Introduction
According to the investigation of the World Health
Organization (2015), cardiovascular disease, a global health
problem, is the leading cause of mortality globally (1,2). In
China, the number of patients with cardiovascular disease is
projected to increase annually (1,2). The
tendency of development of cardiovascular disease in China is
consistent with the global increase (1,2).
Atherosclerosis is a chronic disease that is induced by the
accumulation of fibrous elements and lipids in the larger arteries
and frequently causes severe problems, including blocking the aorta
and aorta rupture bleeding (1–3).
Atherosclerosis that occurs in the coronary artery is termed
coronary atherosclerosis. Coronary atherosclerosis, particularly
atherosclerosis of the external branch of the coronary artery, is
the most common constrictive coronary artery disease (1–3).
Numerous factors, including smoking, lack of exercise and dietary
habits are associated with the production of coronary
atherosclerosis (1–4). Due to the complicated mechanisms
underlying cardiovascular disease, there are currently no
successful treatments; therefore, there is a requirement to
determine the possible mechanism underlying cardiovascular disease,
and future studies should investigate the biomarkers of coronary
atherosclerosis. These studies will provide data, for the early
diagnosis and individual therapy of cardiovascular disease.
MicroRNAs (miRs) are defined as noncoding RNA
molecules involved in regulating and controlling the
post-transcription expression of target mRNAs (5,6). They
have complementary sequences in the 3′-untranslated region (UTR)
and 5′-UTR of target mRNAs and regulate the expression of
protein-coding genes by causing mRNA cleavage (1). Previous studies have demonstrated that
miRs can directly or indirectly regulate the expression of
intercellular adhesion molecules and various inflammatory factors,
including those involved in cell cycle progression, cell
proliferation, lipid metabolism, endothelial cell function,
angiogenesis, and plaque formation and rupture. These processes
involve the regulation of vascular endothelial cells, vascular
smooth muscle cells and macrophage function; therefore, the
pathophysiology of atherosclerosis and cardiovascular disease are
affected (3,5,6). As for
miRs in coronary atherosclerosis or other cardiovascular diseases,
a number of studies have published their outcomes. For example,
Jiang et al (7) demonstrated
that miR is a signature and biomarker in chronic cardiovascular
diseases. It has been reported by Hoekstra et al (8) that miR-370 is notably increased in
PBMCs of patients with coronary atherosclerosis. In addition, Liu
et al (9) revealed that
plasma expression levels of miR-370 were significantly higher in
patients with coronary artery disease compared with patients with
non-coronary artery disease. Also, miR-370 was identified to be
critical in the lipid metabolism, which is a potential biomarker
for the diagnosis of coronary artery disease (7). However, the effect of miR-370 in
critical cellular processes associated with coronary
atherosclerosis remains unknown and requires further study.
Materials and methods
Chemicals and materials
Fetal bovine serum (FBS) was purchased from Gibco
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) and Dulbecco's
modified Eagle's medium (DMEM) was purchased from Dalian Meilun
Biology Technology Co., Ltd (Dalian, China). Smooth muscle cell
medium (SMCM; cat. no. 1101; Sciencell Research Laboratories, Inc.,
San Diego, CA, USA) was also obtained. Antibodies of Forkhead Box 1
(FOXO1; cat. no. 2880), B-cell lymphoma 2 (Bcl-2)-associated X
(Bax; cat. no. 5023), Bcl-2 (cat. no. 4223), cleaved-poly
(ADP-ribose) polymerase (PARP; cat. no. 5625) and β-actin (cat. no.
4970) were purchased from Cell Signaling Technology, Inc. (Danvers,
MA, USA). Antibodies for caspase 3 (cat. no. ab2302) were obtained
from Abcam (Cambridge, MA, USA). Human umbilical vein endothelial
cells HUVECs and 293 cells were obtained from the Type Culture
Collection of the Chinese Academy Sciences (Shanghai, China).
Reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)-associated chemicals were purchased from Thermo Fisher
Scientific, Inc. The miR-370 mimics, miR-370 inhibitor, hsa-miR-370
mimics and miRNA negative control (miR-NC) were provided by
Shanghai GenePharma Co., Ltd. (Shanghai, China). Western blot and
gel analysis instruments were purchased from Bio-Rad Laboratories,
Inc. (Hercules, CA, USA). All other reagents were of the highest
purity and were commercially available from Shenyang LaiBo Science
and Trade Co., Ltd (http://www.11467.com/shenyang/co/75880.htm, Shenyang,
China).
Clinical samples
A total of 20 patients (10 male, 10 female; age,
40–60 years) with coronary atherosclerosis who required coronary
artery bypass surgery, as confirmed by a coronary angiogram, and 20
healthy patients without any abnormal conditions in the coronary
artery and heart, as confirmed by a coronary angiogram, were
treated at Nanjing First Hospital, Nanjing Medical University
(Nanjing, China). Patients with either of the following conditions
were excluded: type 1 diabetes mellitus, autoimmune disease,
malignancy, chronic or acute inflammatory disease, asthma, severe
heart failure and renal and hepatic dysfunction. Clinical samples
used in the present experiment were obtained from the Nanjing First
Hospital, Nanjing Medical University from October 2016 to October
2017. Written informed consent was obtained in all cases. The study
protocol was approved by the Ethics Committee at Nanjing First
Hospital, Nanjing Medical University. PBMCs were obtained from
these volunteers and separated using the Ficoll-Hypaque gradient
separation technique (10). The
samples were collected and immediately stored in a refrigerator at
−80°C. The differential expression of miR-370 between patients with
coronary atherosclerosis and healthy patients was detected using
RT-qPCR. Simultaneously, the association between the expression of
miR-370 and clinical information of patients was also analyzed.
Cell culture and cell
transfection
HUVECs were cultivated in SMCM culture containing
10% FBS in a 5% CO2 incubator at 37°C with 70–80%
humidity. The 293T cells were cultivated in DMEM culture
supplemented with 10% FBS in a 5% CO2 incubator at 37°C
with 70–80% humidity. miR-370 mimics
(5′-GCCUGCUGGGGUGGAACCUGGU-3′), miR-370 inhibitor
(5′-CGGACGACCCCACCUUGGACCA-3′) and miR-NC
(5′-GCCAGCCGUUGUGGCAGAUGGU-3′) were used.
