Introduction
Gastric cancer (GC) is one of the most common
malignant cancers in the world. The incidence of GC has declined
with identification of several risk factors including dietary and
environmental risks in the past several years (1–4).
However, most patients diagnosed with GC at advanced stage have a
low 5-year survival rate (5,6). The main reason for the poor prognosis
in GC is invasion and metastasis (7–9). To
improve the understanding of GC progression, novel oncogenes and
therapeutic targets should be explored.
T-box (TBX)2 is a member of T-box gene family which
is one of the most prominent gene families in the field of
embryonic development (10). Members
in T-box gene family are highly conserved in evolution ranging from
hydra to human (11–13). T-box transcription factors are
characterized by a highly conserved DNA binding domain and have
been shown to function as either transcriptional activators or
repressors (14–17).
Genetic researches showed that T-box proteins are
generally expressed in certain cell types of specific organs in
flies, worms, fish, mice, dogs, and humans and they were reported
to be necessary for the development of these structures (18,19). In
addition, a growing list of studies have reported dysregulation of
T-box factors in different cancers and inappropriate T-box factor
expression was directly linked to oncogenesis (20–23).
TBX2 overexpression was first reported in breast
cancer tissues and cell lines, and high TBX2 level correlated with
poor prognosis (24). TBX2
overexpression was also found in pancreatic cancer, endometrial
cancer, colorectal cancer and cervical cancer (25–29).
However, the effects of TBX2 on GC progression remains unclear.
This study aims to clarify not only the clinical of TBX2 in GC
tissues but also its biological functions and potential
mechanisms.
Materials and methods
Patients and specimens
This study protocol was approved by the ethics
review board of the First Affiliated Hospital of China Medical
University (Shenyang, China). Primary tumor specimens were obtained
from 161 patients diagnosed with GC who underwent resection in the
First Affiliated Hospital of China Medical University between 2010
and 2015. The histological diagnosis was evaluated for sections
stained with hematoxylin and eosin according to the 2004 World
Health Organization (WHO) classification guidelines.
Immunohistochemistry
Immunohistochemistry was performed using the
Elivision plus kit purchased from MaiXin (Fuzhou, China). Samples
were prepared for 4-µm thick tissue sections and deparaffinized
using xylene. Sections were rehydrated with graded alcohol (100%, 5
min for 2 times; 95%, 2 min; 80%, 2 min; 70%, 2 min). Citrate
buffer (pH 6.0; 2 min) was used for antigen retrieval.
H2O2 was used to block the endogenous
peroxidase. Normal goat serum was used to reduce nonspecific
binding. Sections were incubated with TBX2 antibody (rabbit
polyclonal; dilution, 1:60; SAB2102383; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) overnight at 4°C. A biotinylated anti-rabbit
horseradish peroxidase polymer was used as a secondary antibody.
The peroxidase reaction was developed with DAB plus from MaiXin.
Counterstaining was done with hematoxylin, and the sections were
dehydrated in alcohol before mounting.
Two independent investigators, who were blinded to
the patient characteristics, examined all tumor slides randomly. To
evaluate the staining, 400 tumor cells were counted and the
intensity/percentage was calculated. The intensity of TBX2 staining
was scored as 0 (no signal), 1 (moderate) or 2 (strong). Percentage
scores were assigned as 1 (1–25%), 2 (26–50%), 3 (51–75%) or 4
(76–100%). The scores of each tumor sample were multiplied to give
a final score of 0–8; tumor samples that scored 4–8 were considered
as TBX2 overexpression.
Cell culture
BGC-823 and SGC-7901 cells were purchased from
Shanghai Cell Bank (Shanghai, China). Cells were cultivated in DMEM
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific,
Inc.), with the conditions of 37°C and 5% CO2. Cells
were seeded at a density of 1×106 cells/ml. Cells were
treated with paclitaxel (final concentration, 5 µM) after cells
attached to the bottom for 24 h.
Cell transfection
pCMV6 TBX2 plasmid was purchased from OriGene
Technologies, Inc. (Rockville, MD, USA). Transfection was performed
in SGC-7901 cell line using Attractene reagent (Qiagen GmbH,
Hilden, Germany). siGENOME TBX2 siRNAs pool and Negative siRNAs
pool were purchased from GE Healthcare Dharmacon, Inc. (Lafayette,
CO, USA). siRNA was transfected into BGC-823 cell line with
DharmaFECT1 (GE Healthcare Dharmacon, Inc.).
Western blotting
Total protein of cells was extracted using lysis
buffer (Pierce; Thermo Fisher Scientific, Inc.) and quantified by
Bradford method. 30 µg protein was separated by SDS-PAGE. Samples
were transferred to polyvinylidene fluoride membranes (EMD
Millipore, Billerica, MA, USA) and incubated overnight at 4°C with
antibody against cyclin E (20808; Cell Signaling Technology, Inc.,
Danvers, MA, USA), matrix metalloproteinase (MMP)2 (1:1,000; cat.
no. 4022; Cell Signaling Technology, Inc.), MMP9 (1:1,000; cat. no.
3852; Cell Signaling Technology, Inc.), E-cadherin (24E10)
(1:1,000; cat. no. 3195; Cell Signaling Technology, Inc.),
N-cadherin (1:600; cat. no. 13116; Cell Signaling Technology,
Inc.), p21 (Waf1/Cip1;12D1) (1:1,000; cat. no. 2947; Cell Signaling
Technology, Inc.), p-ERK (1:1,000; cat. no. 9101; Cell Signaling
Technology, Inc.), ERK (1:1,000; cat. no. 4695; Cell Signaling
Technology, Inc.), β-actin (1:3,000; cat. no. 4967; Cell Signaling
Technology, Inc.), GAPDH (1:3,000; cat. no. G5262; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). After incubation with
peroxidase-coupled anti-mouse/rabbit IgG (1:1,000; Cell Signaling
Technology, Inc.) at 37°C for 2 h, bound proteins were visualized
using ECL (Pierce; Thermo Fisher Scientific, Inc.) and detected
using a DNR BioImaging System (DNR, Jerusalem, Israel). Relative
protein levels were quantified using ImageJ software.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol Reagent (Thermo
Fisher Scientific, Inc.). Total RNA was reverse transcribed using
PrimeScript RT Master Mix (5X). A total of 10 µl of reverse
transcribed reaction system was consisted of 800 ng RNA template, 2
µl RT master mix and moderate volume of RNase-free H2O.
