Introduction
Colon cancer is the second most common cancer in
women and the third most common in men worldwide (1). A recent systematic review by Gall et
al (2) evaluated the treatments
available for colon cancer and determined the accuracy of
mini-probe endoscopic ultrasound in determining the clinical stage
of colon cancer. The results suggested that screening, treatment
options and prognoses for patients with colon cancer have improved
over time (3). Although the
systematic review included a number of targeted therapies for the
treatment of advanced colorectal cancer and explored the potential
of predictive biomarkers, at present there is no satisfactory
therapy for colon cancer due to local disease migration and long
distance metastasis (4,5). Metastasis and recurrence aggravates
disease progression in patients with stage II and III colon cancer
(6).
Colon cancer cell growth, metastasis and invasion
are difficult to treat and increase the mortality of patients with
colon cancer (7,8). Inhibiting apoptotic resistance and
promoting apoptosis in colorectal cancer cells is an important part
of cancer treatment, as well as the prevention of neoplasm
metastasis (9,10). Previous researchers have developed
targeted therapies, which suppress the underlying mechanisms of
colorectal cancer cell metastasis and invasion (11–13).
Advances in molecular bioinformatics have enabled scientists to
screen for target molecules associated with diagnosis and therapy
protocols, which suggests the potential of individual tailored
treatments for patients with colorectal cancer and other chronic
diseases (14,15).
Regulating apoptosis-associated protein expression
is beneficial for the prevention and treatment of colon cancer as
it may increase the apoptosis of tumor cells (16). Tumor necrosis factor-(TNF)-related
apoptosis-inducing ligand (TRAIL) is a potential anticancer
protein, which lyses various human tumor cells by inducing
apoptosis (17). A previous study
has demonstrated that TRAIL is safe for normal cells, as it
selectively induces apoptosis via binding with death receptors on
tumor cells (18). Previous studies
have demonstrated the inhibitory effects of TRAIL on tumor cells,
which suggests that it is an effective oncolytic agent for the
treatment of different types of human cancer (19–21).
Similar results were demonstrated when binding with
‘death-inducing’ and ‘decoy’ receptors, while further activation of
Fas-associated death domain or other proteins was observed in the
caspase signaling pathway (22). In
conclusion, these results suggest there is potential for the
application of TRAIL in cancer therapy.
In the present study, the anticancer effects and
mechanisms of TRAIL were examined in association with in
vitro colon tumor cell growth, migration, invasion and
apoptosis, as well as in vivo tumor growth inhibition. The
immunoregulatory functions of TRAIL on colon tumors in a xenograft
mouse model were analyzed following a 30-day treatment period.
Exogenous apoptosis signaling pathways induced by TRAIL were also
examined.
Materials and methods
Cells and reagents
Colon tumor cell lines LoVo and HT-29 were purchased
from the American Type Culture Collection (Manassas, VA, USA). All
tumor cells were cultured in Dulbecco's modified Eagle's medium
(Gibco) supplemented with 10% fetal bovine serum (Invitrogen; both
Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cells were
cultured in a 37°C humidified atmosphere containing 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from LoVo and HT-29 cells
and tumors using an RNAeasy Mini kit (Qiagen Sciences, Inc.,
Gaithersburg, MD, USA). cDNA was synthesized with ReverTra Ace
(Toyobo Life Science, Osaka, Japan) at 42°C for 2 h. Fibronectin
(FN), Vimentin and epithelial (E)-cadherin expression was analyzed
using an iCycler thermal cycler (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) using iQ SYBR Green Supermix (Bio-Rad
Laboratories, Inc.). All forward and reverse primers (Table I) were synthesized by Invitrogen
(Thermo Fisher Scientific, Inc.). The following thermocycling
conditions were applied: 45 amplification cycles consisting of
denaturation at 95°C for 30 sec, primer annealing at 62.5°C for 45
sec with touchdown at 56.5°C for 50 sec and extension at 72°C for
60 sec. The relative mRNA expression changes were calculated using
the 2−ΔΔCq method (23).
Results are expressed as fold change compared with the control.
 | Table I.Sequences of primers were used in
this study. |
Table I.
Sequences of primers were used in
this study.
|
| Sequence
(5′-3′) |
|---|
|
|
|
|---|
| Gene name | Reverse | Forward |
|---|
| Fibronectin |
TTCATTATAAATCTAGAGACTCCAGGA |
CTTTGGGACTGGTGGAAGAATC |
| Vimentin |
ACGTCTTGACCTTGAACGCA |
TCTTGGCAGCCACACTTTCA |
| E-cadherin |
GTGGCCCGGATGTGAGAAG |
GGAGCCCTTGTCGGATGATG |
| β-actin |
CGGAGTCAACGGATTTGGTC |
AGCCTTCTCCATGGTCGTGA |
MTT cytotoxicity assays
LoVo and HT-29 cells were incubated with TRAIL
(0–2.5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in
96-well plates for 24, 48, 72 and 96 h in triplicate for each
concentration. PBS was used as the control. Following the indicated
incubation time, 20 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA) in
PBS was added to each well and the plate was further incubated for
4 h. The majority of the medium was removed and 100 µl dimethyl
sulfoxide was added into the wells to solubilize the formazan
crystals. The optical density was measured using a microplate
reader at a wavelength of 450 nm.
Cell invasion and migration
assays
LoVo and HT-29 cells were incubated with TRAIL (0.20
mg/ml) for 12 h at 37°C. For the invasion assay, LoVo and HT-29
cells were suspended at a density of 1×105 in 500 µl
serum-free DMEM. The cells were seeded in the upper chamber of a
BioCoat Matrigel Invasion Chamber (BD Biosciences, Franklin Lakes,
NJ, USA) in DMEM for 24 h at 37°C. The lower chamber contained 10%
FBS in DMEM. For the migration assay, cells were seeded on a
control insert (BD Biosciences) instead of a Matrigel Invasion
Chamber for 24 h at 37°C. The migratory and invasive cells were
fixed with 3% formaldehyde for 15 min at 37°C and stained with 0.5%
crystal violet for 10 min at 37°C. The tumor cell migration and
invasion were counted in a minimum of three randomly-selected
stained fields using a light microscope at a magnification of
×40.
