activity can improve the
prognosis of tumors to some extent, and luteolin has a beneficial
effect on the tumors by affecting Sp1 activity.
For a variety of chemotherapeutic drugs, the
inhibition of tumor cell proliferation and apoptosis is the
ultimate goal. The main manifestations at the early stage of
apoptosis are the phosphorylation of H2AX (γ-H2AX) and the
activation of caspase family proteins (30). The γ-H2AX is an important sign of
early apoptosis, and the activation of Caspase family proteins can
act on DNA repair enzyme, poly ADP-ribose polymerase (PARP), to
deactivate it (31). Our study on
the phosphorylation of H2AX and the expression of PARP shows that
luteolin treatment inhibits the proliferation of cells, promotes
the phosphorylation of H2AX and the activation of caspase, and
induces the expression of PARP, suggesting that the inhibitory
effect of luteolin on breast cancer cells may be achieved by
inducing apoptosis. In conclusion, the present study suggests that
luteolin inhibits the growth of breast cancer by decreasing the
methylation and upregulating the expression of OPCML gene. Although
additional experiments involving OPCML-silenced cells are required
to further confirm that OPMLC mediates the antitumor effects of
luteolin, there are a few pieces of evidence that support our
hypothesis that inactivated OPCML by methylation is associated with
multiple malignancies (32,33). Its effect is similar with that of
5-aza-2-deoxycytidine. However, 5-aza-2-deoxycytidine may also
inhibit human normal gene methylation when suppressing tumor gene
methylation. This limits its clinical application. Luteolin is
originated from natural plants. Although animal experiments show
that luteolin has low toxicity (34,35), its
clinical application still remain to be studied. Therefore, the
application values of luteolin in tumor chemotherapy will be
further enhanced, if its structure is modified to reduce its effect
on normal cells without compromising its antitumor activity.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XD, JianZ and JirenZ collaborated to design the
study. FY, JW, RC and TW were responsible for performing the
experiments. XD and JianZ analyzed the data. All authors
collaborated to interpret the results and develop the manuscript.
The final version of the manuscript was read and approved by all
authors, and each author believes that the manuscript represents
honest work.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sauvaget C, Nishino Y, Konno R, Tase T,
Morimoto T and Hisamichi S: Challenges in breast and cervical
cancer control in Japan. Lancet Oncol. 17:e305–e312. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schoemaker MJ, Jones ME, Wright LB,
Griffin J, McFadden E, Ashworth A and Swerdlow AJ: Psychological
stress, adverse life events and breast cancer incidence: A cohort
investigation in 106,000 women in the United Kingdom. Breast Cancer
Res. 18:722016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yan X, Han R, Zhou J, Yu H, Yang J and Wu
M: Incidence, mortality and survival of female breast cancer during
2003–2011 in Jiangsu province, China. Chin J Cancer Res.
28:321–329. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Golubnitschaja O, Debald M, Yeghiazaryan
K, Kuhn W, Pešta M, Costigliola V and Grech G: Breast cancer
epidemic in the early twenty-first century: Evaluation of risk
factors, cumulative questionnaires and recommendations for
preventive measures. Tumour Biol. 37:12941–12957. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee JY and Kong G: Roles and epigenetic
regulation of epithelial-mesenchymal transition and its
transcription factors in cancer initiation and progression. Cell
Mol Life Sci. 73:4643–4660. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kasai H: What causes human cancer?
Approaches from the chemistry of DNA damage. Genes Environ.
38:192016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song Y, Wu F and Wu J: Targeting histone
methylation for cancer therapy: ENzymes, inhibitors, biological
activity and perspectives. J Hematol Oncol. 9:492016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shui IM, Wong CJ, Zhao S, Kolb S, Ebot EM,
Geybels MS, Rubicz R, Wright JL, Lin DW, Klotzle B, et al: Prostate
tumor DNA methylation is associated with cigarette smoking and
adverse prostate cancer outcomes. Cancer. 122:2168–2177. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Duarte-Pereira S, Paiva F, Costa VL,
Ramalho-Carvalho J, Savva-Bordalo J, Rodrigues A, Ribeiro FR, Silva
VM, Oliveira J, Henrique R and Jerónimo C: Prognostic value of
opioid binding protein/cell adhesion molecule-like promoter
methylation in bladder carcinoma. Eur J Cancer. 47:1106–1114. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou F, Tao G, Chen X, Xie W, Liu M and
Cao X: Methylation of OPCML promoter in ovarian cancer tissues
predicts poor patient survival. Clin Chem Lab Med. 52:735–742.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou F, Ma M, Tao G, Chen X, Xie W, Wang Y
and Cao X: Detection of circulating methylated opioid binding
protein/cell adhesion molecule-like gene as a biomarker for ovarian
carcinoma. Clin Lab. 60:759–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J,
Zhang Q, Jin J, Liu D, Rhim JS, Rha SY, et al: OPCML is a broad
tumor suppressor for multiple carcinomas and lymphomas with
frequently epigenetic inactivation. PLoS One. 3:e29902008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu CW, Lin HW, Yang DJ, Chen SY, Tseng
JK, Chang TJ and Chang YY: Luteolin inhibits viral-induced
inflammatory response in RAW264.7 cells via suppression of STAT1/3
dependent NF-κB and activation of HO-1. Free Radic Biol Med.