Real-time PCR was performed using SYBR-Green master mix kit (ABI,
USA) with a 7500 Real-Time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.). PCR reaction system was consisted of 10
µl SYBR-Green master mix, 4 µl RNase-free H2O, 0.5 µl
forward primer, 0.5 µl reverse primer and 5 µl cDNA template. The
process of PCR was 95°C for 2 min, 40 cycles of 95°C for 2 sec,
annealing/extension at 60°C for 30 sec. A dissociation step was
performed to generate melting curves to confirm the specificity of
the amplification. Expression levels of the analyzed genes were
normalized to the expression of β-actin. The fold change of gene
expression was calculated by the 2−ΔΔCq method. The
sequences of the primer pairs are as follows: TBX2 forward,
5′-GGCTTCAACATCCTAAACTCC-3′; and reverse,
5′-AAGATCGACCAACAACCCGTTT-3′. β-actin forward,
5′-ATAGCACAGCCTGGATAGCAACGTAC-3′; and reverse,
5′-CACCTTCTACAATGAGCTGCGTGTG-3′.
MTT assay
Cells (5×103/well) were plated in 96-well
plates and then cultured overnight. 20 µl of 5 mg/ml MTT
[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide;
Sigma-Aldrich; Merck KGaA] solution was added to each well and
incubated for another 4 h at 37°C. The supernatant was removed and
DMSO (150 µl) was added to dissolve the formazan crystals.
Absorbance was measured at 490 nm. Data was obtained from
triplicate wells per condition and representative of at least three
independent experiments.
Matrigel invasion assay
Matrigel invasion assay was performed using a
24-well Transwell chamber. 48 h after the transfection, cells were
trypsinized and inoculated to the upper chamber with our serum and
incubated for 18 h. Lower chamber was added with medium
supplemented with 10% serum. Non-invaded cells were wiped out and
cells invaded through the filter were fixed with 4%
paraformaldehyde and stained with hematoxylin. The number of
invading cells was counted under electron microscope (Olympus BX53;
Olympus Corporation, Tokyo, Japan).
Statistical analysis
SPSS version 16 for Windows (SPSS, Inc., Chicago,
IL, USA) was used for all statistical analyses. A χ2
test was used to examine possible associations between TBX2
expression and clinicopathological factors. The Kaplan-Meier method
was used to estimate the probability of patient survival, and
differences in the survival of subgroups of patients were compared
by using Mantel's log-rank test. Cox proportional hazards
regressions were applied to estimate the individual hazard ratio
(HR). The Student's t-test was used to compare differences between
control and treatment groups. P<0.05 was considered to indicate
a statistically significant difference.
Results
TBX2 is overexpressed in human GC
To explore expression pattern of TBX2 in human GC,
161 cases of GC tissues and 10 cases of normal gastric tissues were
collected and then TBX2 protein expression was determined using
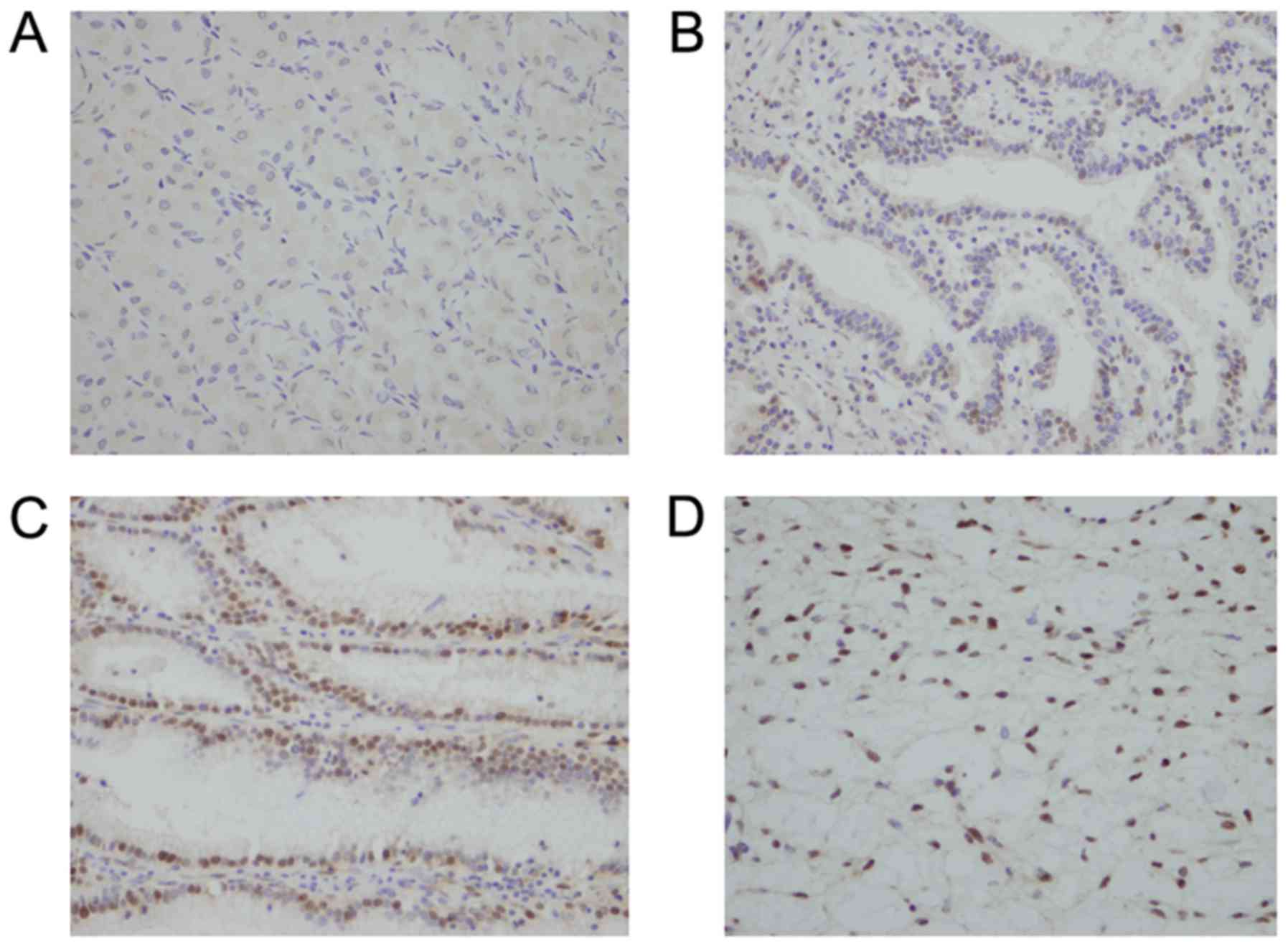
immunohistochemistry. As shown in Fig.