ELISA
96-well plates were incubated with rabbit anti-mouse
TNF-α antibody (cat. no. T8300; 1:200; Sigma-Aldrich; Merck KGAa)
for 24 h at 4°C. Plates were washed with PBS three times at room
temperature and then incubated with albumin (0, 0.25, 0.5, 0.75,
1.0, 1.25 and 1.5 mg/ml) or TRAIL (0, 0.25, 0.5, 0.75, 1.0, 1.25
and 1.5 mg/ml) for 1 h at 37°C. Plates were washed with PBS three
times at room temperature and then incubated with rabbit anti-mouse
TRAIL antibody (1:500; cat. no. ab231063, Abcam) for 24 h at 4°C.
Plates were washed with PBS three times at room temperature and
then incubated for 2 h at room temperature with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibodies
(1:1,000; cat. no. ab6785; Abcam). The optical density value was
detected at 450 nm using a microplate reader.
Cell cycle analysis
LoVo and HT-29 cells were incubated with TRAIL (0.20
mg/ml) for 48 h at 37°C. Cells were harvested, stained with Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodine (PI; BD
Biosciences) and analyzed as previously described (24). The cell cycle was analyzed using a
flow cytometer and ModFit LT™ software (Version 4;
Verity Software House Inc., Topsham, ME, USA).
Transfection of short interfering
(si)RNA
All siRNAs were synthesized by Invitrogen (Thermo
Fisher Scientific, Inc.) including the siRNA-Fas ligand (Si-FasL
sense, 5′-CGCGGATCCGCGTTGCAGAA-3′ and antisense,
5′-TCCCCGCGGGGAGCGACACTAA-3′) and the Si-RNA-vector (sense,
5′-TCCCCGCGGGGAAGGTCTGTCTTATT-3′ and antisense,
5′-CCATCGATGGTATACCGC-3′). LoVo or HT-29 cells (1×106)
were transfected with 100 pmol of Si-FasL targeting FasL with
si-RNA-vector as the control (both Thermo Fisher Scientific, Inc.)
using a Cell Line Nucleofector kit L (Lonza Group, Ltd., Basel,
Switzerland). Subsequent experiments were performed after a 72-h
transfection. Si-Fas transfected cells were treated with TRAIL
(0.20 mg/ml) for 48 h at 37°C.
Animal studies
A total of 40 male Balb/c nude mice (age, 8 weeks;
body weight, 22–28 g) were purchased from Shanghai SLAC laboratory
Animal Co., Ltd. (Shanghai, China). All mice were housed at 23±1°C,
50±5% humidity with a 12-h light/dark cycle and free access to food
and water. Mice were inoculated with LoVo or HT-29 tumor cells
(1×106) into the subcutaneous tissue and divided into
four groups: Control-LoVo, TRAIL-LoVo, Control-HT-29 and
TRAIL-HT-29 (n=10 per group). Treatment was initiated on day 8
following inoculation, when tumor diameters reached 5–6 mm.
Tumor-bearing mice were intravenously injected with TRAIL (0.20
mg/kg) or PBS (0.20 mg/kg) as the control. The treatment was
performed once per day for 7 days. Tumor volumes were calculated
every 3 days according to a previous study (25). The present study was approved by the
Committee on the Ethics of Yantaishan Hospital (Yantai, China).
Immunohistochemistry
Mice (n=3 per group) were anesthetized with 60 mg/kg
IP pentobarbital (Sigma-Aldrich; Merck KGaA) and sacrificed using
cervical dislocation on day 31. Colon tumors from the xenograft
mice were fixed using 10% formaldehyde for 2 h at room temperature
and embedded in paraffin. Tumor tissues were fabricated to
5-µm-thick tumor sections. Antigen retrieval was performed on the
tumor sections using Lab Vision™ Tris-HCl buffer for
heat-induced epitope retrieval (cat no. AP-9005-050; Thermo Fisher
Scientific, Inc.) for 15 min at 65°C. The tumor sections were
incubated with primary antibodies against P53 (cat. no. ab131442)
and B-cell lymphoma-2 (Bcl-2; ab182858; 1:200; Abcam, Cambridge,
UK) for 12 h at 4°C. The membranes were then incubated with goat
anti-rabbit horseradish peroxidase (HRP)-conjugated immunoglobulin
G secondary antibodies (1:2,000; cat. no. PV-6001; OriGene
Technologies Inc., Rockville, MD, USA) at 37°C for 2 h. Blots were
imaged using WesternBright ECL Chemiluminescent HRP Substrate
(Advansta, Menlo Park, CA, USA).
Apoptosis assay
LoVo or HT-29 cells were grown at 37°C in an
atmosphere containing 5% CO2 until 90% confluence was
reached. Cells were incubated with TRAIL (0.20 mg/ml) for 48 h at
37°C, following which the tumor cells were trypsinized and
collected. Cells were washed in cold PBS, adjusted to a
concentration of 1×106 cells/ml with PBS and labeled
with Annexin V-FITC and PI from the Annexin V-FITC kit (BD
Biosciences) for 2 h at 4°C. The results were analyzed using a
FACScan flow cytometer and BD FACSDIVA™ software (version 1.2; BD
Biosciences). The treatments were performed in triplicate and the
percentage of apoptotic cells in each group was measured.
Western blotting
Colorectal tumors and cells were homogenized in a
10% RIPA buffer (Sigma-Aldrich; Merck KGaA) and centrifuged at
6,000 × g at 4°C for 10 min to collect the supernatant.
Transmembrane proteins were extracted using a Transmembrane Protein
Extraction kit (Qiagen Sciences, Inc.) according to the
manufacturer's protocol. The nuclear and cytoplasmic proteins were
extracted using a Nuclear and Cytoplasmic Protein Extraction kit
(Qiagen Sciences, Inc.). Protein concentration was measured using a
BCA protein assay kit (Thermo Fisher Scientific Inc.). Protein
samples (30 µg/lane) were separated on 12.5% SDS-PAGE and
transferred onto polyvinylidene difluoride membranes (EMD
Millipore, Billerica, MA, USA) as previously described (26). Membranes were blocked with 5% skimmed
milk for 1 h at 37°C and then incubated with the following primary
antibodies for 24 h at 4°C: FN (cat. no. ab2413), Vimentin (cat.
no. ab8978), E-cadherin (cat. no. ab1416), P53 (cat. no. ab131442),
Bcl-2 (cat. no. ab182858; 1:1,000), caspase-3 (cat. no. ab13847),
caspase-8 (cat. no. ab25901), NF-κB (cat. no. ab220803; all
1:1,000) and β-actin (cat. no. ab8226; 1:2,000; both Abcam). The
membranes were then incubated with horseradish
peroxidase-conjugated goat anti-rabbit immunoglobulin G monoclonal
secondary antibodies (cat. no. PV-6001; 1:1,000; OriGene
Technologies, Inc., Beijing, China) for 24 h at 4°C. The results
were visualized using WesternBright ECL Chemiluminescent HRP
Substrate (Advansta, Menlo Park, CA, USA).
