Introduction
Gastric cancer is the fourth most common malignancy
and the second leading cause of cancer-associated mortality
worldwide (1). An estimated 60% of
patients with advanced gastric cancer succumb to peritoneal
metastasis, which accounts for 50% of recurrences, thus making it
the leading cause of mortality associated with this disease
(2,3). Peritoneal metastasis may be regarded as
a complex and multi-step process that consists of a series of
independent stages, whereby cancer cells migrate from the primary
neoplasm to a distant location. To date, the mechanisms of
peritoneal metastasis of gastric cancer have remained elusive, and
molecular markers for gastric cancer metastasis and tumor
progression remain to be identified. Data from available studies
are difficult to interpret due to the complexity arising from tumor
heterogeneity. Thus, identification of molecular and biological
alterations that occur during the peritoneal metastasis of gastric
cancer may facilitate the investigation of the pathology of the
disease and lead to the discovery of novel prognostic markers to
more accurately predict clinical outcomes and to individualize
treatments for patients with gastric cancer.
MicroRNAs (miRNAs/miRs) are a series of non-coding
RNAs of 19–25 nt in length that regulate gene expression at the
post-transcriptional level (4,5). miRNAs
may silence their target genes by inhibiting mRNA translation or
degrading the mRNA molecules by binding to their 3-untranslated
region, thereby having a crucial role in cancer biology. Abundant
evidence has demonstrated that dysregulated expression of certain
miRNAs is involved in cancer progression and metastasis, and that
these miRNAs may serve as novel biomarkers or therapeutic targets
(6–8). With the recent development of miRNA
detection techniques, identification of miRNAs associated with
cancer progression/metastasis may complement and enhance the
current understanding of the origins and evolution of peritoneal
metastasis, and may revolutionize the pre-operative prediction of
the status of peritoneal metastases from gastric cancer. However,
despite such great promise, the role of miRNAs in gastric cancer,
particularly during peritoneal metastasis, remains to be
elucidated. Therefore, in the present study, miRNA expression
profiles were examined in gastric cancer cell lines with different
metastatic potential by miRNA microarray analysis. The biological
functions of miR-21-5p, a miRNA whose downregulation was identified
to be associated with peritoneal metastasis, were subsequently
examined by cell migration and invasion assays.
Materials and methods
Cell lines and culture
The human gastric cancer cell line GC9811 and its
derived sub-cell line GC9811-P with high peritoneal metastatic
potential were obtained from the State Key Laboratory of Cancer
Biology at Xijing Hospital of Digestive Diseases (The Fourth
Military Medical University, Xi'an, China). The GC9811 cell line
was originally purchased from American Type Culture Collection
(Manassas, VA, USA). The GC9811-P cell line was established and
characterized as having a high metastatic potential in the
peritoneum by orthotopic tumor cell implantation (9). Cells were cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS; both Gibco; Thermo
Fisher Scientific, Inc. Thermo Fisher Scientific, Inc., Waltham,
MA, USA), 100 units/ml penicillin and 0.1 mg/ml streptomycin (both
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in 6-well plates
(Corning Costar; Corning Inc., Corning, NY, USA) at 37°C in a
humidified atmosphere containing 5% CO2.
RNA extraction and small RNA
isolation
When the cells reached 80–90% confluence, they were
washed with ice-cold PBS three times and homogenized immediately in
TRK-100 lysis solution (LC Sciences, Houston, TX, USA). Small RNA
was extracted from each sample using an RNAiso for Small RNA kit
(cat. no. 9753A; Takara Biotechnology Co., Ltd., Dalian, China)
according to the manufacturer's protocol with certain
modifications; the extracted small RNA was resuspended and
incubated at −80°C overnight to enhance the precipitation
efficiency of RNAs with a low molecular weight (20–200 nt). The
concentration and purity of the small RNA were determined with an
ultraviolet spectrophotometer at 260 and 280 nm. Only the RNA
samples with a 260/280 nm absorbance ratio of >1.8 were used for
the subsequent analysis.
miRNA microarray assay
Microarray assays were performed by an external
service provider (LC Sciences). The assay began with a 4–8-µg total
RNA sample, which was 3-extended with a poly(A) tail using poly(A)
polymerase. An oligonucleotide tag was ligated to the poly(A) tail
for subsequent fluorescent dye staining. Hybridization was
performed overnight on a µParaflo microfluidic chip using a
micro-circulation pump (both Atactic Technologies, Inc., Houston,
TX, USA). On the microfluidic chip, each detection probe consisted
of a chemically modified nucleotide-coding segment complementary to
the target miRNA (from miRbase; http://www.mirbase.org/) or other RNA (control or
customer-defined sequences), and a spacer segment of polyethylene
glycol to extend the coding segment away from the substrate. The
detection probes were generated by in situ synthesis using a
PCR-generated template method. The melting temperatures of
hybridized products were balanced by chemical modifications of the
detection probes. Hybridization was performed using 100 l formamide
solution (25%) in 6X buffer (0.90 M NaCl, 60 mM
Na2HPO4, 6 mM EDTA; pH 6.8) at 34°C for 1 h.