1A, negative staining was observed in normal gastric tissue.
Positive nuclear staining was observed in 90 of 161 (55.9%) cases
of GCs (Fig. 1B-D). The relationship
between TBX2 expression and various clinicopathological parameters
was analyzed and the results were listed in Table I. It was observed that TBX2
overexpression was significantly associated with deep tumor
invasion (T1+T2 45.9% vs. T3+T4 62%; P=0.0460), advanced TNM stage
(I+II 36.8% vs. III 72.9%; P<0.0001) and presence of nodal
metastasis (absent 41.7% vs. present 64.4%; P=0.0051). To
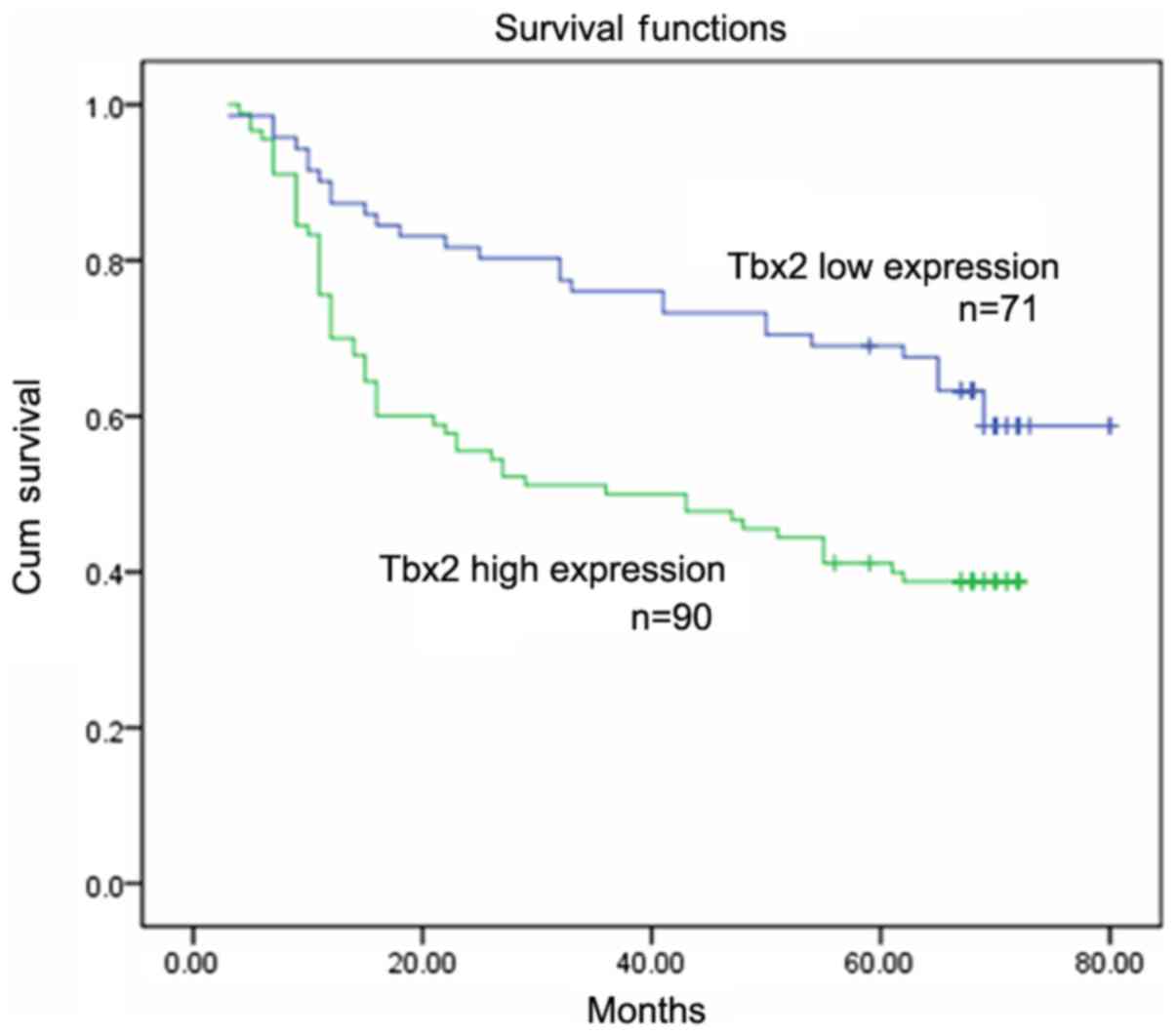
investigate the association between TBX2 expression and prognosis,
Kaplan-Meier analysis was performed and the results showed that
patients with higher TBX2 levels showed poorer survival than those
with low TBX2 levels (P<0.05; Fig.
2). In addition, Cox regression analysis revealed that TBX2 was
predictor for overall survival of patients with GCs using
univariate analysis (Table II).
 | Table I.Distribution of TBX2 status in
gastric cancer according to clinicopathological
characteristics. |
Table I.
Distribution of TBX2 status in
gastric cancer according to clinicopathological
characteristics.
|
Characteristics | Number of
patients | TBX2 low
expression | TBX2 high
expression | P-value |
|---|
| Age |
|
|
| 0.7901 |
|
<60 | 82 | 37 | 45 |
|
|
≥60 | 79 | 34 | 45 |
|
| Sex |
|
|
| 0.2644 |
|
Male | 116 | 48 | 68 |
|
|
Female | 45 | 23 | 22 |
|
|
Differentiation |
|
|
| 0.6369 |
|
Poor | 76 | 35 | 41 |
|
|
Well-moderate | 85 | 36 | 49 |
|
| Tumor invasion
(T) |
|
|
| 0.0460 |
|
T1+T2 | 61 | 33 | 28 |
|
|
T3+T4 | 100 | 38 | 62 |
|
| Lymph node
metastasis |
|
|
| 0.0051 |
|
Absent | 60 | 35 | 25 |
|
|
Present | 101 | 36 | 65 |
|
| TNM stage |
|
|
| <0.0001 |
|
I+II | 76 | 48 | 28 |
|
|
III | 85 | 23 | 62 |
|
 | Table II.Multivariate analysis for predictive
factors in patients with gastric cancer (Cox regression model). |
Table II.