Statistical analysis
Data are presented as the mean ± standard deviation
of three independent experiments. The results were analyzed using
Student t-tests or one-way analysis of variance with Tukey's post
hoc test. All the data were analyzed using SPSS Statistics 19.0
(IBM Corp., Armonk, NY, USA) and GraphPad Prism version 5.0
(GraphPad Software, Inc., La Jolla, CA, USA) with Microsoft Excel
(Microsoft Corporation, Redmond, WA, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
TRAIL inhibits colon cancer cell
growth and induces cell cycle arrest
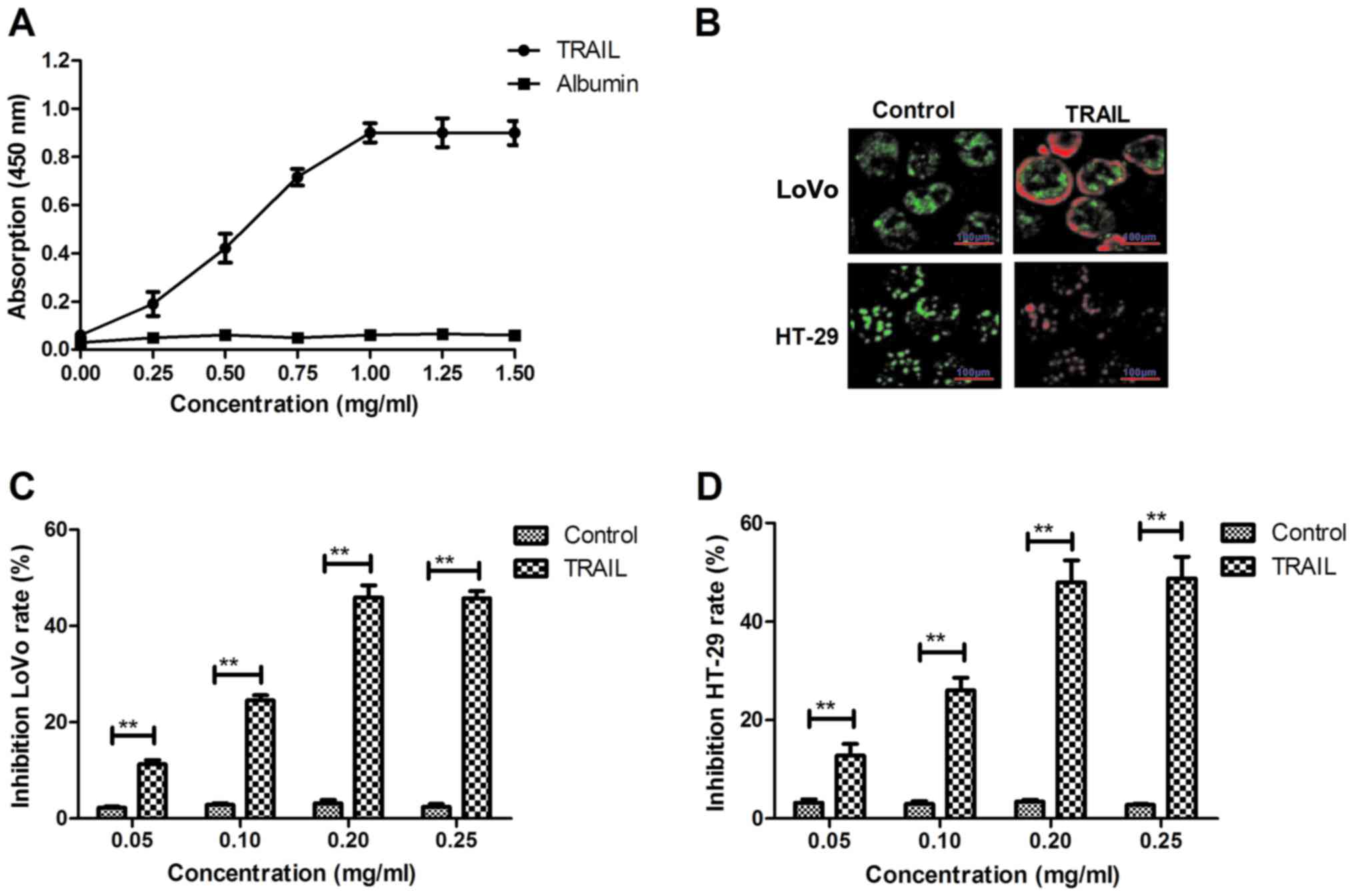
The affinity of TRAIL with TNF-α was analyzed by an
ELISA assay. TRAIL could bind to TNF-α (Fig. 1A). An immunofluorescence assay
revealed that TRAIL (0.20 mg/ml) could integrate with the surface
of LoVo and HT-29 cells (Fig. 1B),
while an in vitro assay demonstrated that TRAIL
significantly inhibited colon cancer cell growth in a
dose-dependent manner (Fig. 1C and
D). Cell growth was most inhibited following treatment with
0.20 mg/ml TRAIL, and so this dosage was selected for use in
further experiments. TRAIL inhibited LoVo (Fig. 1E) and HT-29 (Fig. 1F) cell growth in a time-dependent
manner. The results also revealed that TRAIL treatment
significantly arrested cell cycle progression in LoVo and HT-29 at
the S phase (Fig. 1G and H). These
results suggest that TRAIL may inhibit colon cancer cell growth by
inducing cell cycle arrest.