Following RNA hybridization, tag-conjugating Cy3 dye was circulated
through the microfluidic chip for dye staining. Fluorescence images
were generated using a laser scanner (GenePix 4000B; Molecular
Devices LLC, Sunnyvale, CA, USA) and digitized using Array-Pro 3.0
image analysis software (Media Cybernetics, Inc., Rockville, MD,
USA). Data were analyzed by first subtracting the background and
subsequently normalizing the signals using locally-weighted scatter
plot smoothing regression.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
miRNA quantification was performed by RT-qPCR
analysis. A total of 40 ng total RNA containing miRNA was
polyadenylated by poly(A) polymerase (Takara Biotechnology Co.,
Ltd.) and reverse transcribed to complementary (c)DNA using a
PrimeScript miRNA cDNA Synthesis kit (Takara Biotechnology Co.,
Ltd.), according to the manufacturer's protocol. qPCR was performed
using the miScript SYBR-Green PCR kit (Takara Biotechnology, Co.,
Ltd.) with the included miScript Universal primer and the
miRNA-specific forward primers in an ABI PRISM 7900 Real-time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
miRNA-specific primer sequences were miR-146a-5p,
5-CGGTGAGAACTGAATTCCATGGGTT-3′; miR-181a-5p,
5-AACATTCAACGCTGTCGGTGAGT-3′; miR-106b-5p,
5-CGGTAAAGTGCTGACAGTGCACGAT-3′; miR-199a-3p,
5-CGGACAGTAGTCTGCACATTGGTTA-3′; miR-148a-3p,
5-CGGTCAGTGCACTACAAGAACTTTGT-3′; miR-21-5p,
5-CGGTAGCTTATCAGACTGATGTTGA-3; miR-222-3p,
5-AGCTACATCTGGCTACTGGGTC-3′; and miR-221-3p,
5-AGCTACATTGTCTGCTGGGTTTC-3. Sequences were designed based on the
miRNA sequences obtained from the miRBase database (http://microrna.sanger.ac.uk/). Each reaction was
performed in a final volume of 20 µl containing 2 µl cDNA, 0.5
mmol/l of each primer and SYBR-Green PCR Master mix (Takara
Biotechnology Co., Ltd.). Following an initial incubation at 95°C
for 15 min, 40 cycles of denaturation at 94°C for 15 sec, annealing
at 55°C for 30 sec and extension at 70°C for 30 sec were performed.
PCRs were performed in triplicate for each sample and experiments
were repeated a minimum of three times. At the end of the PCR
cycles, melting curve analyses were performed in addition to
electrophoresis of the products on 3.5% agarose gels. U6 small
nuclear RNA 6, pseudogene was used as an endogenous control
(primers: U6 stem-loop forward, 5′-AGCGGGAAATCGTGCGTGACA-3′ and
reverse, 5′-GTGGACTFGGGAGAGGACTGG-3). Relative expression of the
target gene was calculated using the formula RQ=2−∆∆Ct
as described previously (10).
Lentiviral transfection
To silence miR-21-5p expression, validated specific
small interfering RNA oligonucleotides (5-UCAACAUCAGUCUGAUAAGCUA-3)
were cloned into the lentiviral GV369/GV280 vector (GeneChem Inc.,
Shanghai, China). Lentiviral vector construction and transfection
were performed according to the manufacturer's protocols. Cells
were seeded in antibiotic-free medium at 1 day prior to
transfection using Lipofectamine 2000 reagent (GE Healthcare Life
Sciences, Little Chalfont, UK). At 24 and 48 h after transfection,
cells were collected and the transfection and gene silencing
efficiency was determined by detecting the fluorescence intensity
of green fluorescent protein and RT-qPCR analysis,
respectively.
Cell invasion assay
In vitro invasion assays were performed using
24-well Transwell chambers (Corning, Inc.) as described previously
(11,12). Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA) was diluted 1:8 with cold serum-free DMEM and
coated onto the Transwell inserts. Cells were plated into the upper
chambers at 2×104 cells per well in 100 µl serum-free
DMEM. The bottom chamber was filled with DMEM containing 10% FBS.
Following incubation at 37°C for 24 h, non-invasive cells in the
upper chamber were removed. Invasive cells on the lower surface of
the inserts were fixed with 4% paraformaldehyde at room temperature
for 30 min, stained with 0.2% crystal violet at room temperature
for 2 h and counted under a light microscope (magnification, ×100).
Each experiment was performed three times in triplicate wells.
Wound healing assay
In brief, cells were seeded in 6-well plates at a
density of 2×105 per well. After 12 h, cells achieved
100% confluence. Two parallel line-shaped wounds of 1 mm in width
were generated using 200 µl pipette microtips (Corning, Inc.).
Subsequently, the cells were washed with PBS twice and cultured in
serum-free medium at 37°C. The size of the wound was measured under
phase-contrast microscopy (Carl Zeiss AG, Oberkochen, Germany) at 0
and 24 h post-wounding.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism version 5.02 (GraphPad Software, Inc., La Jolla, CA, USA).