Multivariate analysis for predictive
factors in patients with gastric cancer (Cox regression model).
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Factors | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| TBX2
overexpression | 2.027
(1.284–3.201) | 0.0024 | 1.049
(0.636–1.730) | 0.8524 |
| Stage | 2.127
(1.560–2.906) | <0.0001 | 2.285
(1.553–3.363) | <0.0001 |
|
Differentiation | 1.103
(0.716–1.697) | 0.6567 | 0.874
(0.560–1.363) | 0.5519 |
| Relapse | 70.552
(21.978–226.486) | 90 | 83.884
(26.938–306.633) | <0.0001 |
Expression pattern of TBX2 in GC
cells
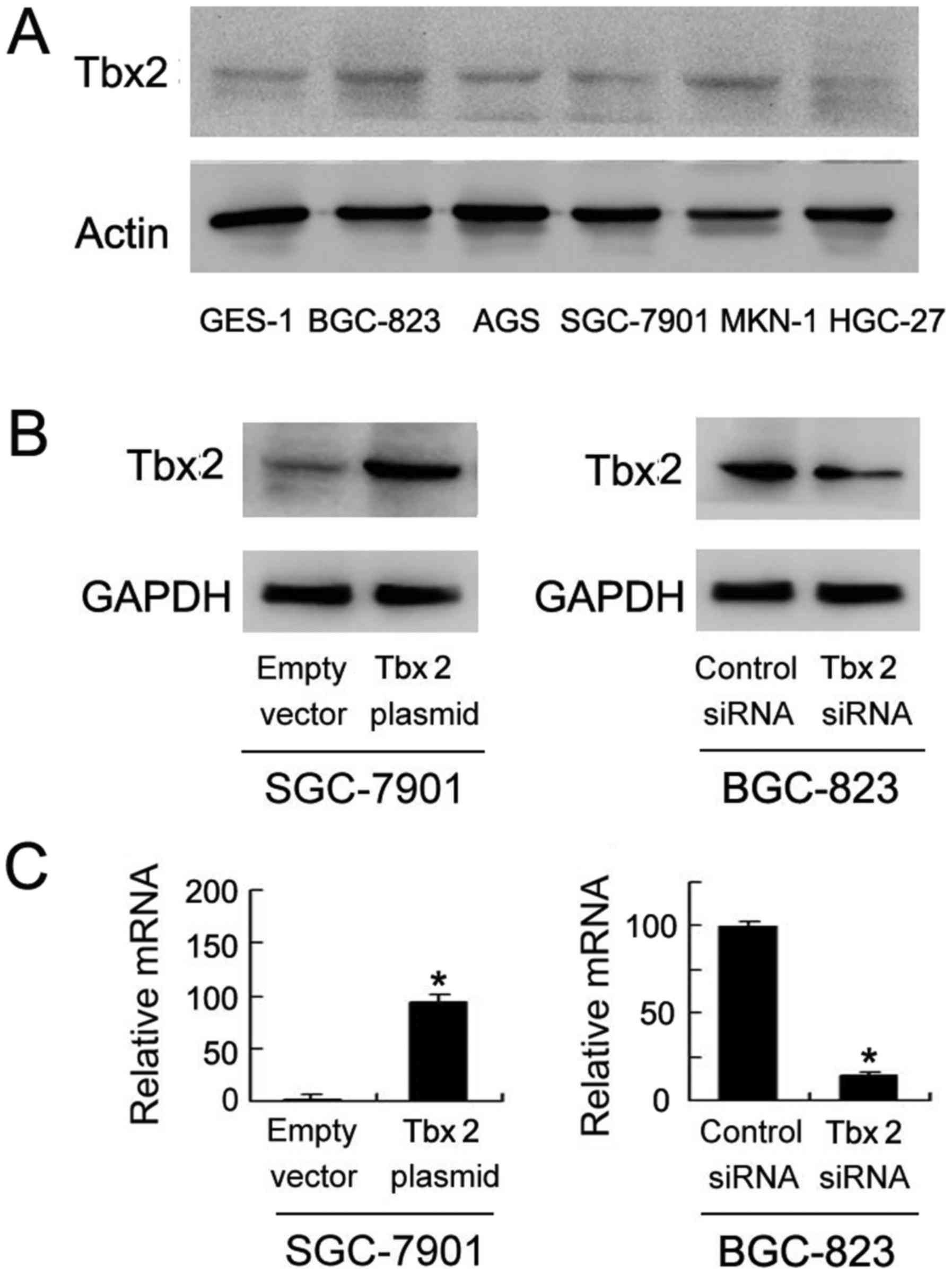
TBX2 protein expression in normal and cancerous
gastric cells (GES-1, BGC-823, AGS, SGC-7901, MKN-1 and HGC-27) was
detected by western blot. As shown in Fig. 3A, relative high level of TBX2 was
found in BGC-823 cells while low TBX2 expression was found in
SGC-7901 cells. In order to confirm effect of TBX2 on GC cells, we
adopted SGC-7901 and BGC-823 to perform TBX2 transfection and siRNA
interference. Western blot results showed that TBX2 protein
expression was significantly upregulated in SGC-7901 cells when
transfected with TBX2 plasmid while downregulated in BGC-823 cells
when treated with TBX2 siRNA (Fig.
3B). Quantitative PCR showed similar results.