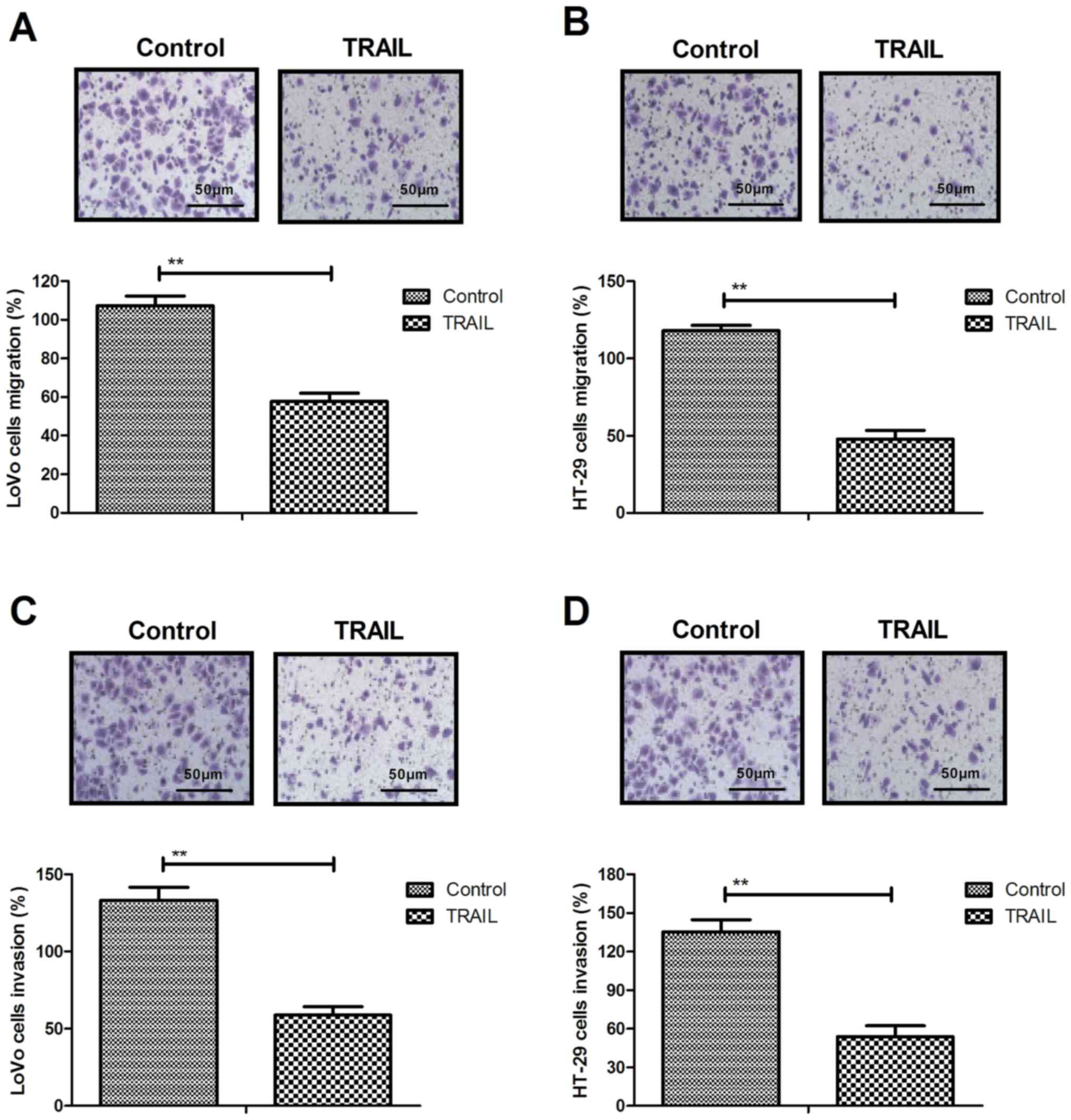
TRAIL inhibits the migration and
invasion of colon cancer cells by downregulating neoplasm
metastasis-associated protein expression levels
Treatment with 0.02 mg/ml TRAIL for 48 h
significantly inhibited the migration of LoVo (Fig. 2A) and HT-29 (Fig. 2B) compared with the control. An
invasion assay demonstrated that TRAIL administration (0.20 mg/ml)
significantly inhibited the invasion of LoVo (Fig. 2C) and HT-29 (Fig. 2D) cells compared with the control,
while RT-qPCR revealed that FN, Vimentin and E-cadherin expression
was significantly downregulated following TRAIL administration in
LoVo (Fig. 2E) and HT-29 (Fig. 2F) cells compared with the control.
Western blotting results demonstrated that TRAIL administration
significantly decreased FN, Vimentin and E-cadherin protein
expression in LoVo (Fig. 2G) and
HT-29 (Fig. 2H) cells compared with
the control. These results suggest that TRAIL inhibits the
migration and invasion of colon cancer cells by downregulating the
expression of FN, Vimentin and E-cadherin.
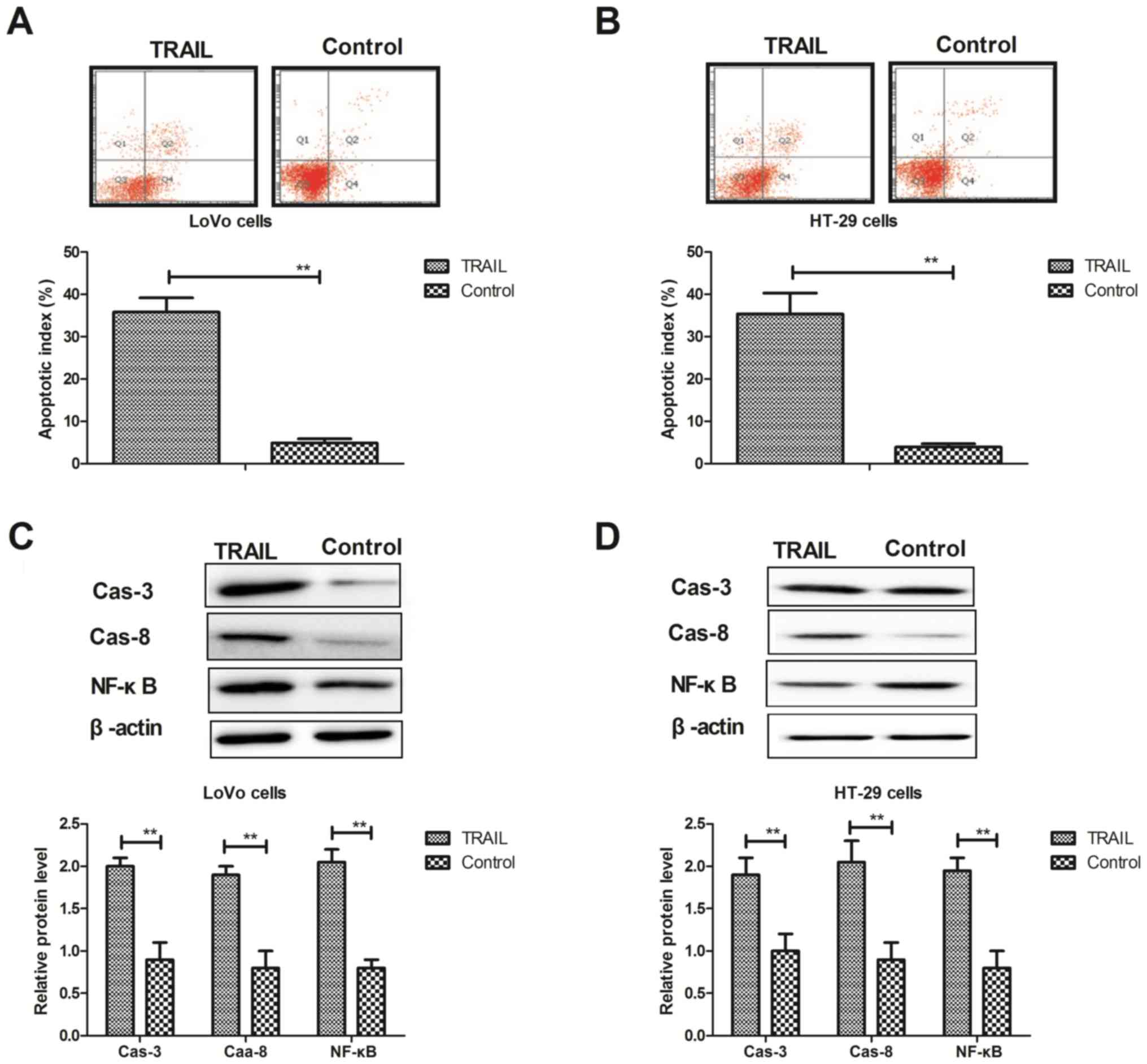
TRAIL treatment promotes the apoptosis
of colon cancer cells via the exogenous apoptosis pathway
The effects of TRAIL treatment on the apoptosis of
colon cancer cells were analyzed in vitro. Flow cytometry
revealed that TRAIL treatment (0.20 mg/ml) significantly promoted
apoptosis in LoVo (Fig. 3A) and
HT-29 (Fig. 3B) cells compared with
the control. Western blotting also demonstrated that TRAIL
treatment significantly increased the expression of caspase-8,