Student's t-tests, and one-way analysis of variance followed by the
Student-Newman-Keuls post-hoc test were employed. P<0.05 was
considered to indicate a statistically significant difference.
Results
Deregulated miRNAs associated with the
peritoneal metastatic potential of gastric cancer cells
The initial step of the present study was to search
for miRNAs that are potentially involved in the peritoneal
metastasis of gastric cancer cells by means of a microarray
analysis. By using the expression levels of miRNAs in GC9811 cells
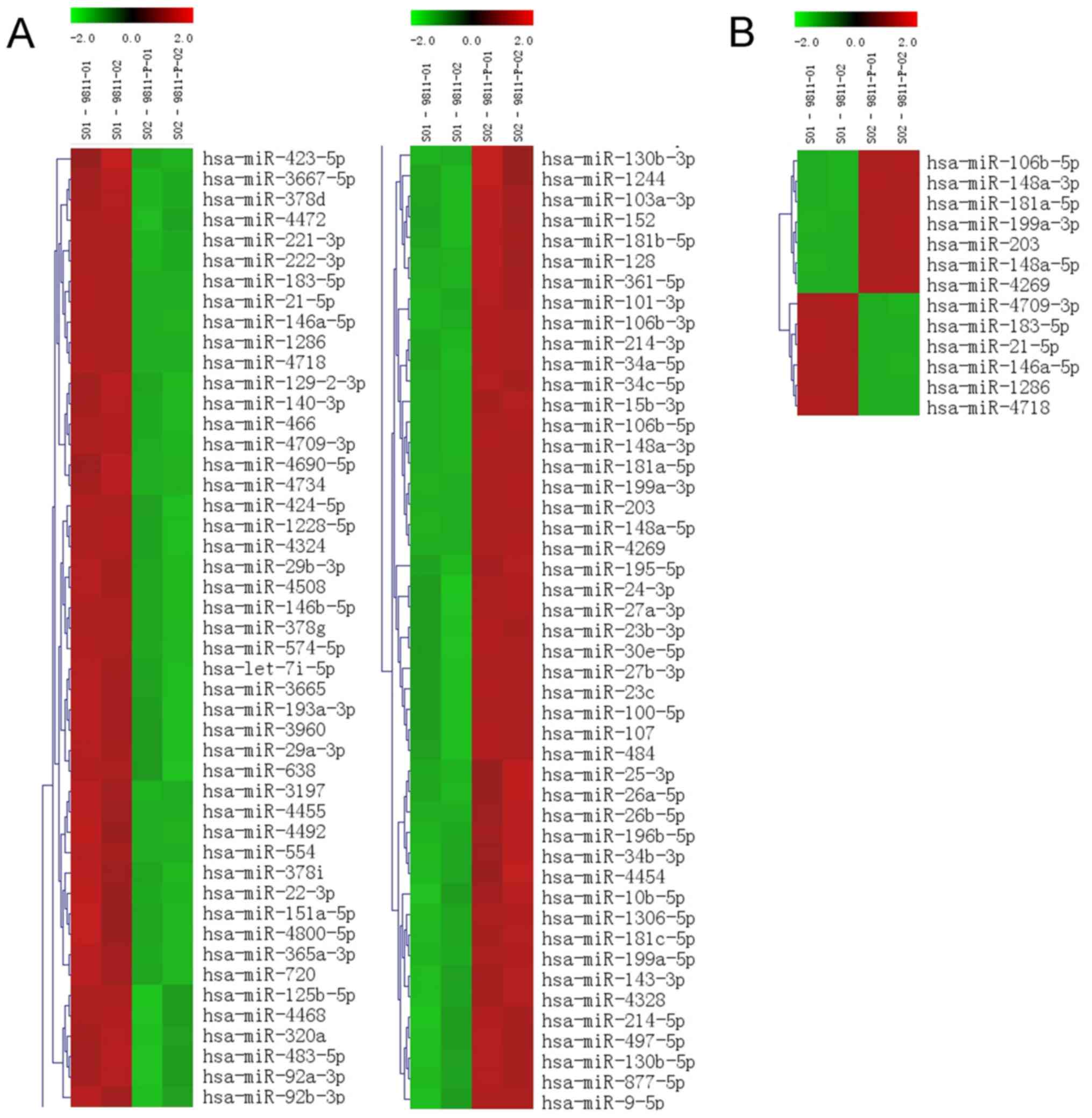
as a control, 153 miRNAs were identified to be differentially
expressed in GC9811-P cells with high metastatic potential via
hierarchical clustering analysis, including 74 upregulated miRNAs
and 79 downregulated miRNAs (Fig.
1A). The absolute value of the fold change was defined as ≥2.