TBX2 promotes cell proliferation and
invasion in GC cells
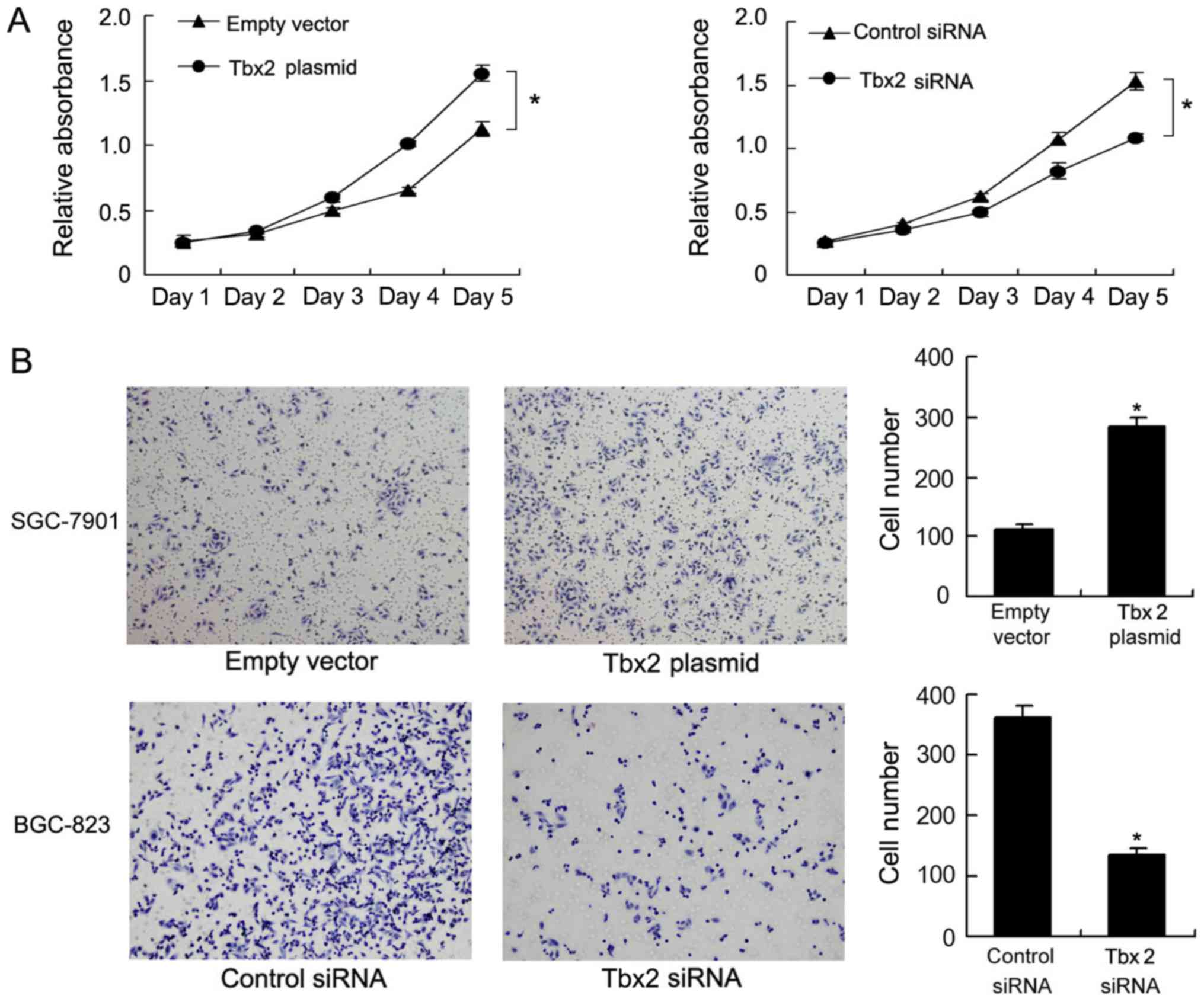
MTT assay was employed to measure the effect of TBX2
on cell viability. TBX2 overexpression induced cell growth rate
significantly while TBX2 depletion decreased cell growth rate
(Fig. 4A). Matrigel invasion assay
was performed to explore change of cell invasion. It was observed
that number of invading cells increased significantly after TBX2
overexpression while decreased after TBX2 depletion (Fig. 4B). These results demonstrated that
TBX2 enhanced the ability of proliferation and invasion in GC
cells.
TBX2 promotes EMT and ERK signaling
pathway
To investigate the potential mechanism responsible
for the effect of TBX2 on cell biological behaviors, related
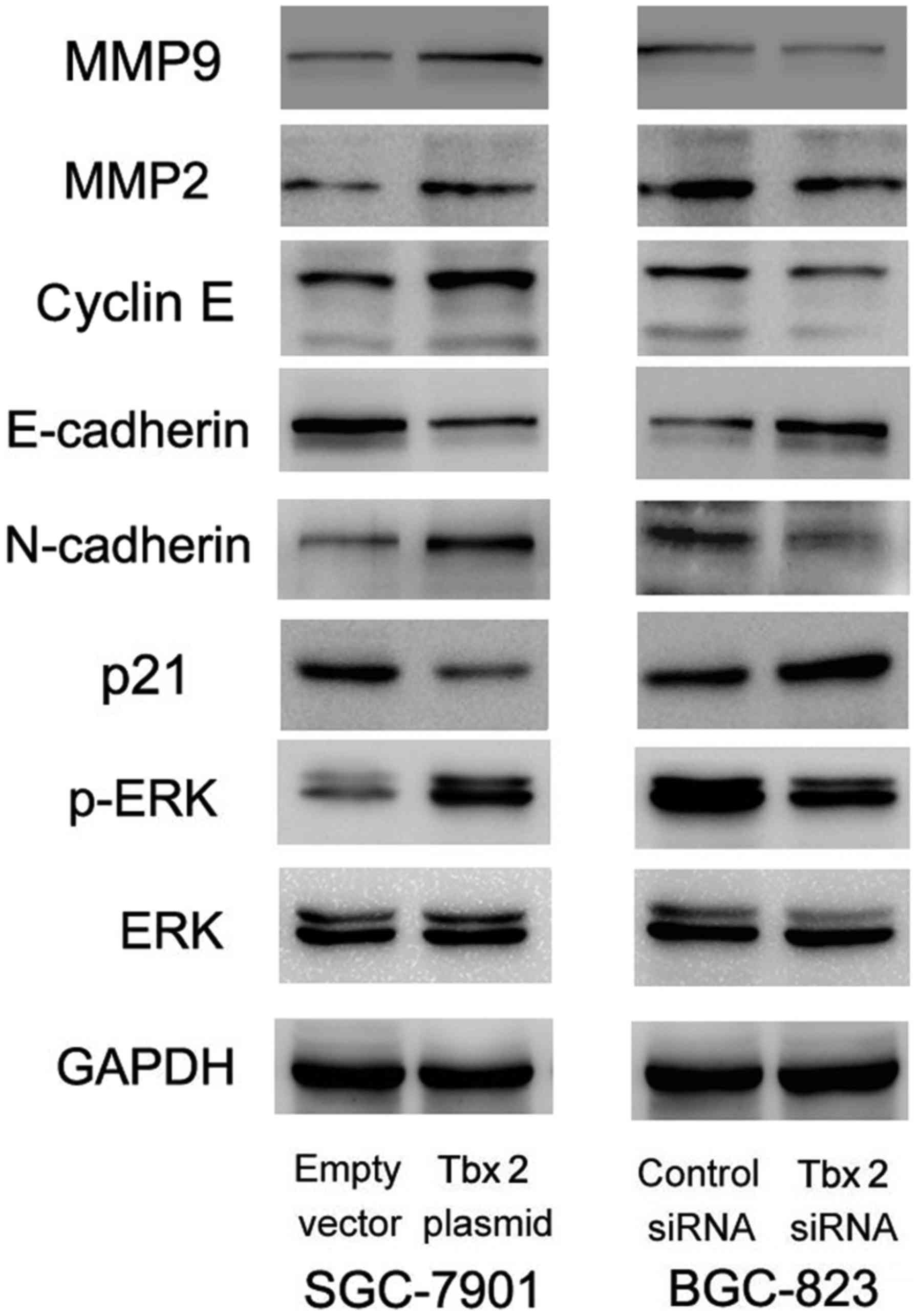
factors were detected using western blot. As shown in Fig. 5, levels of MMP2, MMP9, cyclin E,
N-cadherin were increased while E-cadherin and p21 levels decreased
in SGC-7901 cells transfected with TBX2 plasmid. In BGC-823 cells,
TBX2 depletion enhanced expression of E-cadherin and p21 while
suppressed expression of MMP2, MMP9, N-cadherin and cyclin E.