caspase-3 and NF-κB proteins in LoVo (Fig. 3C) and HT-29 (Fig. 3D) cells compared with the control.
However, the expression of P53 and Bcl-2 proteins was significantly
downregulated by TRAIL treatment in LoVo (Fig. 3E) and HT-29 (Fig. 3F) cells compared with the control. In
addition, TRAIL treatment significantly increased apoptosis in
FasL-inhibited LoVo (Fig. 3G) and
HT-29 (Fig. 3H) cells compared with
FasL-inhibited cells. These results suggest that TRAIL treatment
may promote the apoptosis of LoVo and HT-29 colon cancer cells via
the exogenous apoptosis pathway.
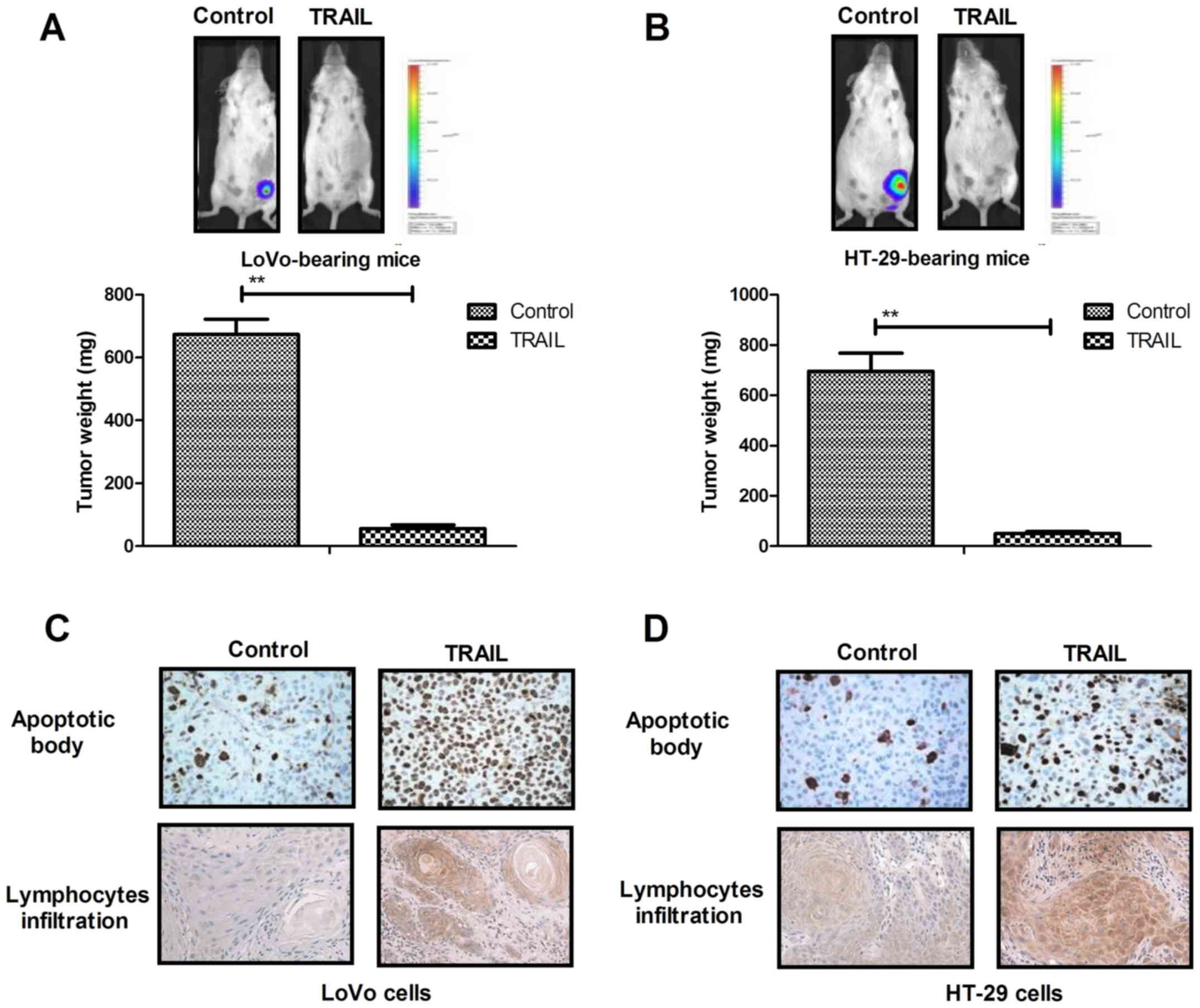
TRAIL treatment inhibits tumors growth
and prolongs the survival of LoVo and HT-29 tumor-bearing mice
TRAIL treatment significantly inhibited tumor growth
in LoVo (Fig. 4A) and HT-29
tumor-bearing (Fig. 4B) mice
compared with the PBS group. Immunohistochemistry revealed that
TRAIL treatment notably increased the infiltration of apoptotic
bodies and lymphocytes in tumors compared with the control group
(Fig. 4C and D). P53 and Bcl-2
expression was markedly downregulated following TRAIL treatment in
LoVo (Fig. 4E) and HT-29
tumor-bearing (Fig. 4F) mice
compared with the PBS group. The survival rate of LoVo (Fig. 4G) and HT-29 tumor-bearing (Fig. 4H) mice was significantly increased
following treatment with TRAIL, compared with the PBS group. These
results indicate that TRAIL treatment inhibits tumor growth and
prolongs survival in LoVo and HT-29 tumor-bearing mice by promoting
apoptosis in colon tumor cells.