Among the differentially expressed miRNAs, 67 had a signal
intensity of >500. Of the 13 miRNAs obtained based on the
criteria of a signal intensity of >500 and P≤0.01 (Fig. 1B), 8 miRNAs with significant fold
changes were selected for further analysis, comprising 4
downregulated miRNAs (miR-146a-5p, miR-21-5p, miR-221-3p and
miR-222-3p) and 4 upregulated miRNAs (miR-181a-5p, miR106b-5p,
miR-199a-3p and miR-148a-3p; Table
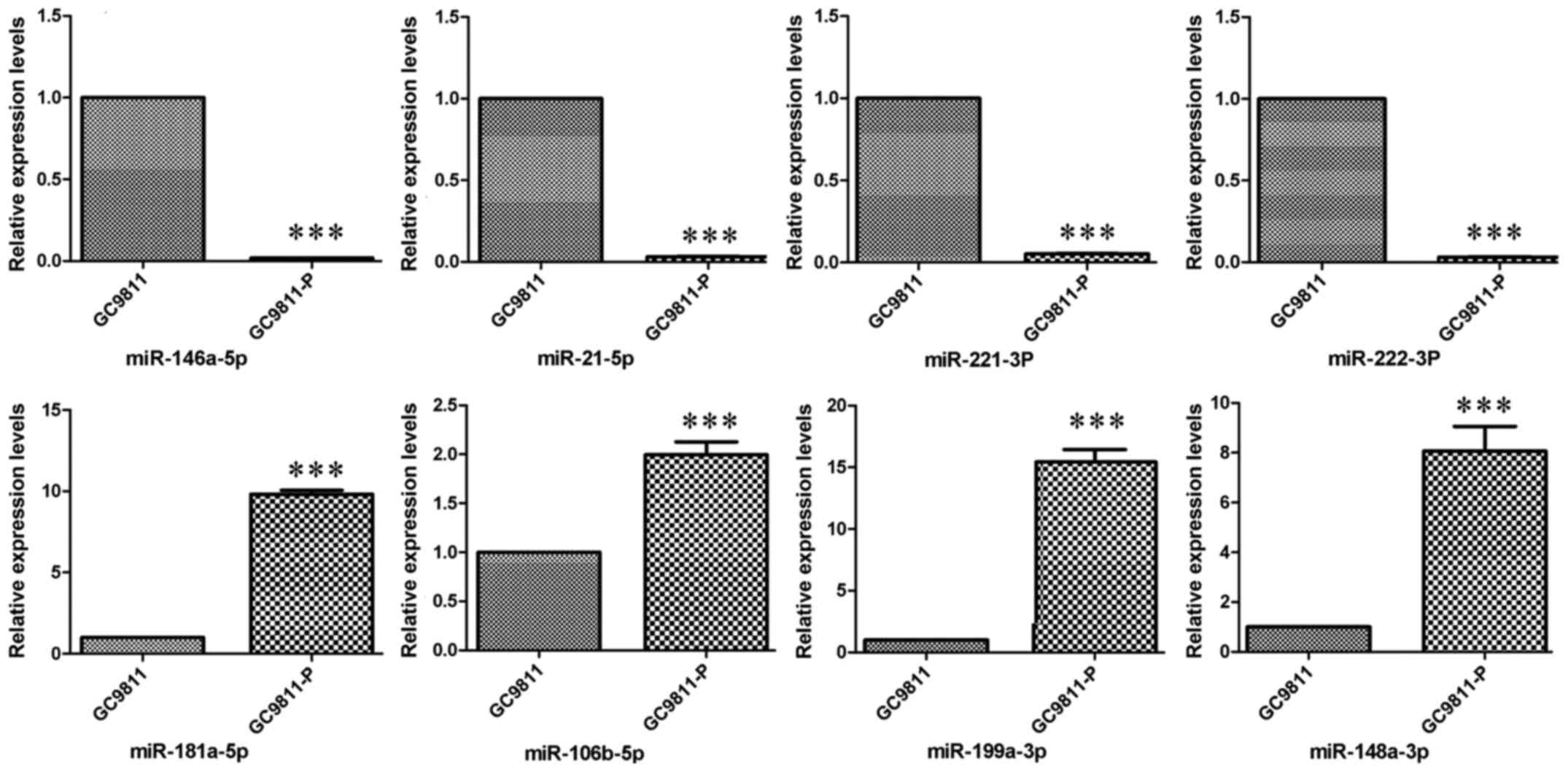
I). The expression levels of these miRNAs were confirmed in
GC9811 and GC9811-P cells by RT-qPCR analysis (Fig. 2).
 | Table I.Differentially expressed miRNAs
detected by microarray analysis. |
Table I.