To further explore the possible molecular mechanism,
MAPKs signaling pathway was examined using western blot. ERK
phosphorylation level was increased in SGC-7901 cells transfected
with TBX2 plasmid and decreased in BGC-823 cells transfected with
TBX2 siRNA.
Discussion
In the present study, we examined TBX2 expression
pattern in GC tissues. The results revealed that TBX2 was
upregulated in GC tissues, with significant association with local
invasion, advanced TNM stage and presence of nodal metastasis.
These results suggested TBX2 correlated with malignant phenotype of
GCs. We also observed that patients with high TBX2 showed worse
prognosis compared with those with low TBX2 expression, suggesting
that TBX2 is a potential diagnostic or prognostic cancer
marker.
We also found that TBX2 directly regulated the
progression of GC cells. Transfection or knocking down of TBX2 was
sufficient to change the ability of cell proliferation, invasion.
Previous studies reported that TBX2 functioned as a potent
growth-promoter in malignant tumors partly due to its ability to
bypass senescence and repress key negative regulators of the cell
cycle (30,31). We observed that cell proliferation
rate was increased after TBX2 overexpression while decreased after
TBX2 depletion. Matrigel invasion results showed that invading
cells was increased transfected with TBX2 plasmid while decreased
when transfected with TBX2 siRNA, suggesting that TBX2 promoted the
ability of invasion.
To clarify the potential mechanism responsible for
the regulatory effect of TBX2 on cell biological behaviors in GC
cells, we examined expression of related protein including MMPs,
cyclins and p21. It was observed that expression of MMP2, MMP9 and
cyclin E was increased after TBX2 overexpression while decreased
when TBX2 depletion. MMPs are calcium-dependent zinc-containing
endopeptidases, which are capable of degrading all kinds of
extracellular matrix proteins and processing a number of bioactive
molecules (32–34). Tumor invasion is a phenomenon that
requires increased motility and proteolysis and MMPs have been
suggested to be critical for the invasive and metastatic potential
in varieties of malignant tumors (35–37). In
addition, we observed that TBX2 reduced the expression of
epithelial-mesenchymal transition marker E-cadherin while
upregulated N-cadherin. Epithelial-to-mesenchymal transition (EMT)
activation is a process that involves the transcriptional
repression of epithelial markers such as E-cadherin, which is
pivotal during cancer invasion and metastasis (38,39).
Thus TBX2 promote cell invasion partly through regulating MMP2,
MMP9 and E-cadherin levels.
TBX2 has been confirmed to function as
transcriptional repressors and promote the bypass of senescence by
downregulating expression of the negative cell cycle regulators p21
(40). Another reports showed that
TBX2 protein is a direct repressor of the p21 promoter through an
element located close to the p21 transcription start site (40,41). Our
study was consistent with previous conclusions, showing that TBX2
overexpression downregulated p21 level in SGC-7901 cells while TBX2
depletion upregulated p21 level in BGC-823 cells. We also found
that TBX2 positively regulated cyclin E expression. Cyclin E
normally accumulates at the G1/S phase transition and then
accelerates the speed of cell proliferation (42,43).
According to above findings, we speculated that TBX2 affected cell
proliferation partly through regulating p21 and cyclin E expression
in GC cells. TBX2 was reported to represses Connexin43, which
functions as gap junction protein (44). TBX2 could also represses CST6, which
induced sustained breast cancer proliferation (45). It is also reported that TBX2
promoting transcription of the canonical WNT3A promoter
(46). Thus TBX2 is a
multifunctional gene regulator which could either inhibit or
activate transcription.
To further investigate potential molecule mechanism,
MAPK signaling pathway was examined. Level of p-ERK increased in
SGC-7901 cells transfected with TBX2, while p-ERK level decreased
in BGC-823 cells interferenced with TBX2 siRNA. The ERK cascade
functions in cellular proliferation, differentiation, and survival,
and its activation promotes tumor progression (47–50). We
speculated that regulatory effect of TBX2 in GC may partly depend
on ERK signaling pathway.
In conclusion, TBX2 is upregulated in GCs and
correlates with lymph node metastasis, local invasion, TNM stage
and poor prognosis. TBX2 promotes proliferation, invasion through
regulation of MMP2, MMP9, cyclin E, E-cadherin and p21 through ERK
signaling pathway. TBX2 might serve as an effective biomarker and
therapeutic target in human GCs.