95:180–189. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Lee IM, Zhang SM, Blumberg JB,
Buring JE and Sesso HD: Dietary intake of selected flavonols,
flavones, and flavonoid-rich foods and risk of cancer in
middle-aged and older women. Am J Clin Nutr. 89:905–912. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gates MA, Tworoger SS, Hecht JL, De Vivo
I, Rosner B and Hankinson SE: A prospective study of dietary
flavonoid intake and incidence of epithelial ovarian cancer. Int J
Cancer. 121:2225–2232. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Landis-Piwowar KR, Milacic V and Dou QP:
Relationship between the methylation status of dietary flavonoids
and their growth-inhibitory and apoptosis-inducing activities in
human cancer cells. J Cell Biochem. 105:514–523. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Z, Liu S, Xie Z, Blum W, Perrotti D,
Paschka P, Klisovic R, Byrd J, Chan KK and Marcucci G:
Characterization of in vitro and in vivo hypomethylating effects of
decitabine in acute myeloid leukemia by a rapid, specific and
sensitive LC-MS/MS method. Nucleic Acids Res. 35:e312007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sellar GC, Watt KP, Rabiasz GJ, Stronach
EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B,
Scott D, et al: OPCML at 11q25 is epigenetically inactivated and
has tumor-suppressor function in epithelial ovarian cancer. Nat
Genet. 34:337–343. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li C, Tang L, Zhao L, Li L, Xiao Q, Luo X,
Peng W, Ren G, Tao Q and Xiang T: OPCML is frequently methylated in
human colorectal cancer and its restored expression reverses EMT
via downregulation of smad signaling. Am J Cancer Res. 5:1635–1648.
2015.PubMed/NCBI
|
|
20
|
Blum W and Marcucci G: Targeting
epigenetic changes in acute myeloid leukemia. Clin Adv Hematol
Oncol. 3:855–865, 882. 2005.PubMed/NCBI
|
|
21
|
Guerrero L, Castillo J, Quiñones M,
Garcia-Vallvé S, Arola L, Pujadas G and Muguerza B: Inhibition of
angiotensin-converting enzyme activity by flavonoids:
Structure-activity relationship studies. PLoS One. 7:e494932012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu YC, Leung SW, Yeung DK, Hu LH, Chen GH,
Che CM and Man RY: Structure-activity relationships of flavonoids
for vascular relaxation in porcine coronary artery. Phytochemistry.
68:1179–1188. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lin Y, Shi R, Wang X and Shen HM:
Luteolin, a flavonoid with potential for cancer prevention and
therapy. Curr Cancer Drug Targets. 8:634–646. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seelinger G, Merfort I, Wölfle U and
Schempp CM: Anti-carcinogenic effects of the flavonoid luteolin.
Molecules. 13:2628–2651. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jeltsch A: Molecular enzymology of
mammalian DNA methyltransferases. Curr Top Microbiol Immunol.
301:203–225. 2006.PubMed/NCBI
|
|
26
|
Sankpal UT, Goodison S, Abdelrahim M and
Basha R: Targeting Sp1 transcription factors in prostate cancer
therapy. Med Chem. 7:518–525. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li L and Davie JR: The role of Sp1 and Sp3
in normal and cancer cell biology. Ann Anat. 192:275–283. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu J, Peng Y, Wu LC, Xie Z, Deng Y, Hughes
T, He S, Mo X, Chiu M, Wang QE, et al: Curcumin down-regulates DNA
methyltransferase 1 and plays an anti-leukemic role in acute
myeloid leukemia. PLoS One. 8:e559342013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Du L, Xie Z, Wu LC, Chiu M, Lin J, Chan
KK, Liu S and Liu Z: Reactivation of RASSF1A in breast cancer cells
by curcumin. Nutr Cancer. 64:1228–1235. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chiu SJ, Chao JI, Lee YJ and Hsu TS:
Regulation of gamma-H2AX and securin contribute to apoptosis by
oxaliplatin via a p38 mitogen-activated protein kinase-dependent
pathway in human colorectal cancer cells. Toxicol Lett. 179:63–70.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Simbulan-Rosenthal CM, Rosenthal DS, Iyer
S, Boulares H and Smulson ME: IInvolvement of PARP and poly
(ADP-ribosyl)ation in the early stages of apoptosis and DNA
replication. Mol Cell Biochem. 193:137–148. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sellar GC, Watt KP, Rabiasz GJ, Stronach
EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B,
Scott D, et al: OPCML at 11q25 is epigenetically inactivated and
has tumor-suppressor function in epithelial ovarian cancer. Nat
Genet. 34:337–343. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J,
Zhang Q, Jin J, Liu D, Rhim JS, Rha SY, et al: OPCML is a broad
tumor suppressor for multiple carcinomas and lymphomas with
frequently epigenetic inactivation. PLoS One. 3:e29902008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu H, Zeng Z, Wang S, Li T, Mastriani E,
Li QH, Bao HX, Zhou YJ, Wang X, Liu Y, et al: Main components of
pomegranate, ellagic acid and luteolin, inhibit metastasis of
ovarian cancer by down-regulating MMP2 and MMP9. Cancer Biol Ther.
18:990–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Katalinić M, Rusak G, Domaćinović Barović
J, Sinko G, Jelić D, Antolović R and Kovarik Z: Structural aspects
of flavonoids as inhibitors of human butyrylcholinesterase. Eur J
Med Chem. 45:186–192. 2010. View Article : Google Scholar : PubMed/NCBI
|