HUVECs in the logarithmic phase were obtained and
transfected with miR-370 mimics, miR-370 inhibitors and miR-NC with
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
The 10 nM miR-370 mimics (11) were
used for miR-370 overexpression and an equal quantity of miR-370
inhibitors was used for miR-370 downregulation. Simultaneously,
miR-NC was used in the present study. miR-370 mimics and inhibitors
are chemically modified small RNAs, and miR-370 mimics are
double-stranded RNAs; however, miR-370 inhibitors are
single-stranded RNAs (12). miR-370
was used to upregulate miR-370 activity and mimic endogenous
miR-370. Additionally, miR-370 inhibitors were used to inhibit
endogenous miR-370 molecules and empower miR-370 functional
analysis by downregulating of miR-370. At 48 h post-transfection,
total RNA were extracted with a TRIzol assay and analyzed with
RT-qPCR to detect the ratio of transfection.
Cell proliferation assay [Cell
counting kit-8 (CCK-8) assay]
HUVECs were selected for the cell proliferation
assay. The cell proliferation value was measured by CCK-8 (Thermo
Fisher Scientific, Inc.) at 0, 24, 48 and 72 h post-transfection.
In brief, the cell viability of cells collected from different time
points post-transfection was determined with CCK-8 in triplicate.
CCK-8 reagent (10 µl) was added to 1×103 cells and
incubated for 1 h at 37°C. Subsequently, the absorbance of the
resulting product was measured at 450 nm using a microplate reader
(L-117; Thermo Fisher Scientific, Inc.).
miR analysis using RT-qPCR
HUVECs were cultured in 6-well plates and harvested
at 80% cell density. The total RNA were extracted according to the
manufacturer's protocols using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). In brief, cells were centrifuged at 2,000
× g for 5 min at room temperature (RT). Subsequently, the
supernatant was removed and the residue was combined with 1 ml
TRIzol. Following 5 min at RT, it was transferred into a new 1.5 ml
Eppendorf tube. Following the addition of 200 µl chloroform, the
mixture was placed at RT for 10 min. Subsequently, the mixture was
centrifuged at 12,000 × g for 15 min at 4°C. The upper supernatant
was collected and combined with 200 µl pre-cold isopropanol.
Following an incubation for 10 min at 4°C, the mixture was
centrifuged at 12,000 × g for 12 min at 4°C and the supernatant was
discarded. The residue was washed with 1 ml 75% ethanol (prepared
with fresh diethyl pyrocarbonate water). Subsequently, the
suspension was centrifuged at 10,800 × g for 5 min at 4°C twice,
and then the supernatant was discarded. Once the total RNA was
obtained, reverse transcription into cDNA was performed according
to the manufacturer's protocols of the High-Capacity cDNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc.). U6 was selected
as the internal control. The sequences of miR-370 and U6 were as
follows: miR-370, forward 5′-TAGCCTGCTGGGGTGGAA-3′ and reverse
5′-TATGGTTTTGACGACTGTGTGAT-3′; and U6, forward
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse
5′-GGAACGCTTCACGAATTTG-3′. In order to quantify the expression of
miR, the cycle quantification (Cq) was determined using
the TaqMan small RNA assay (Thermo Fisher Scientific, Inc.) with
miR-specific primers, according to manufacturer's protocols. In
brief, 7.6 µl nuclease-free water and 10 µl probe qPCR mix were
added to 1.4 µl cDNA. The TaqMan small RNA assay primer of
has-miR-9-5p was used. RT-qPCR reactions were conducted with a
QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific,
Inc.). Samples were run in triple for each experiment. PCR cycling
procedures were as follows: 95°C for 5 min, followed by 35 cycles
of 95°C for 40 sec, 60°C for 40 sec, 72°C for 40 sec and 72°C for
10 min.
The expression of miR-370 in PBMCs was also
determined with RT-qPCR. The method of sample preparation was
performed as mentioned above.
Wound healing assay
The influence of miR-370 on the wound healing of
HUVECs was measured using a wound healing assay. In brief, HUVECs
were cultured in 6-well plates and transfected according to the
aforementioned method. At 24 h post-transfection, the ratio of
transfection was ~90%. The vertically lineation was scratched on
the cell culture plate with a 200 µl pipette tip. Following washing
with PBS three times, cells were cultured in SMCM without serum for
24 h at 37°C. The width of the scratch at different time points (0
and 24 h) was recorded under a light microscope (magnification,
×200). Samples were run in triplicate for each experiment.
Matrigel invasion assay
HUVEC invasion was conducted using BioCoat Matrigel
Invasion Chambers (cat. no. 354480; Corning Incorporated, Corning,
NY, USA), according to the manufacturer's protocols. The upper
chamber was pre-coated with 100 µl Matrigel and exposed to ultra
violet light for 2 h. DMEM medium containing with 10% FBS (500 µl)
was added to the lower wells. Following transfection with miR-370
mimics, the HUVECs, in serum-free SMCM medium, were added to the
upper well (4×104 cells/ml). Subsequently, the cells
were incubated at 37°C for 24 h. Following this, the cells on the
surface of the upper chamber membrane were discarded with cotton
swabs. The cells underside of the membrane were fixed in 100%
methanol and stained with a solution containing 50% isopropanol, 1%
formic acid and 0.5% crystal violet for 20 min at RT. Subsequently,
the cells were counted under a light microscope (views were
randomly selected, at least 5 views/well; magnification, ×200).
Results were presented by mean ± standard deviation (SD). Samples
were run in sextuple for each experiment.
Cell apoptosis assay
The collected HUVECs at 48 h post-transfection were
washed two times with cold PBS, centrifuged at 1,000 × g for 5 min
at 4°C and then the supernatant was discarded. The residue was
resuspended in 100 µl binding buffer (Thermo Fisher Scientific,
Inc.). Subsequently, 4 µl Annexin V-fluorescein isothiocyanate and
3 µl propidium iodide (Thermo Fisher Scientific, Inc.) were added
in the dark. Following incubating for 15 min at RT, 200 µl binding
buffer was added and measured using a flow cytometer (BD
Biosciences, San Jose, CA, USA). The same experiments were
performed in triplicate.