References
|
1
|
Castro C, Peleteiro B, Bento MJ and Lunet
N: Trends in gastric and esophageal cancer incidence in northern
Portugal (1994–2009) by subsite and histology and predictions for
2015. Tumori. 103:155–163. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leylabadlo HE, Kafil HS and Yousefi M:
Gastric cancer mortality in a high-incidence area (Ardabil
Province, Northwest Iran): What risk factors are causative? Eur J
Cancer Prev. 25:573–574. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yoon SB, Park JM, Lim CH, Kim JS, Cho YK,
Lee BI, Lee IS, Kim SW and Choi MG: Incidence of gastric cancer
after endoscopic resection of gastric adenoma. Gastrointest Endosc.
83:1176–1183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nicolas C, Sylvain M, Come L, Jean F,
Anne-Marie B and Valerie J: Trends in gastric cancer incidence: A
period and birth cohort analysis in a well-defined French
population. Gastric Cancer. 19:508–514. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu H, Cai Z, Lu G, Cao S, Huang H, Jiang Y
and Sun W: Impact of c-erbB-2 protein on 5-year survival rate of
gastric cancer patients after surgery: A cohort study and
meta-analysis. Tumori. 103:249–254. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greenhill C: Gastric cancer. Metformin
improves survival and recurrence rate in patients with diabetes and
gastric cancer. Nat Rev Gastroenterol Hepatol. 12:1242015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie R, Wang X, Qi G, Wu Z, Wei R, Li P and
Zhang D: DDR1 enhances invasion and metastasis of gastric cancer
via epithelial-mesenchymal transition. Tumour Biol. 37:12049–12059.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deng QJ, Xie LQ and Li H: Overexpressed
MALAT1 promotes invasion and metastasis of gastric cancer cells via
increasing EGFL7 expression. Life Sci. 157:38–44. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ara H, Takagishi M, Enomoto A, Asai M,
Ushida K, Asai N, Shimoyama Y, Kaibuchi K, Kodera Y and Takahashi
M: Role for Daple in non-canonical Wnt signaling during gastric
cancer invasion and metastasis. Cancer Sci. 107:133–139. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Armas P, Margarit E, Mouguelar VS, Allende
ML and Calcaterra NB: Beyond the binding site: In vivo
identification of tbx2, smarca5 and wnt5b as molecular targets of
CNBP during embryonic development. PLoS One. 8:e632342013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miskolczi-McCallum CM, Scavetta RJ,
Svendsen PC, Soanes KH and Brook WJ: The Drosophila melanogaster
T-box genes midline and H15 are conserved regulators of heart
development. Dev Biol. 278:459–472. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takabatake Y, Takabatake T, Sasagawa S and
Takeshima K: Conserved expression control and shared activity
between cognate T-box genes TBX2 and Tbx3 in connection with Sonic
hedgehog signaling during Xenopus eye development. Dev Growth
Differ. 44:257–271. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takabatake Y, Takabatake T and Takeshima
K: Conserved and divergent expression of T-box genes TBX2-Tbx5 in
Xenopus. Mech Dev. 91:433–437. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Papaioannou VE: The T-box gene family:
Emerging roles in development, stem cells and cancer. Development.
141:3819–3833. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takahashi H: Functional evolution of T-box
gene family on animal development. Tanpakushitsu Kakusan Koso.
46:1349–1357. 2001.(In Japanese). PubMed/NCBI
|
|
16
|
Papaioannou VE and Silver LM: The T-box
gene family. Bioessays. 20:9–19. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yi CH, Terrett JA, Li QY, Ellington K,
Packham EA, Armstrong-Buisseret L, McClure P, Slingsby T and Brook
JD: Identification, mapping and phylogenomic analysis of four new
human members of the T-box gene family: EOMES, TBX6, TBX18 and
TBX19. Genomics. 55:10–20. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brend T and Holley SA: Expression of the
oscillating gene her1 is directly regulated by Hairy/Enhancer of
Split, T-box and Suppressor of Hairless proteins in the zebrafish
segmentation clock. Dev Dyn. 238:2745–2759. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Butz NV, Gronostajski RM and Campbell CE:
T-box proteins differentially activate the expression of the
endogenous interferon gamma gene versus transfected reporter genes
in non-immune cells. Gene. 377:130–139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wan Z, Jiang D, Chen S, Jiao J, Ji L, Shah
AS, Wei H, Yang X, Li X, Wang Y and Xiao J: T-box transcription
factor brachyury promotes tumor cell invasion and metastasis in
non-small cell lung cancer via upregulation of matrix
metalloproteinase 12. Oncol Rep. 36:306–314. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu H, Liu BO, Liu A, Li K and Zhao H:
T-box 2 expression predicts poor prognosis in gastric cancer. Oncol
Lett. 10:1689–1693. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen S, Jiao J, Jiang D, Wan Z, Li L, Li
K, Xu L, Zhou Z, Xu W and Xiao J: T-box transcription factor
Brachyury in lung cancer cells inhibits macrophage infiltration by
suppressing CCL2 and CCL4 chemokines. Tumour Biol. 36:5881–5890.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Abrahams A, Parker MI and Prince S: The
T-box transcription factor TBX2: Its role in development and
possible implication in cancer. IUBMB Life. 62:92–102.
2010.PubMed/NCBI
|
|
24
|
Douglas NC and Papaioannou VE: The T-box
transcription factors TBX2 and TBX3 in mammary gland development
and breast cancer. J Mammary Gland Biol Neoplasia. 18:143–147.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Boskovic G and Niles RM: T-box binding
protein type two (TBX2) is an immediate early gene target in
retinoic-acid-treated B16 murine melanoma cells. Exp Cell Res.
295:281–289. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu WK, Jiang XY and Zhang ZX: Expression
of PSCA, PIWIL1 and TBX2 in endometrial adenocarcinoma. Onkologie.
33:241–245. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu WK, Jiang XY and Zhang ZX: Expression
of PSCA, PIWIL1 and TBX2 and its correlation with HPV16 infection
in formalin-fixed, paraffin-embedded cervical squamous cell
carcinoma specimens. Arch Virol. 157:657–663. 2010. View Article : Google Scholar
|
|
28
|
Han Y, Tu WW, Wen YG, Yan DW, Qiu GQ, Peng
ZH and Zhou CZ: Increased expression of TBX2 is a novel independent
prognostic biomarker of a worse outcome in colorectal cancer
patients after curative surgery and a potential therapeutic target.
Med Oncol. 30:6882013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duo S, Tiao-Dong T, Lei Z, Wei W, Hong-Li
S and Xian-Wei D: Expression and clinical significance of tbx2 in
pancreatic cancer. Asian Pac J Cancer Prev. 10:118–122.