Differentially expressed miRNAs
detected by microarray analysis.
| miRNA | Fold change | P-value |
|---|
| miR-146a-5p | −208.3 |
2.70×10−3 |
| miR-21-5p | −10.48 |
7.40×10−3 |
| miR-221-3p | −13.98 |
1.26×10−2 |
| miR-222-3p | −9.74 |
1.01×10−2 |
| miR-181a-5p | 13.20 |
4.28×10−3 |
| miR-106b-5p | 7.02 |
5.98×10−3 |
| miR-199a-3p | 14.52 |
6.19×10−3 |
| miR-148a-3p | 85.44 |
6.49×10−3 |
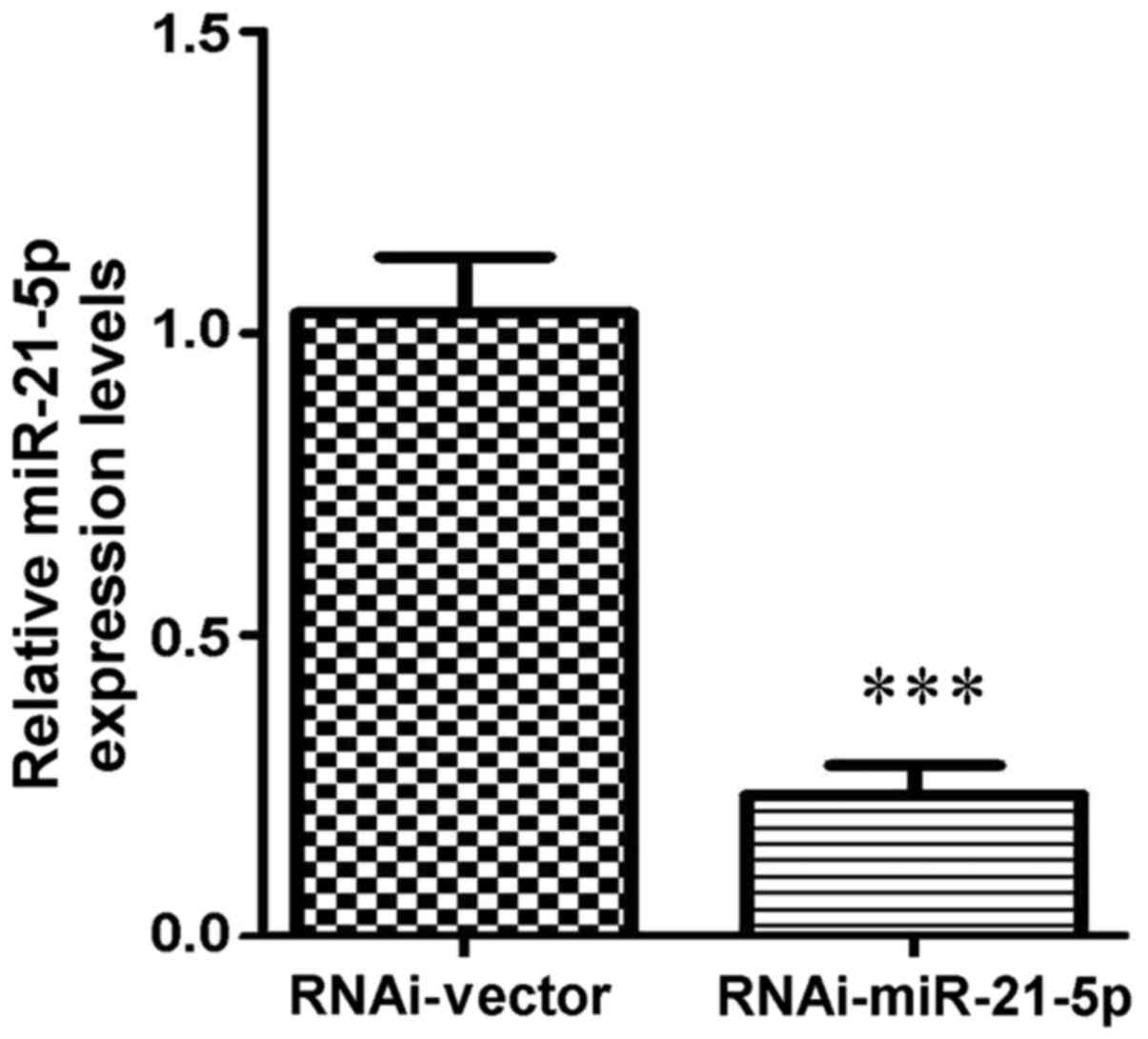
Knockdown of miR-21-5p promotes the
migration of GC9811 cells
Based on the above data, miR-21-5p was selected for
further analysis. Lentiviral transfection was performed to achieve
knockdown of miR-21-5p in GC9811 cells, in which miR-21-5p
expression was increased compared with that in GC9811-P cells.