The dual luciferase reporter system
assay
A total of three types of internet-based
bioinformatics online software, TargetScan 6.0 (http://www.targetscan.org/vert_71/), miRanda
(http://www.microrna.org/microrna/home.do) and miRbase
(http://www.mirbase.org/), were used to search the
specific targets between miR-370 and the 3′-untranslated region
(UTR) of FOXO1. Based on the complementarity nucleotide sequence to
the 3′-UTR region of FOXO1 mRNA, the specific targets were
recognized. According to the information recorded on microRNA.org, miR expression data were obtained. The
target gene sequence and the mutation gene sequence were
synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The dual
luciferase reporter system assay kit was provided by Promega
Corporation (Madison, WI, USA) and contained the luciferase
reporter plasmid. The gene sequence was bound with the carrier
psiCHECK-2. Gene sequences were digested using Xho I and
Not I enzymes and the sequences were authenticated by
electrophoresis and sequencing. 293 cells in the exponential phase
were selected for the present assay. The psiCHECK2-FOXO1-wild type
(WT) and psiCHECK2-FOXO1-mutation (MT) were treated with
hsa-miR-370 mimics separately. Lipofectamine 2000 was used in the
transfection procedure according to the manufacturer's protocols.
Following incubation in 5% CO2 for 24 h at 37°C, the
medium was replaced with fresh SMCM medium. Subsequently, the 293T
cells were transfected for 48 h. Three samples were contained in
each group. Following transfection, the 293 cells in WT and MT
groups were digested using tritonX-100. The lysate was treated with
Firefly luciferase and Renilla luciferase buffer solution
and their relative substrate separately. Firefly luciferase was
used as the internal reference of psiCHECK-2 and the expression of
the carrier psiCHECK-2+miR-NC was used as the control. Luciferase
activities were determined with a dual-luciferase assay kit 48 h
following transfection.
Western blot analysis
HUVECs were lysed with radioimmunoprecipitation
assay buffer containing protease inhibitor cocktail (Dalian Meilun
Biotech Co., Ltd., Dalian, China). Subsequently, the lysate was
centrifuged for 10 min at 10,800 × g at 4°C. The supernatant was
obtained for further analysis. Total protein was measured using a
Pierce BCA protein assay kit (Thermo Fisher Scientific, Inc). Equal
amounts of total protein (40 µg) were subjected to SDS-PAGE (10%
gels) and transferred onto polyvinylidene fluoride membranes (PVDF;
Dalian Meilun Biotech Co., Ltd.) with a wet transmembrane device
(Thermo Fisher Scientific, Inc.). The following primary antibodies
were used: Anti-FOXO1 (1:1,000 dilution), anti-Bax (1:1,000
dilution), anti-Bcl-2 (1:1,000 dilution), anti-cleaved PARP
(1:1,000 dilution) anti-active caspase 3 (1:1,000 dilution) and
anti-β-actin (1:1,000). Following blocking with 5% non-fat milk at
RT for 1 h, the membranes were incubated overnight with primary
antibodies and incubated with the appropriate horseradish
peroxidase (HRP)-conjugated secondary antibodies (HRP-anti-rabbit,
cat. no. 0208, 1:5,000 dilution; HRP-anti-goat, cat. no. 0216,
1:5,000 dilution; both Beyotime Institute of Biotechnology,
Shanghai, China) for 2 h at RT. Subsequently, the PVDF membranes
were incubated with enhanced chemiluminescence reagent (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), in order to develop the
blots. All values were normalized to those of β-actin. Protein
densitometry was determined using Image-Pro Plus 6.0 software
(Media Cybernetics, Inc., Rockville, MD, USA).
Statistical analysis
Values were expressed as the mean ± SD. All
experiments were conducted a minimum of three times. All
statistical analyses were calculated using SPSS 19.0 software (IBM
Corp., Armonk, NY, USA). One-way analysis of variance followed by
Dunnett's post hoc test was used to compare the differences between
multiple groups. Statistical differences between two groups were
determined using the Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-370 enhances the cell
proliferation of HUVECs
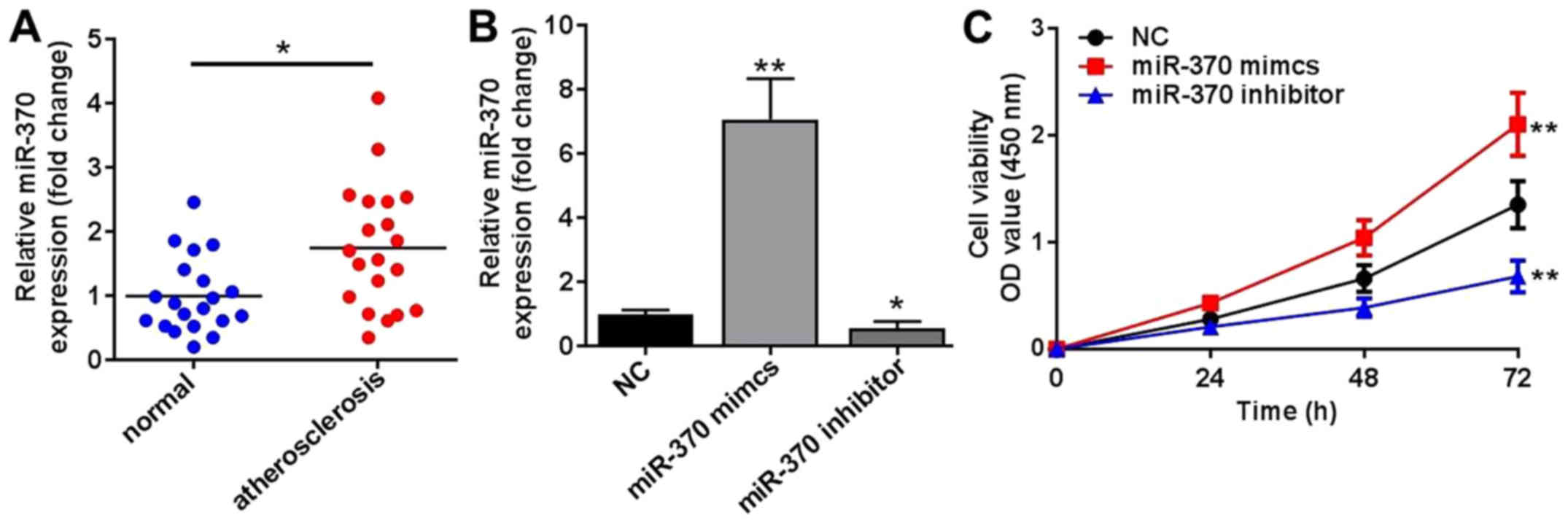
Compared with healthy patients, the expression
levels of miR-370 in PBMCs of patients with atherosclerosis were
significantly increased, which indicated that the abnormal
expression of miR-370 was associated with atherosclerosis (Fig. 1A). Following transfection with
miR-370 mimics, the expression levels of miR-370 in HUVECs were
significantly increased compared with the NC group (Fig. 1B); however, the expression levels of
miR-370 in HUVECs were significantly reduced compared with the NC
group, following transfection with miR-370 inhibitors (Fig. 1C). These results demonstrated that
the transfection model was successful. Following transfection with
miR-370 mimics or inhibitors, HUVEC proliferation was affected in
two different directions. Accompanied with the enhanced expression
of miR-370 induced by transfection with miR-370 mimics, HUVEC
proliferation was significantly increased compared with the NC
group (P<0.01; Fig. 1C). miR-370
enhanced the cell proliferation, which was also confirmed by the
decreased cell viability in the miR-370 inhibitor group compared
with the NC group (Fig. 1C).