2009.PubMed/NCBI
|
|
30
|
Ludtke TH, Farin HF, Rudat C,
Schuster-Gossler K, Petry M, Barnett P, Christoffels VM and Kispert
A: TBX2 controls lung growth by direct repression of the cell cycle
inhibitor genes Cdkn1a and Cdkn1b. PLoS Genet. 9:e10031892013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bilican B and Goding CR: Cell cycle
regulation of the T-box transcription factor tbx2. Exp Cell Res.
312:2358–2366. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Figueira RC, Gomes LR, Neto JS, Silva FC,
Silva ID and Sogayar MC: Correlation between MMPs and their
inhibitors in breast cancer tumor tissue specimens and in cell
lines with different metastatic potential. BMC Cancer. 9:202009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nutt JE, Durkan GC, Mellon JK and Lunec J:
Matrix metalloproteinases (MMPs) in bladder cancer: The induction
of MMP9 by epidermal growth factor and its detection in urine. BJU
Int. 91:99–104. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bachmeier BE, Nerlich AG, Lichtinghagen R
and Sommerhoff CP: Matrix metalloproteinases (MMPs) in breast
cancer cell lines of different tumorigenicity. Anticancer Res.
21:3821–3828. 2001.PubMed/NCBI
|
|
35
|
Roh MR, Zheng Z, Kim HS, Kwon JE, Jeung
HC, Rha SY and Chung KY: Differential expression patterns of MMPs
and their role in the invasion of epithelial premalignant tumors
and invasive cutaneous squamous cell carcinoma. Exp Mol Pathol.
92:236–242. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jung SA, Yang SK, Kim JS, Shim KN, Im SA,
Myung SJ, Jung HY, Yu CS, Kim JC, Hong WS, et al: The expression of
matrix metalloproteinases (MMPs), tissue inhibitor of
metalloproteinases (TIMPs) and angiogenesis in relation to the
depth of tumor invasion and lymph node metastasis in submucosally
invasive colorectal carcinoma. Korean J Gastroenterol. 45:401–408.
2005.(In Korean). PubMed/NCBI
|
|
37
|
Hotary KB, Yana I, Sabeh F, Li XY,
Holmbeck K, Birkedal-Hansen H, Allen ED, Hiraoka N and Weiss SJ:
Matrix metalloproteinases (MMPs) regulate fibrin-invasive activity
via MT1-MMP-dependent and -independent processes. J Exp Med.
195:295–308. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matos ML, Lapyckyj L, Rosso M, Besso MJ,
Mencucci MV, Briggiler CI, Giustina S, Furlong LI and Vazquez-Levin
MH: Identification of a Novel Human E-cadherin splice variant and
assessment of its effects upon EMT-related events. J Cell Physiol.
232:1368–1386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rogers CD, Saxena A and Bronner ME: Sip1
mediates an E-cadherin-to-N-cadherin switch during cranial neural
crest EMT. J Cell Biol. 203:835–847. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Prince S, Carreira S, Vance KW, Abrahams A
and Goding CR: TBX2 directly represses the expression of the p21
(WAF1) cyclin-dependent kinase inhibitor. Cancer Res. 64:1669–1674.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang Y, Li Z, Zhong Q, Li G, Zhang Y and
Huang Z: Association of TBX2 and P21 expression with
clinicopathological features and survival of laryngeal squamous
cell carcinoma. Int J Clin Exp Med. 7:5394–5402. 2014.PubMed/NCBI
|
|
42
|
Zheng X, Wang Y, Liu B, Liu C, Liu D, Zhu
J, Yang C, Yan J, Liao X, Meng X and Yang H: Bmi-1-shRNA inhibits
the proliferation of lung adenocarcinoma cells by blocking the G1/S
phase through decreasing cyclin D1 and increasing p21/p27 levels.
Nucleic Acid Ther. 24:210–216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bartkova J, Lukas J, Strauss M and Bartek
J: Cyclin D3: Requirement for G1/S transition and high abundance in
quiescent tissues suggest a dual role in proliferation and
differentiation. Oncogene. 17:1027–1037. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen JR, Chatterjee B, Meyer R, Yu JC,
Borke JL, Isales CM, Kirby ML, Lo CW and Bollag RJ: TBX2 represses
expression of Connexin43 in osteoblastic-like cells. Calcif Tissue
Int. 74:561–573. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
D'Costa ZC, Higgins C, Ong CW, Irwin GW,
Boyle D, McArt DG, McCloskey K, Buckley NE, Crawford NT,
Thiagarajan L, et al: TBX2 represses CST6 resulting in uncontrolled
legumain activity to sustain breast cancer proliferation: A novel
cancer-selective target pathway with therapeutic opportunities.
Oncotarget. 5:1609–1620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nandana S, Tripathi M, Duan P, Chu CY,
Mishra R, Liu C, Jin R, Yamashita H, Zayzafoon M, Bhowmick NA, et
al: Bone metastasis of prostate cancer can be therapeutically
targeted at the TBX2-WNT signaling axis. Cancer Res. 77:1331–1344.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ma JW, Zhang Y, Ye JC, Li R, Wen YL, Huang
JX and Zhong XY: Tetrandrine exerts a radiosensitization effect on
human glioma through inhibiting proliferation by attenuating ERK
phosphorylation. Biomol Ther (Seoul). 25:186–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Selvaraj P, Xiao L, Lee C, Murthy SR,
Cawley NX, Lane M, Merchenthaler I, Ahn S and Loh YP: Neurotrophic
Factor-α1: A key wnt-β-catenin dependent anti-proliferation factor
and ERK-Sox9 activated inducer of embryonic neural stem cell
differentiation to astrocytes in neurodevelopment. Stem Cells.
35:557–571. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu X, Tian S, Liu M, Jian L and Zhao L:
Wogonin inhibits the proliferation and invasion and induces the
apoptosis of HepG2 and Bel7402 HCC cells through NFκB/Bcl-2, EGFR
and EGFR downstream ERK/AKT signaling. Int J Mol Med. 38:1250–1256.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hamaoka Y, Negishi M and Katoh H: EphA2 is
a key effector of the MEK/ERK/RSK pathway regulating glioblastoma
cell proliferation. Cell Signal. 28:937–945. 2016. View Article : Google Scholar : PubMed/NCBI
|