Following 72 h of transfection followed by puromycin selection,
RT-qPCR analysis confirmed that the expression levels of miR-21-5p
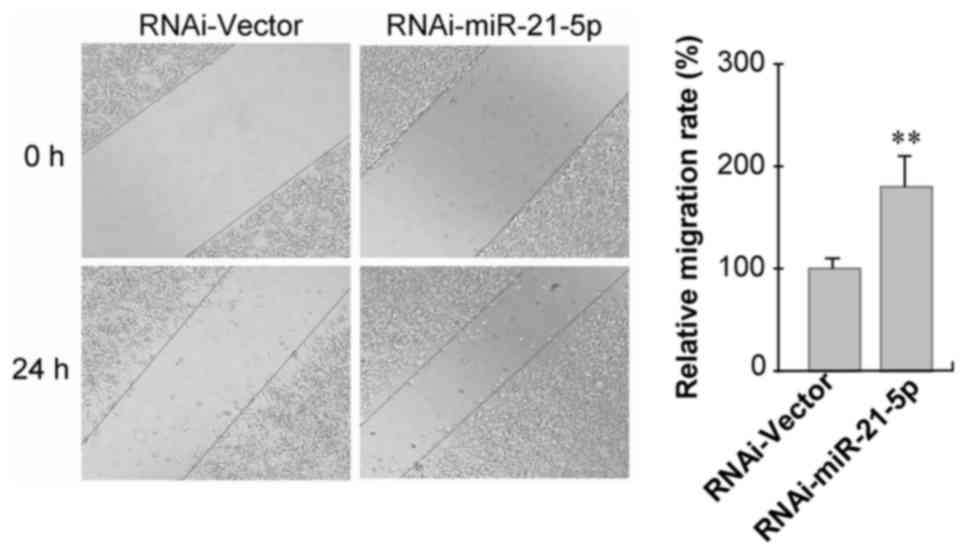
were significantly downregulated in GC9811 cells (Fig. 3). To clarify the functions of
miR-21-5p in gastric cancer progression and metastasis, the effect
of miR-21-5p knockdown on GC9811 cell migration and invasion was
examined. Confluent GC9811 cells with or without knockdown of
miR-21-5p were scratched and cell migration was observed. It was
observed that GC9811 cells with miR-21-5p knockdown exhibited a
statistically significantly accelerated closure of the wound area
compared with that of the empty vector-infected control cells
(Fig. 4).
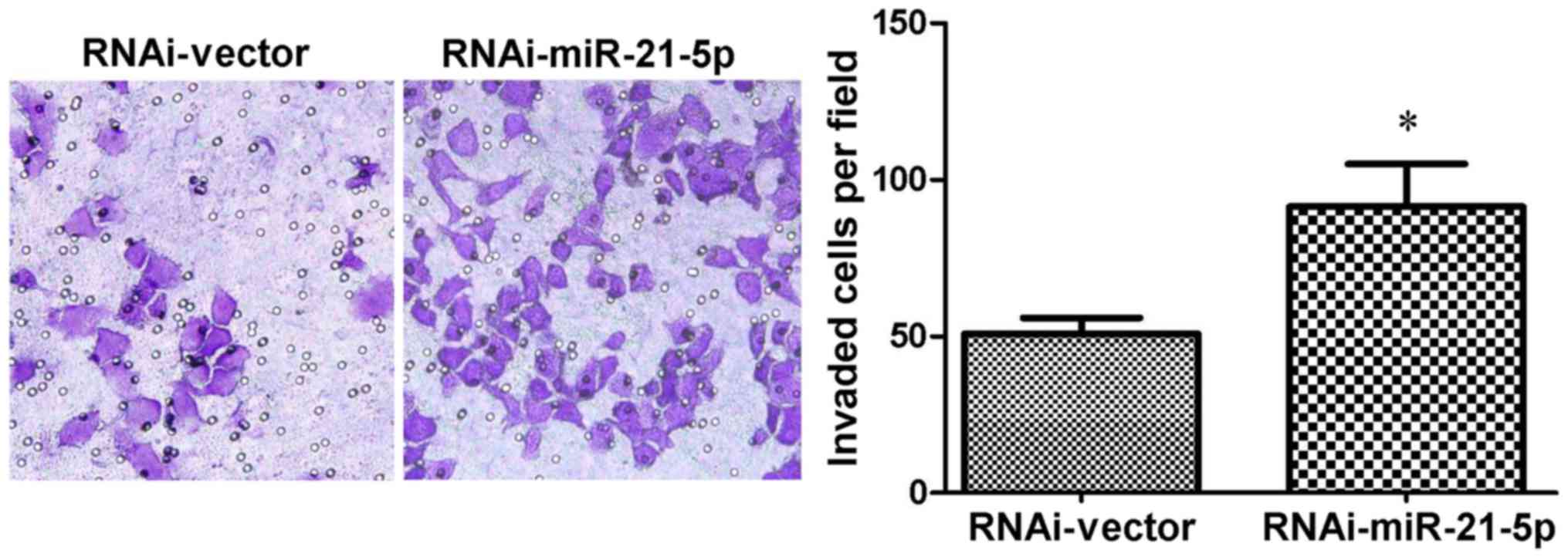
Knockdown of miR-21-5p elevates the
invasiveness of GC9811 cells
Matrigel invasion assays were subsequently performed
to confirm the effect of miR-21-5p on the metastatic progression of
gastric cancer cells. As expected, it was observed that GC9811
cells with miR-21-5p knockdown exhibited a significant increase in
their invasive capacity compared with that of the
vector-transfected control cells (Fig.
5).