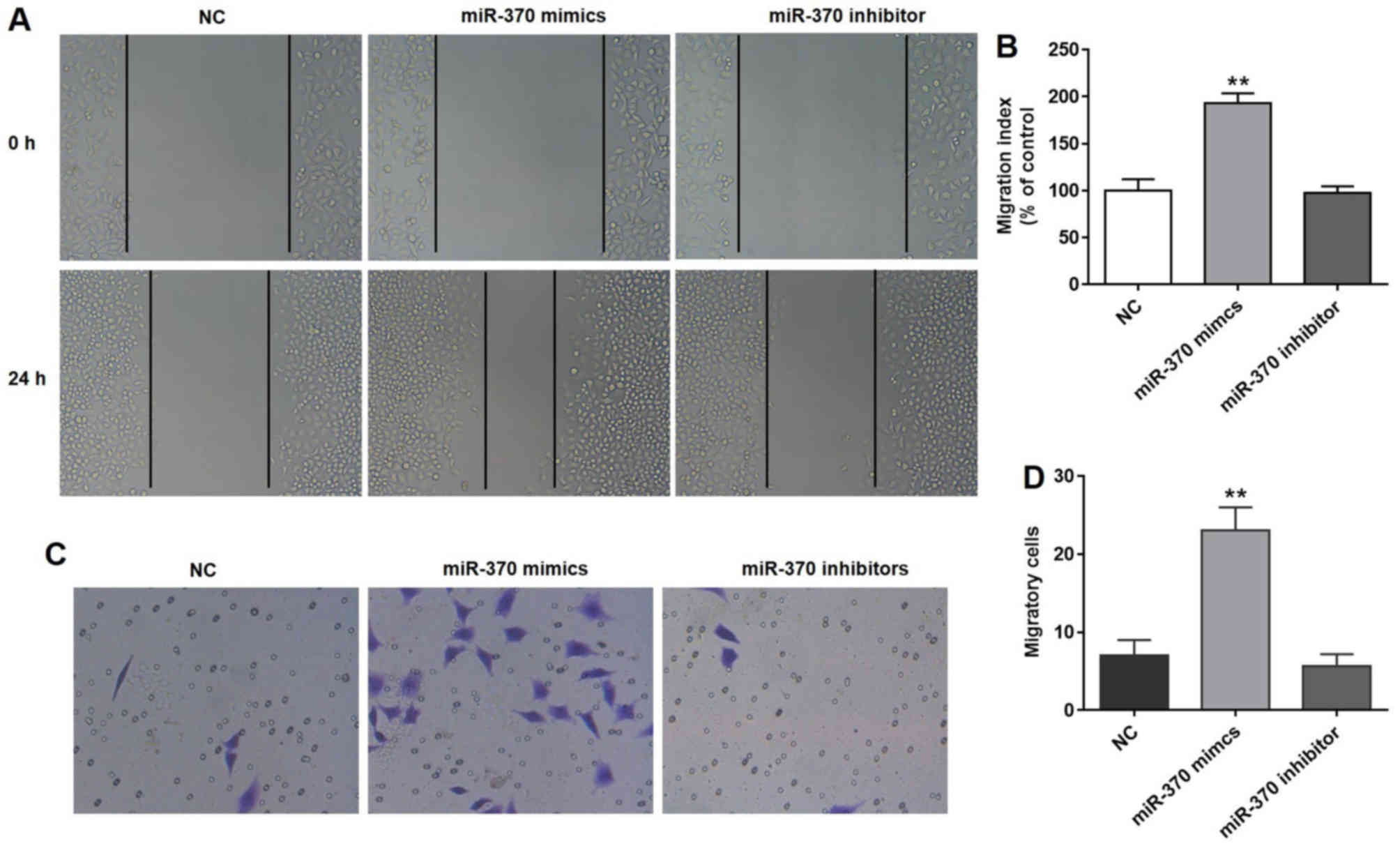
miR-370 promotes cell migration and
invasion
As depicted in Fig.
2A, the distance in the NC, miR-370 mimics and miR-370
inhibitor groups was similar at 0 h; however, the distance in the
miR-370 mimics group was reduced at 24 h compared with the NC
group. According to the results observed in the wound healing
assay, overexpression of miR-370 significantly promoted the
activity of cell migration compared with the NC group (Fig. 2B). The invasion capacity of HUVECs,
which were transfected with the miR-370 mimics, was evaluated by
performing the Matrigel assay. As indicated in Fig. 2C and D, upregulation of miR-370 with
miR-370 mimics significantly promoted cell migration compared with
the NC group. Notably, miR-370 inhibitors did not affect cell
migration and invasion (Fig.