Discussion
Colon cancer is one of the most common types of
gastrointestinal cancer worldwide, which can be highly invasive and
is characterized by rapid local invasion of the lymphatic system
(27). Death receptor 5 (DR5) and
TRAIL have been reported to induce the apoptosis of human tumor
cells and may represent a novel approach to cancer therapy by
increasing the apoptotic sensitivity of cells (28). Numerous studies have demonstrated the
effects of TRAIL by targeting the metabolic signaling pathway in
human colon cancer (29–32). However, the exogenous apoptosis
signaling pathway and TRAIL apoptotic death pathway are not well
understood within colon cancer cells (33,34). In
the present study, the inhibitory effects and underlying mechanisms
of TRAIL in LoVo and HT-29 cells in vitro and in vivo
were examined. The results revealed that TRAIL treatment
significantly inhibited the growth and invasion of colon cancer
cells. In addition, the results demonstrated that TRAIL treatment
significantly promoted the apoptosis of colon cancer cells by
increasing the expression of caspase-3, FasL and caspase-8
proteins, which are components of the exogenous apoptosis
pathway.
TRAIL is a member of the TNF superfamily, which
interacts with DRs on tumor cells and leads to the apoptosis of
cancer cells (35). Gupta et
al (32) demonstrated that TRAIL
induces apoptosis through the extracellular signal regulated
kinase-dependent upregulation of DRs p53 and Bcl-2-associated X
protein. The increased expression of TRAIL receptors may enhance
the apoptotic response induced by TRAIL and cause an increase in
the apoptosis rate of tumor cells (36,37). The
results of the present study revealed that TRAIL inhibited the
growth of LoVo and HT-29 cancer cells and also promoted their
apoptosis. Caspase-3 and caspase-8 expression was upregulated in
TRAIL-treated colon tumor cells, which further increases apoptosis
(38). The results also suggest that
TRAIL-induced apoptosis was achieved via the exogenous apoptosis
pathway via the downregulation of P53 and Bcl-2.
A previous study indicated that TRAIL may mediate
apoptosis through the upregulation of DR5 by zerumbone and
celecoxib (39). The results of the
present study demonstrated that TRAIL treatment significantly
increased the expression of caspase-8, caspase-3 and NF-κB proteins
in LoVo and HT-29 cells. The current study indicated that the
inhibition of FasL expression ameliorates the TRAIL-induced
apoptosis of LoVo and HT-29 cells. In addition, a previous study
has suggested that FasL-induced apoptosis serves a role in the
elimination of tumor cells by natural killer cells (40). Contassot et al (41) have suggested that FasL is an
important molecule in TRAIL-mediated apoptosis, which is associated
with impaired DR and FLICE-inhibitory protein expression. The
results of the present study demonstrated that TRAIL-induced colon
tumor apoptosis occurred via the FasL-mediated exogenous apoptosis
signaling pathway.
The results of the present study also revealed that
TRAIL treatment markedly inhibits the invasion of colon tumor cells
by downregulating FN, Vimentin and E-cadherin expression in LoVo
and HT-29 cells. Kamoshida et al (42) reported that decreased matrix
metalloproteinase production and FN expression may downregulate
tumor cell invasion. Previous studies have also demonstrated that
variable E-cadherin overexpression is a risk marker for the
development of multiple tumors in animal models of colon tumors
(43,44). Additionally, Vimentin mediates the
regulation of cell motility through the modulation of integrin β4
protein expression in various tumor cells (45). The results of the present study
suggest that TRAIL treatment may decrease FN, Vimentin and
E-cadherin expression in LoVo and HT-29 cells, which inhibits the
migration and invasion of colon tumor cells. It was also observed
that TRAIL treatment inhibited tumor growth and prolonged the
survival of tumor-bearing mice compared with the controls. However,
further studies should be perform to explore the anticancer effects
of TRAIL on other types of human cancer.
In conclusion, TRAIL treatment significantly
promotes apoptosis and inhibits the growth and aggressiveness of
colon tumor cells. In addition, TRAIL treatment had a
tumor-suppressing effect on colon tumor cells in vitro and
in vivo, which suggests that TRAIL may be a promising
anticancer agent for the treatment of colon cancer.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The analyzed datasets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HG and WC designed the study. and performed the
experiments. YW analyzed the data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lopez NE, Weiss AC, Robles J, Fanta P and
Ramamoorthy SL: A systematic review of clinically available gene
expression profiling assays for stage II colorectal cancer: Initial
steps toward genetic staging. Am J Surg. 212:700–714. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gall TM, Markar SR, Jackson D, Haji A and
Faiz O: Mini-probe ultrasonography for the staging of colon cancer:
A systematic review and meta-analysis. Colorectal Dis. 16:O1–O8.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim HD, Ha KS, Woo IS, Jung YH, Han CW and
Kim TJ: Tumor lysis syndrome in a patient with metastatic colon
cancer after treatment with 5-fluorouracil/leucovorin and
oxaliplatin: Case report and literature review. Cancer Res Treat.