Discussion
Accumulating evidence suggests that miRNAs serve
important roles as either oncogenes or tumor suppressor genes in
the initiation and progression of cancer. The prognostic and
therapeutic potential of a number of miRNAs that have been examined
in various cancer types, including gastric cancer (13), but remain insufficiently elucidated.
In particular, studies examining the miRNA expression status
specifically associated with the peritoneal metastasis of gastric
cancer are currently lacking. The principal reasons for this
include unavailability or difficulty in obtaining model systems,
including proper cell lines and gene-engineered animals. The
present study focused on the miRNA expression profiles in a pair of
gastric cancer cell lines with different peritoneal metastatic
potential using microarray analysis. Among the 13 miRNAs selected
according to the criteria of signal intension >500 and P≤0.01, 8
significantly altered miRNAs with fold changes in expression of
>2 in the GC9811-P cell line as compared with that in the GC9811
cell line were selected. In line with previous studies on other
cancer types (14–22), the miRNAs with an altered expression
status may be associated with the metastatic potential of gastric
cancer.
It is conceivable that aberrant miRNA expression may
contribute to the progression of cancer and that these miRNAs may
serve as regulators in peritoneal metastasis (23). To this end, the expression level of
miR-21-5p, one transcript of miR-21, was further investigated
regarding its biological functional relevance to cell invasion and
metastasis of gastric cancer. Previous studies have demonstrated
that miR-21 may act as an oncogene that is overexpressed and
hyperactivated in a variety of cancer types, and that its
upregulation promotes the proliferation, apoptosis, invasion and
chemoresistance by targeting numerous tumor suppressor genes,
including phosphatase and tensin homologue, programmed cell death
4, tissue inhibitor of metalloproteinases 3, tropomyosin 1, ras
homolog gene family member B and maspin (24–28). It
appears that the majority of studies has considered miR-21-5p a
cancer-promoting ‘oncomiRs’ that drives tumor progression and
development (29,30). The present study, however, indicated
that downregulated miR-21-5p expression may be associated with the
peritoneal metastatic potential of gastric cancer cells.
Furthermore, in vitro functional assays demonstrated that
silencing of miR-21-3p expression in the native GC9811 cell line
markedly enhanced its migratory and invasive potential, suggesting
that miR-21-3p may act as a tumor suppressor to prevent gastric
cancer peritoneal metastasis. The intrinsic discrepancies between
different cancer types and cancer cell lines may be a possible
explanation. These results provide novel clues regarding the role
of miR-21-5p and indicate its potential clinical value by rendering
it a novel target for therapeutic interventions in gastric cancer
with peritoneal metastasis.
The target genes of miR-21-5p remain to be
completely elucidated. It was speculated that miR-21-5p may inhibit
gastric cancer metastasis by regulating its specific target genes.
A bioinformatics analysis using the MiRanda online prediction tool
(http://www.microrna.org/microrna/home.do) identified
five genes that are associated with miR-21-5p in the GC9811-P cell
line, including stromal antigen 2, nuclear factor I/B,
methylthioadenosine phosphorylase, activity-dependent
neuroprotector homeobox and chromodomain helicase DNA binding
protein 7. Based on this, further examination and validation of the
precise signaling pathways and molecular mechanisms underlying the
involvement of miR-21-5p in the peritoneal metastasis of gastric
cancer is warranted.
In conclusion, the present study identified altered
miRNA expression patterns associated with peritoneal metastasis in
gastric cancer cells. The results also suggested that
downregulation of miR-21-5p may be associated with the peritoneal
metastatic potential of gastric cancer cells. These data further
support the notion that altered miRNA expression contributes to the
peritoneal metastasis of gastric cancer, and provide molecular
biomarkers that may be of therapeutic value for patients with
gastric cancer metastasis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81760440 and
81860426), the Natural Science Foundation of Ningxia, China (grant
no. 2018AAC02016), and the Regional Science and Technology
Development Program Conducted by the Central Government of China
(grant no. YDZX20176400004650).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YF, FaB, CW, XL and RX performed the experiments,
and YF and FaB analyzed and interpreted the data. YY, FeB and YN
conceived the study, designed the experiment and drafted the
manuscript. All authors have read and approved the final version of
the manuscript.
Ethics approval and informed consent
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yonemura Y, Endou Y, Sasaki T, Hirano M,
Mizumoto A, Matsuda T, Takao N, Ichinose M, Miura M and Li Y:
Surgical treatment for peritoneal carcinomatosis from gastric
cancer. Eur J Surg Oncol. 36:1131–1138. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bozzetti F, Yu W, Baratti D, Kusamura S
and Deraco M: Locoregional treatment of peritoneal carcinomatosis
from gastric cancer. J Surg Oncol. 98:273–276. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans. Cell.
75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takei Y, Takigahira M, Mihara K, Tarumi Y
and Yanagihara K: The metastasis-associated microRNA miR-516a-3p is
a novel therapeutic target for inhibiting peritoneal dissemination
of human scirrhous gastric cancer. Cancer Res. 71:1442–1453. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nishida N, Mimori K, Fabbri M, Yokobori T,
Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y and Mori M:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang S, He L, Zhao X, Miao Y, Gu Y, Guo
C, Xue Z, Dou W, Hu F, Wu K, et al: MicroRNA let-7f inhibits tumor
invasion and metastasis by targeting MYH9 in human gastric cancer.