2A-D).
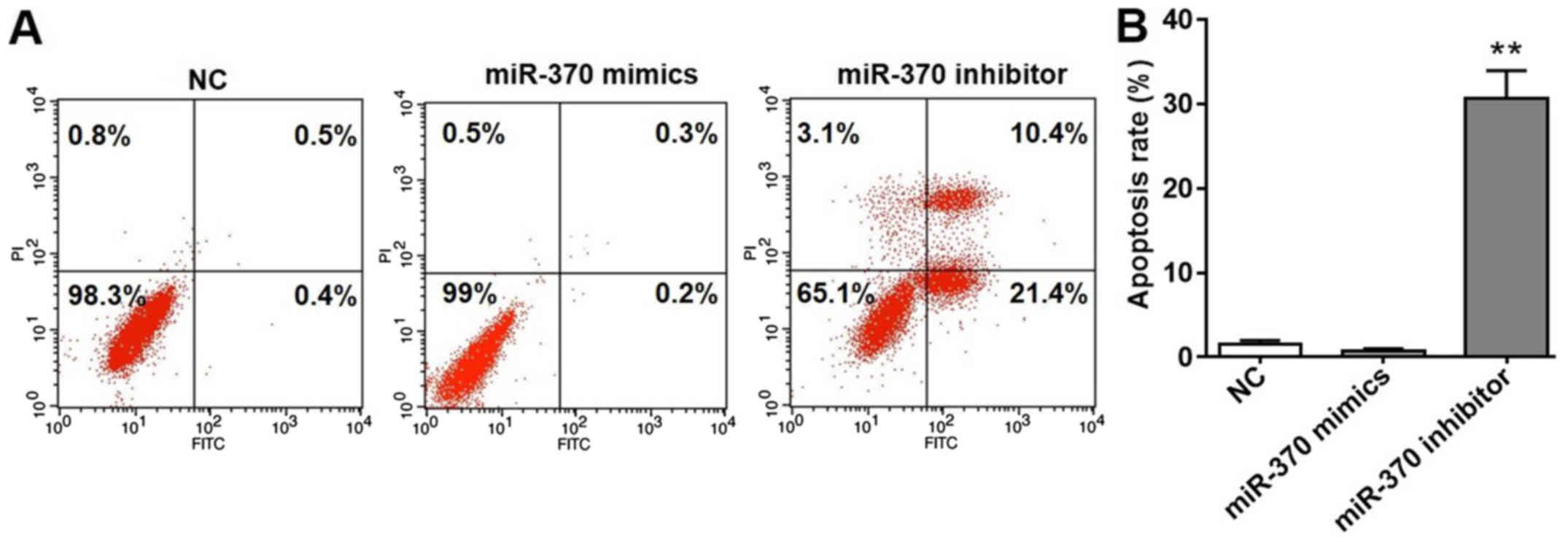
Downregulation of miR-370 promotes
cell apoptosis
As depicted in Fig.
3A, a notable number of apoptotic cells were observed in HUVECs
transfected with miR-370 inhibitors. Based on the results obtained
with flow cytometry, the apoptosis rate was calculated and depicted
in Fig. 3B. The apoptotic rate in
the NC and miR-370 mimics groups were low. According to the results
depicted in Fig. 1B, the expression
of miR-370 was significantly reduced following transfection with
miR-370 inhibitors; however, the apoptotic rate of HUVECs in the
miR-370 inhibitor group was significantly increased compared with
the NC group. Collectively, the results demonstrated that
downregulation of miR-370 promoted cell apoptosis. Notably, miR-370
mimics could not induce HUVEC apoptosis.
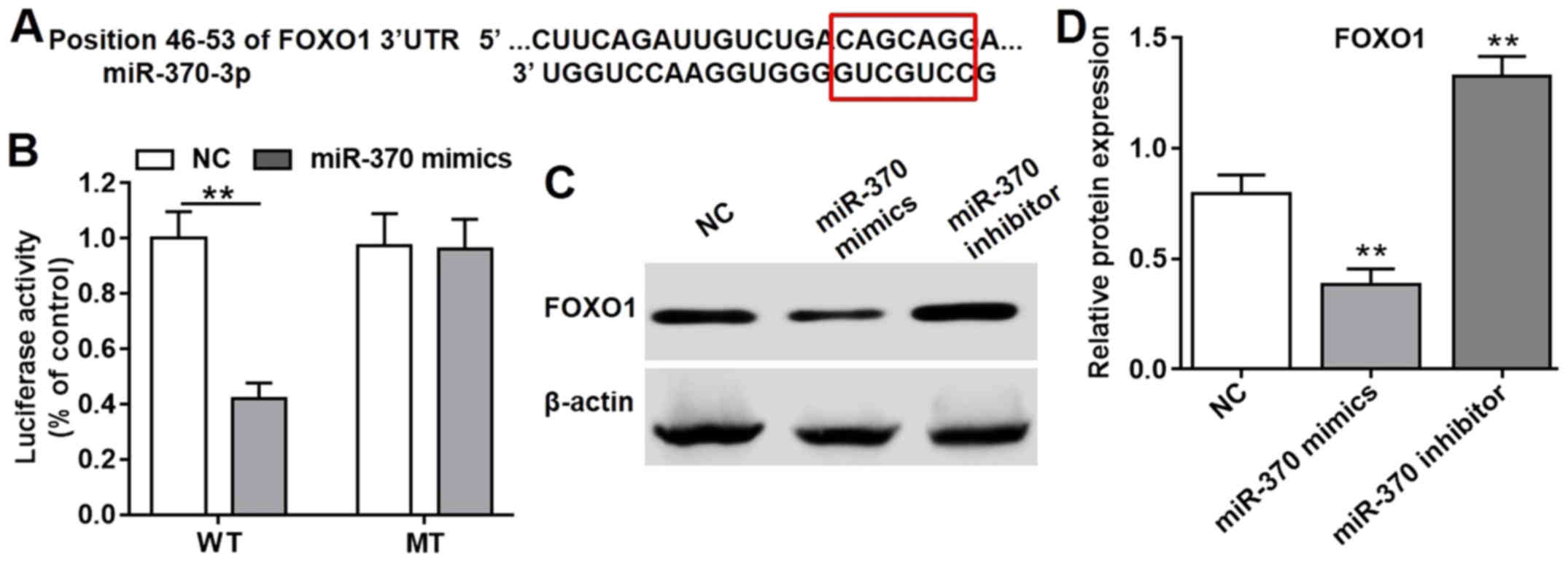
miR-370 is a regulator of FOXO1
The miRNA sequence in the bioinformatics platforms
was used to search for potential targets, which were weighted and a
list of the optimal potential targets are generated. According to
the three most commonly used bioinformatics prediction tools
(TargetScan 6.0, miRanda and miRbase), protein FOXO-1 was projected
as the most possible target of miR-370; the specific combined
targets of miR-370 on FOXO1 gene was focused from 46–53 of FOXO1
3′-UTR (Fig. 4A); therefore, FOXO1
was indicated as the target gene of miR-370. In order to confirm
the result, the dual luciferase reporter assay was performed. 293T
cells, the most commonly used tool in this assay, were selected.