46:204–207. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tudyka V, Blomqvist L, Beets-Tan RG,
Boelens PG, Valentini V, van de Velde CJ, Dieguez A and Brown G:
EURECCA consensus conference highlights about colon & rectal
cancer multidisciplinary management: The radiology experts review.
Eur J Surg Oncol. 40:469–475. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sheets N, Powers J and Richmond B:
Cutaneous metastasis of colon cancer: Case report and literature
review. W V Med J. 110:22–24. 2014.PubMed/NCBI
|
|
6
|
Bockelman C, Engelmann BE, Kaprio T,
Hansen TF and Glimelius B: Risk of recurrence in patients with
colon cancer stage II and III: A systematic review and
meta-analysis of recent literature. Acta Oncol. 54:5–16. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moilanen JM, Löffek S, Väyrynen JP,
Syväniemi E, Hurskainen T, Mäkinen M, Klintrup K, Mäkelä J,
Sormunen R, et al: Collagen XVII expression correlates with the
invasion and metastasis of colorectal cancer. Hum Pathol.
46:434–442. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan Z, Cui H, Xu X, Lin Z, Zhang X, Kang
L, Han B, Meng J, Yan Z, Yan X and Jiao S: MiR-125a suppresses
tumor growth, invasion and metastasis in cervical cancer by
targeting STAT3. Oncotarget. 6:25266–25280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tian M, Wan Y, Tang J, Li H, Yu G, Zhu J,
Ji S, Guo H, Zhang N, Li W, et al: Depletion of tissue factor
suppresses hepatic metastasis and tumor growth in colorectal cancer
via the downregulation of MMPs and the induction of autophagy and
apoptosis. Cancer Biol Ther. 12:896–907. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu K: Role of apoptosis resistance in
immune evasion and metastasis of colorectal cancer. World J
Gastrointest Oncol. 2:399–406. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang XB, Song L, Wen HJ, Bai XX, Li ZJ
and Ma LJ: Upregulation of microRNA-31 targeting integrin α5
suppresses tumor cell invasion and metastasis by indirectly
regulating PI3K/AKT pathway in human gastric cancer SGC7901 cells.
Tumour Biol. 37:8317–8325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo J, Yu X, Gu J, Lin Z, Zhao G, Xu F, Lu
C and Ge D: Regulation of CXCR4/AKT-signaling-induced cell invasion
and tumor metastasis by RhoA, Rac-1, and Cdc42 in human esophageal
cancer. Tumour Biol. 37:6371–6378. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang N, Deng JY, Liu Y, Ke B, Liu HG and
Liang H: Incorporation of perineural invasion of gastric carcinoma
into the 7th edition tumor-node-metastasis staging system. Tumour
Biol. 35:9429–9436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bukurova IuA, Khankin SL, Krasnov GS,
Grigor'eva ES, Mashkova TD, Lisitsin NA, Karpov VL and Beresten'
SF: Comparison of 2D analysis and bioinformatics search efficiency
for colon cancer marker identification. Mol Biol. 44:375–381.
2010.(In Russian). View Article : Google Scholar
|
|
15
|
Thompson BA, Goldgar DE, Paterson C,
Clendenning M, Walters R, Arnold S, Parsons MT, Michael DW,
Gallinger S, Haile RW, et al: A multifactorial likelihood model for
MMR gene variant classification incorporating probabilities based
on sequence bioinformatics and tumor characteristics: A report from
the Colon Cancer Family Registry. Hum Mutat. 34:200–209. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JS, Jung WK, Jeong MH, Yoon TR and Kim
HK: Sanguinarine induces apoptosis of HT-29 human colon cancer
cells via the regulation of Bax/Bcl-2 ratio and caspase-9-dependent
pathway. Int J Toxicol. 31:70–77. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grosse-Wilde A, Voloshanenko O, Bailey SL,
Longton GM, Schaefer U, Csernok AI, Schütz G, Greiner EF, Kemp CJ
and Walczak H: TRAIL-R deficiency in mice enhances lymph node
metastasis without affecting primary tumor development. J Clin
Invest. 118:100–110. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ndebele K, Gona P, Jin TG, Benhaga N,
Chalah A, Degli-Esposti M and Khosravi-Far R: Tumor necrosis factor
(TNF)-related apoptosis-inducing ligand (TRAIL) induced
mitochondrial pathway to apoptosis and caspase activation is
potentiated by phospholipid scramblase-3. Apoptosis. 13:845–856.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ivanov VN, Bhoumik A and Ronai Z: Death
receptors and melanoma resistance to apoptosis. Oncogene.
22:3152–3161. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang XD, Franco A, Myers K, Gray C,
Nguyen T and Hersey P: Relation of TNF-related apoptosis-inducing
ligand (TRAIL) receptor and FLICE-inhibitory protein expression to
TRAIL-induced apoptosis of melanoma. Cancer Res. 59:2747–2753.
1999.PubMed/NCBI
|
|
21
|
Qiu B, Sun X, Zhang D, Wang Y, Tao J and
Ou S: TRAIL and paclitaxel synergize to kill U87 cells and
U87-derived stem-like cells in vitro. Int J Mol Sci. 13:9142–9156.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang XD, Franco AV, Nguyen T, Gray CP and
Hersey P: Differential localization and regulation of death and
decoy receptors for TNF-related apoptosis-inducing ligand (TRAIL)
in human melanoma cells. J Immunol. 164:3961–3970. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Skinner SO, Xu H, Nagarkar-Jaiswal S,
Freire PR, Zwaka TP and Golding I: Single-cell analysis of
transcription kinetics across the cell cycle. Elife. 5:e121752016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bai FL, Yu YH, Tian H, Ren GP, Wang H,
Zhou B, Han XH, Yu QZ and Li DS: Genetically engineered Newcastle
disease virus expressing interleukin-2 and TNF-related
apoptosis-inducing ligand for cancer therapy. Cancer Biol Ther.