PLoS One. 6:e184092011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai F, Guo X, Yang L, Wang J, Shi Y, Zhang
F, Zhai H, Lu Y, Xie H, Wu K and Fan D: Establishment and
characterization of a high metastatic potential in the peritoneum
for human gastric cancer by orthotopic tumor cell implantation. Dig
Dis Sci. 52:1571–1578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
You Y, Liu J, Wang Z, Zhang Y, Ran Y, Guo
X, Liu H and Wang H: The enhancement of radiosensitivity in human
esophageal squamous cell carcinoma cells by zoledronic acid and its
potential mechanism. Cytotechnology. 66:17–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
You Y, Yang W, Qin X, Wang F, Li H, Lin C,
Li W, Gu C, Zhang Y and Ran Y: ECRG4 acts as a tumor suppressor and
as a determinant of chemotherapy resistance in human nasopharyngeal
carcinoma. Cell Oncol(Dordr). 38:205–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tseng CW, Lin CC, Chen CN, Huang HC and
Juan HF: Integrative network analysis reveals active microRNAs and
their functions in gastric cancer. BMC Syst Biol. 5:992011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu K, LI G, Fan C, Diao Y, Wu B and LI J:
Increased expression of microRNA-221 in gastric cancer and its
clinical significance. J Int Med Res. 40:467–474. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kogo R, Mimori K, Tanaka F, Komune S and
Mori M: Clinical significance of miR-146a in gastric cancer cases.
Clin Cancer Res. 17:4277–4284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Vandenboom TG II, Wang Z, Kong D,
Ali S, Philip PA and Sarkar FH: miR-146a suppresses invasion of
pancreatic cancer cells. Cancer Res. 70:1486–1495. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bhaumik D, Scott GK, Schokrpur S, Patil
CK, Campisi J and Benz CC: Expression of microRNA-146 suppresses
NF-kappaB activity with reduction of metastatic potential in breast
cancer cells. Oncogene. 27:5643–5647. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hurst DR, Edmonds MD, Scott GK, Benz CC,
Vaidya KS and Welch DR: Breast cancer metastasis suppressor 1
up-regulates miR-146, which suppresses breast cancer metastasis.
Cancer Res. 69:1279–1283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin SL, Chiang A, Chang D and Ying SY:
Loss of mir-146a function in hormone-refractory prostate cancer.
RNA. 14:417–424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Magrelli A, Azzalin G, Salvatore M,
Viganotti M, Tosto F, Colombo T, Devito R, Di Masi A, Antoccia A,
Lorenzetti S, et al: Altered microRNA expression patterns in
hepato-blastoma patients. Transl Oncol. 2:157–163. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song G, Zeng H, LI J, Xiao L, He Y, Tang Y
and Li Y: miR-199a regulates the tumor suppressor mitogen-activated
protein kinase 11 in gastric cancer. Biol Pharm Bull. 33:1822–1827.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV and Cheng JQ:
MicroRNA expression profling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ichimi T, Enokida H, Okuno Y, Kunimoto R,
Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama
K, et al: Identifcation of novel microRNA targets based on microRNA
signatures in bladder cancer. Int J Cancer. 125:345–352. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feng YH and Tsao CJ: Emerging role of
microRNA-21 in cancer. Biomed Rep. 5:395–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Motoyama K, Inoue H, Mimori K, Tanaka F,
Kojima K, Uetake H, Sugihara K and Mori M: Clinicopathological and
prognostic significance of PDCD4 and microRNA-21 in human gastric
cancer. Int J Oncol. 36:1089–1095. 2010.PubMed/NCBI
|
|
26
|
Ou H, Li Y and Kang M: Activation of
miR-21 by STAT3 induces proliferation and suppresses apoptosis in
nasopharyngeal carcinoma by targeting PTEN gene. PLoS One.
9:e1099292014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei X, Wang W, Wang L, Zhang Y, Zhang X,
Chen M, Wang F, Yu J, Ma Y and Sun G: MicroRNA-21 induces
5-fluorouracil resistance in human pancreatic cancer cells by
regulating PTEN and PDCD4. Cancer Med. 5:693–702. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song Y, Zuo Y, Qian XL, Chen ZP, Wang SK,
Song L and Peng LP: Inhibition of microRNA-21-5p promotes the
radiation sensitivity of non-small cell lung cancer through HMSH2.
Cell Physiol Biochem. 43:1258–1272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai L, Wang W, Li X, Dong T, Zhang Q, Zhu
B, Zhao H and Wu S: MicroRNA-21-5p induces the metastatic phenotype
of human cervical carcinoma cells in vitro by targeting the von
Hippel-Lindau tumor suppressor. Oncol Lett. 15:5213–5219.
2018.PubMed/NCBI
|