The luciferase activity was significantly reduced following
transfection with psiCHECK-2-FOXO1-WT and miR-370 mimics compared
with the NC group (Fig. 4B);
however, the luciferase activity was not significantly affected
when the specific target was mutated (Fig. 4B). Therefore, miR-370 was indicated
as a vital regulator of the FOXO1 gene. Furthermore, the western
blot analysis result of the FOXO1 protein performed on HUVECs also
supported this conclusion. As depicted in Fig. 4C and D, the protein expression levels
of FOXO1 in HUVECs following transfection with miR-370 mimics was
significantly reduced compared with the NC group. However, HUVECs
in the presence of miR-370 inhibitors exhibited increased
expression levels of FOXO1 protein compared with the NC group.
miR-370 regulates cell apoptosis by
affecting relative target proteins
FOXO1, as a transcription factor, serves a vital
role in numerous crucial cellular processes, including cell
apoptosis and cell cycle progression (13). FOXO1 is considered to be involved in
the process of inducing cell apoptosis through its transcription of
a number of genes in the apoptotic pathway and its location in the
cell nucleus (13,14). A total of four apoptotic proteins,
Bax, Bcl-2, cleaved-PARP and active caspase 3, were selected to
evaluate the association between miR-370 and cell apoptosis
(Fig. 5). Following transfection
with miR-370 mimics, protein expression levels of pro-apoptotic
proteins, including Bax, cleaved-PARP and active caspase 3, were
reduced compared with the NC group (Fig.
5A and B). However, the expression levels of Bcl-2, a type of
inhibitor of apoptotic proteins, was significantly decreased
following transfection with miR-370 inhibitors compared with the NC
group (Fig. 5A and C). In addition,
miR-370 inhibitors significantly increased the expression levels of
Bax, cleaved-PARP and active caspase 3 in HUVECs following
transfection (Fig. 5A, B, D and
E).
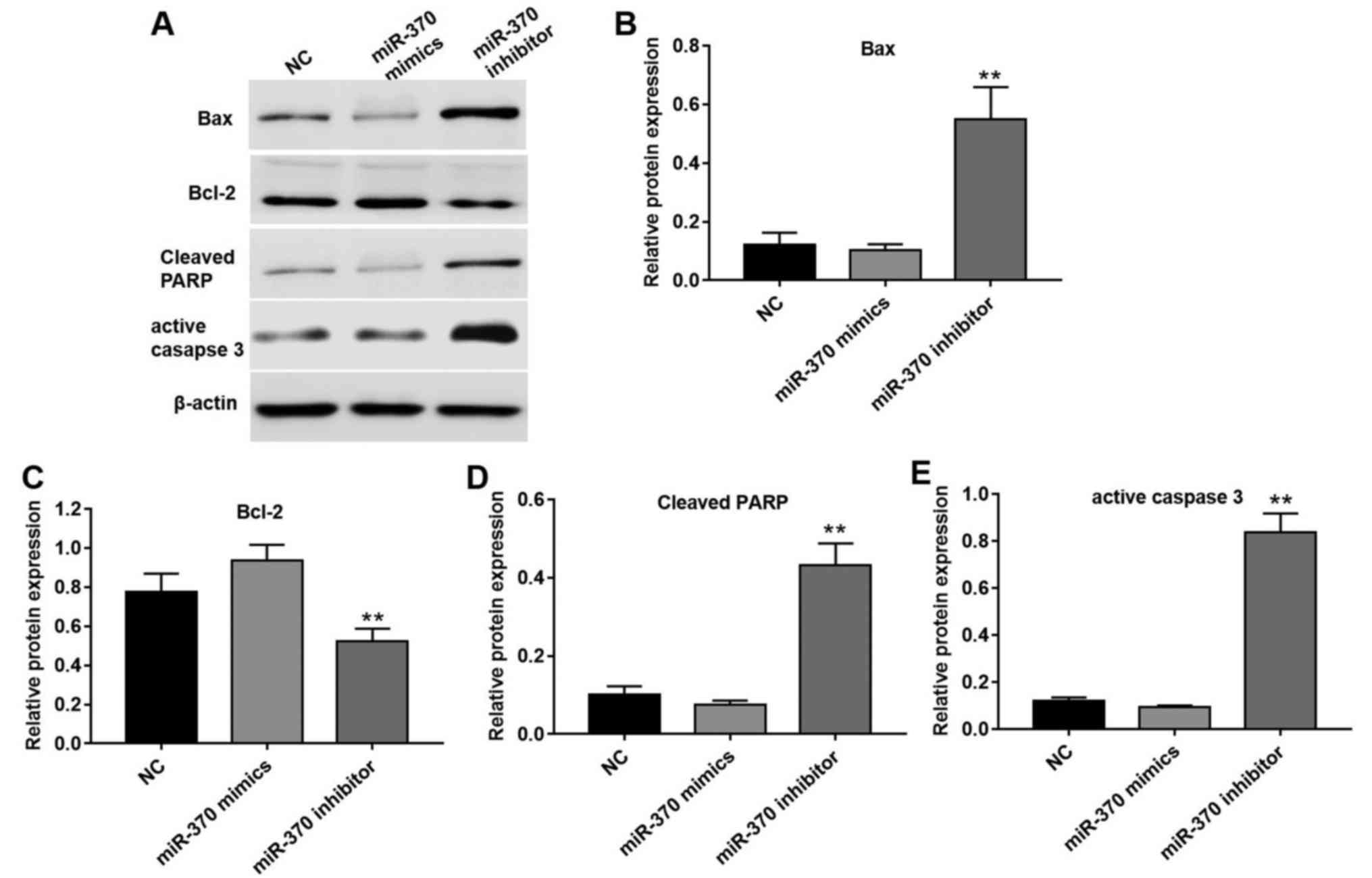 | Figure 5.miR-370 regulated cell apoptosis by
affecting relative target proteins. (A) Effect of overexpression
and downregulation of miR-370 on apoptotic proteins, including Bax,
Bcl-2, cleaved-PARP and active caspase 3. Effect of overexpression
and downregulation of miR-370 on apoptotic relative proteins (B)
Bax, (C) Bcl-2, (D) cleaved-PARP and (E) active caspase 3 in HUVECs
quantified by Image pro plus 6.0. Values were presented as the mean
± standard deviation. **P<0.05 vs. NC group. NC, negative
control; miR, microRNA; Bcl-2, B-cell lymphoma-2; Bax,
Bcl-2-associated X; HUVEC, human umbilical vein endothelial cells;
PARP, poly(ADP-ribose) polymerase. |
Discussion
miR-370 is considered to be a key miR in lipid
metabolism that downregulates the expression of the carnitine
palmitoyl transferase 1α gene, which controls fatty acid oxidation
(15). Liu et al (9) revealed that plasma expression levels of
miR-370 were significantly higher in the patients with coronary
artery disease compared with those without coronary artery disease
(9). Consistent with a previous
report (9), the expression levels of
miR-370 in the PBMCs of patients with atherosclerosis were
significantly increased. In addition, it has been reported that
miR-370 expression levels are potential diagnostic tools for
discriminating coronary artery disease to facilitate with the
management of patient care (9).
However, to the best of our knowledge, there is no report on the
function of miR-370 in atherosclerosis.