15:1226–1238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wai-Hoe L, Wing-Seng L, Ismail Z and
Lay-Harn G: SDS-PAGE-based quantitative assay for screening of
kidney stone disease. Biol Proced Online. 11:145–160. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saito T, Morohashi H, Hasebe T, Sakamoto
Y, Koyama M, Murata A and Hakamada K: A review of stereotactic
radiotherapy (SRT) for lung metastasis of colon cancer. Gan To
Kagaku Ryoho. 41:1462–1464. 2014.(In Japanese). PubMed/NCBI
|
|
28
|
Kojima Y, Nakayama M, Nishina T, Nakano H,
Koyanagi M, Takeda K, Okumura K and Yagita H: Importin β1
protein-mediated nuclear localization of death receptor 5 (DR5)
limits DR5/tumor necrosis factor (TNF)-related apoptosis-inducing
ligand (TRAIL)-induced cell death of human tumor cells. J Biol
Chem. 286:43383–43393. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen L, Meng Y, Guo X, Sheng X, Tai G,
Zhang F, Cheng H and Zhou Y: Gefitinib enhances human colon cancer
cells to TRAIL-induced apoptosis of via autophagy- and JNK-mediated
death receptors upregulation. Apoptosis. 21:1291–1301. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo X, Meng Y, Sheng X, Guan Y, Zhang F,
Han Z, Kang Y, Tai G, Zhou Y and Cheng H: Tunicamycin enhances
human colon cancer cells to TRAIL-induced apoptosis by
JNK-CHOP-mediated DR5 upregulation and the inhibition of the EGFR
pathway. Anticancer Drugs. 28:66–74. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen L, Meng Y, Sun Q, Zhang Z, Guo X,
Sheng X, Tai G, Cheng H and Zhou Y: Ginsenoside compound K
sensitizes human colon cancer cells to TRAIL-induced apoptosis via
autophagy-dependent and -independent DR5 upregulation. Cell Death
Dis. 7:e23342016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gupta SC, Reuter S, Phromnoi K, Park B,
Hema PS, Nair M and Aggarwal BB: Nimbolide sensitizes human colon
cancer cells to TRAIL through reactive oxygen species- and
ERK-dependent up-regulation of death receptors, p53, and Bax. J
Biol Chem. 291:169252016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bousserouel S, Le Grandois J, Gossé F,
Werner D, Barth SW, Marchioni E, Marescaux J and Raul F: Methanolic
extract of white asparagus shoots activates TRAIL apoptotic death
pathway in human cancer cells and inhibits colon carcinogenesis in
a preclinical model. Int J Oncol. 43:394–404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee SC, Cheong HJ, Kim SJ, Yoon J, Kim HJ,
Kim KH, Kim SH, Kim HJ, Bae SB, Kim CK, et al: Low-dose
combinations of LBH589 and TRAIL can overcome TRAIL-resistance in
colon cancer cell lines. Anticancer Res. 31:3385–3394.
2011.PubMed/NCBI
|
|
35
|
Hwang JS, Lee HC, Oh SC, Lee DH and Kwon
KH: Shogaol overcomes TRAIL resistance in colon cancer cells via
inhibiting of survivin. Tumour Biol. 36:8819–8829. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Woo JK, Kang JH, Jang YS, Ro S, Cho JM,
Kim HM, Lee SJ and Oh SH: Evaluation of preventive and therapeutic
activity of novel non-steroidal anti-inflammatory drug, CG100649,
in colon cancer: Increased expression of TNF-related
apoptosis-inducing ligand receptors enhance the apoptotic response
to combination treatment with TRAIL. Oncol Rep. 33:1947–1955. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Moriwaki K, Shinzaki S and Miyoshi E:
GDP-mannose-4,6-dehydratase (GMDS) deficiency renders colon cancer
cells resistant to tumor necrosis factor-related apoptosis-inducing
ligand (TRAIL) receptor- and CD95-mediated apoptosis by inhibiting
complex II formation. J Biol Chem. 286:43123–43133. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Skender B, Hofmanová J, Slavík J,
Jelínková I, Machala M, Moyer MP, Kozubík A and Hyršlová Vaculová
A: DHA-mediated enhancement of TRAIL-induced apoptosis in colon
cancer cells is associated with engagement of mitochondria and
specific alterations in sphingolipid metabolism. Biochim Biophys
Acta. 1841:1308–1317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Edagawa M, Kawauchi J, Hirata M, Goshima
H, Inoue M, Okamoto T, Murakami A, Maehara Y and Kitajima S: Role
of activating transcription factor 3 (ATF3) in endoplasmic
reticulum (ER) stress-induced sensitization of p53-deficient human
colon cancer cells to tumor necrosis factor (TNF)-related
apoptosis-inducing ligand (TRAIL)-mediated apoptosis through
up-regulation of death receptor 5 (DR5) by zerumbone and celecoxib.
J Biol Chem. 289:21544–21561. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Screpanti V, Wallin RP, Grandien A and
Ljunggren HG: Impact of FASL-induced apoptosis in the elimination
of tumor cells by NK cells. Mol Immunol. 42:495–499. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Contassot E, Kerl K, Roques S, Shane R,
Gaide O, Dupuis M, Rook AH and French LE: Resistance to FasL and
tumor necrosis factor-related apoptosis-inducing ligand-mediated
apoptosis in Sezary syndrome T-cells associated with impaired death
receptor and FLICE-inhibitory protein expression. Blood.
111:4780–4787. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kamoshida G, Matsuda A, Miura R, Takashima
Y, Katsura A and Tsuji T: Potentiation of tumor cell invasion by
co-culture with monocytes accompanying enhanced production of
matrix metalloproteinase and fibronectin. Clin Exp Metastasis.
30:289–297. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tuncel H, Shimamoto F, Cagatay P and
Kalkan MT: Variable E-cadherin expression in a MNU-induced colon
tumor model in rats which exposed with 50 Hz frequency sinusoidal
magnetic field. Tohoku J Exp Med. 198:245–249. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Muller N, Reinacher-Schick A, Baldus S,
van Hengel J, Berx G, Baar A, van Roy F, Schmiegel W and
Schwarte-Waldhoff I: Smad4 induces the tumor suppressor E-cadherin
and P-cadherin in colon carcinoma cells. Oncogene. 21:6049–6058.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dmello C, Sawant S, Alam H, Gangadaran P,
Tiwari R, Dongre H, Rana N, Barve S, Costea DE, Chaukar D, et al:
Vimentin-mediated regulation of cell motility through modulation of
beta4 integrin protein levels in oral tumor derived cells. Int J
Biochem Cell Biol. 70:161–172. 2016. View Article : Google Scholar : PubMed/NCBI
|