In the present study, the function and target gene
of miR-370 in HUVECs were investigated. HUVECs are the most
commonly used model for the study of coronary atherosclerosis, as
the proliferation and migration of endothelial cells (ECs) are
vital processes involved in atherosclerosis (2). The present study results suggested that
overexpression of miR-370 enhanced cell migration and invasion. The
enhanced invasion of assorted exogenous cells across a stiffened
extracellular matrix and an increase in the migratory behavior of
resident cells are considered to characterize atherosclerosis
(16). For example, as an RNA
aptamer, Apt-alphavbeta3 has been identified to reduce
platelet-derived growth factor-stimulated tube formation and to
increase HUVEC apoptosis through inhibition of focal adhesion
kinase phosphorylation pathway. In addition, human endothelial cell
proliferation and survival were significantly inhibited by
Apt-alphavbeta3, which in turn resulted in reduced angiogenesis
(17). Similarly, the present study
indicated that downregulation of miR-370 promoted cell
apoptosis.
In order to further investigate the underlying
molecular target involved in miR-370-mediated atherosclerosis,
three tools, namely TargetScan 6.0, miRanda and miRbase, were used.
According to the result of these three tools, protein FOXO-1 was
considered to be the possible target of miR-370. FOXO-1 was
selected in the present study to investigate its association with
miR-370. The dual luciferase reporter system assay suggested that
miR-370 could bind to the 3′-UTR of FOXO1 mRNA, which resulted in
the inhibition of FOXO1 expression.
FOXOs have been confirmed to be essential regulators
of cellular homeostasis, including oxidative stress response and
redox signaling, lipid metabolism, apoptosis and cell cycle
progression (18,19). FOXO1 has been considered to be
important in different types of cellular action (18,19).
Deng et al (20) demonstrated
that FOXO1 served an important role in vascular smooth muscle cell
calcification and the ubiquitination of PTEN/Akt-modulated
Runt-associated transcription factor 2. Li et al (21) demonstrated that FOXO1 and the
pro-apoptotic protein Bcl-2 like 11 (Bim) served vital roles in
regulating endothelial cell apoptosis induced by oxidative stress;
however, endothelial cell apoptosis caused by oxidative stress is
an early event in the development of atherosclerosis. Furthermore,
miR-370 has been reported to be effective in regulating the
proliferation of human prostate cancer cells through directly
inhibiting the tumor suppressor FOXO1 (12,18,19).
According to the present results, miR-370 was a specific regulator
of FOXO1, which was consistent with previous reports (12,18,19).
Apoptosis has critical role in maintaining tissue
homeostasis and eliminating damaged cells, and has been confirmed
to be a programmed cell suicide mechanism (21). The Bcl-2 family and caspases are the
two major regulators of apoptosis (21). According to reports published
previously, FOXO1 is inhibited by the phosphorylation of Akt, while
the downstream protein Bim is activated by the phosphorylation of
FOXO1 (18–21). Additionally, Bim can directly
activate the pro-apoptotic protein Bax, which is downstream of Bim
and inhibits the anti-apoptotic protein Bcl-2 (21). Furthermore, PARP is inhibited by
caspase 3 and cleaved PARP is an important protein in promoting
cell apoptosis (22). These proteins
are all involved in the mitochondrial apoptotic pathways (22).
Collectively, as a specific regulator of FOXO1, the
present findings suggested that miR-370 serves a vital role in
coronary atherosclerosis. Overexpression of miR-370 inhibited the
expression of FOXO1. Furthermore, Bax, Bcl-2 and cleaved PARP
expression levels were affected in the process of cell apoptosis;
therefore, miR-370 has the potential to be used as a biomarker in
identifying coronary atherosclerosis and may be used in treatment
to improve coronary atherosclerosis. Further research on the
function between these markers in vivo will provide an
essential insight into basic and clinical processes. However, many
miRNAs may be associated with atherosclerotic disease. Thus, the
assessment of other miRNAs and their functional effects in
atherosclerosis is required, which may offer a potential target for
coronary artery disease therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Jiangsu
Provincial Special Program of Medical Science (grant. no.
BE2015612).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XS performed experiments, analyzed data and was the
major contributor in developing the manuscript. XC collected
tissues, interpreted the patient data and reviewed the final
version of the manuscript.
Ethics approval and consent to
participate
Ethical approval for the present study was received
from Nanjing First Hospital, Nanjing Medical University. All
patients provided written informed consent to participate in the
study.
Patient consent for publication
All patients provided written informed consent for
the publication of all associated information.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu Z, Han Y, Liu J, Jiang F, Hu H, Wang Y,
Liu Q, Gong Y and Li X: MiR-135b-5p and MiR-499a-3p promote cell
proliferation and migration in atherosclerosis by directly
targeting MEF2C. Sci Rep. 5:122762015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ndrepepa G, Colleran R and Kastrati A:
Gamma-glutamyl transferase and the risk of atherosclerosis and
coronary heart disease. Clin Chim Acta. 476:130–138. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang H, Jia K, Sun D and Yang M:
Protective effect of HSP27 in atherosclerosis and coronary heart
disease by inhibiting reactive oxygen species. J Cell Biochem. Dec
12–2017.(Epub ahead of print). View Article : Google Scholar
|
|
5
|
Zhu N, Zhang D, Chen S, Liu X, Lin L,
Huang X, Guo Z, Liu J, Wang Y, Yuan W and Qin Y: Endothelial
enriched microRNAs regulate angiotensin II-induced endothelial
inflammation and migration. Atherosclerosis. 215:286–293. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Quan X, Ji Y, Zhang C, Guo X, Zhang Y, Jia
S, Ma W, Fan Y and Wang C: Circulating MiR-146a may be a potential
biomarker of coronary heart disease in patients with subclinical
hypothyroidism. Cell Physiol Biochem. 45:226–236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang Y, Wang HY, Li Y, Guo SH, Zhang L
and Cai JH: Peripheral blood miRNAs as a biomarker for chronic
cardiovascular diseases. Sci Rep. 4:50262014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hoekstra M, van der Lans CA, Halvorsen B,
Gullestad L, Kuiper J, Aukrust P, van Berkel TJ and Biessen EA: The
peripheral blood mononuclear cell microRNA signature of coronary
artery disease. Biochem Biophys Res Commun. 394:792–797. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu H, Yang N, Fei Z, Qiu J, Ma D, Liu X,
Cai G and Li S: Analysis of plasma miR-208a and miR-370 expression
levels for early diagnosis of coronary artery disease. Biomed Rep.
5:332–336. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Märkl B, Wilhelms N, Anthuber M,
Schenkirsch G, Schlimok G and Oruzio D: Circulating
cytokeratin-positive cells and tumor budding in colorectal cancer.
World J Clin Oncol. 7:433–440. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeng L, Chen Y, Wang Y, Yu LR, Knox B,
Chen J, Shi T, Chen S, Ren Z, Guo L, et al: MicroRNA hsa-miR-370-3p
suppresses the expression and induction of CYP2D6 by facilitating
mRNA degradation. Biochem Pharmacol. 140:139–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu Z, Sun H, Zeng W, He J and Mao X:
Upregulation of MircoRNA-370 induces proliferation in human
prostate cancer cells by downregulating the transcription factor
FOXO1. PLoS One. 7:e458252012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xing YQ, Li A, Yang Y, Li XX, Zhang LN and
Guo HC: The regulation of FOXO1 and its role in disease
progression. Life Sci. 193:124–131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pan CW, Jin X, Zhao Y, Pan Y, Yang J,
Karnes RJ, Zhang J, Wang L and Huang H: AKT-phosphorylated FOXO1
suppresses ERK activation and chemoresistance by disrupting
IQGAP1-MAPK interaction. EMBO J. 36:995–1010. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iliopoulos D, Drosatos K, Hiyama Y,
Goldberg IJ and Zannis VI: MicroRNA-370 controls the expression of
microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid
Res. 51:1513–1523. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kai F, Laklai H and Weaver VM: Force
Matters: Biomechanical regulation of cell invasion and migration in
disease. Trends Cell Biol. 26:486–497. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mi J, Zhang X, Giangrande PH, McNamara JO
II, Nimjee SM, Sarraf-Yazdi S, Sullenger BA and Clary BM: Targeted
inhibition of alphavbeta3 integrin with an RNA aptamer impairs
endothelial cell growth and survival. Biochem Biophys Res Commun.
338:956–963. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berry E, Hardt JL, Clardy J, Lurain JR and
Kim JJ: Induction of apoptosis in endometrial cancer cells by
psammaplysene A involves FOXO1. Gynecol Oncol. 112:331–336. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsuchiya K and Ogawa Y: Forkhead box class
O family member proteins: The biology and pathophysiological roles
in diabetes. J Diabetes Investig. 8:726–734. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng L, Huang L, Sun Y, Heath JM, Wu H and
Chen Y: Inhibition of FOXO1/3 promotes vascular calcification.
Arterioscler Thromb Vasc Biol. 35:175–183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li Y, Ren M, Wang X, Cui X, Zhao H, Zhao
C, Zhou J, Guo Y, Hu Y, Yan C, et al: Glutaredoxin 1 mediates the
protective effect of steady laminar flow on endothelial cells
against oxidative stress-induced apoptosis via inhibiting Bim. Sci
Rep. 7:155392017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu C, Zhu Q, Wu Z, Yin Y, Kang D, Lu S
and Liu P: Isorhapontigenin induced cell growth inhibition and
apoptosis by targeting EGFR-related pathways in prostate cancer. J
Cell Physiol. 233:1104–1119. 2018. View Article : Google Scholar : PubMed/NCBI
|