Introduction
According to the Chinese Society of Hepatology, the
most common cause of liver fibrosis is chronic viral hepatitis B
(CHB) infection (1). Globally, 30%
of cirrhosis cases are caused by hepatitis B virus (HBV) (2). Cirrhosis is the most advanced stage of
liver fibrosis, which may develop into hepatocellular carcinoma
(HCC). Liver cirrhosis is considered to be the final pathological
stage of fibrosis, and fibrosis is the precursor of cirrhosis
(3).
The early detection of fibrosis would provide
additional time for drug treatment, which may reverse fibrosis. At
present, liver biopsy is considered the ‘gold standard’ for
identifying liver fibrosis. However, this procedure is invasive,
painful and incurs the risk of complications (4); non-invasive diagnostic methods would
resolve these issues. However, current, non-invasive procedures are
costly and not readily accessible in developing countries,
including China (5). There is an
urgent requirement for a precise predictive biomarker for liver
fibrosis.
MicroRNAs (miRNAs) are a series of short noncoding
regulatory RNA molecules that contain 20–25 nucleotides, bind to
the 3′-untranslated region of target miRNAs, and interfere with the
output of a number of protein-coding genes at the
post-translational level by inducing translational arrest, which in
turn decreases or prevents protein synthesis (6,7). It has
been established that they serve critical roles in plant and animal
development, cell lineage differentiation, virus-host interactions,
oncogenesis and immune responses (7). Plant miRNAs introduced through diet
have been identified in the sera and tissues of various animals,
which indicates that exogenous plant miRNAs may modulate the
expression of target genes in mammals. Previously, miRNAs were
identified to act in cross-kingdom manner (i.e., the same miRNAs
were identified to be active in a number of different species and
kingdoms), which is an important function (8). miRNAs affect gene expression primarily
through epigenetic mechanisms and additionally regulate the
epigenetic network in different pathological conditions and
diseases (9). miRNAs are stable in
plasma and may serve as critical diagnostic biomarkers and in
disease therapy development. In addition, the number of studies
examining the association between the expression of miRNAs and
liver fibrosis has increased, which has highlighted the role of
miRNAs in the development of liver fibrosis staging tools.
Patients and methods
Patient selection
All patients were selected based on medical and
pathological data. In addition, all patients were positive for
HBsAg and did not have any other liver diseases, including
cholangiocarcinoma, hepatitis C virus (HCV) infection, or
alcoholic, autoimmune or metabolic liver diseases. Written informed
consent was obtained from each participant, and the study was
approved by the Ethics Review Committee of Beijing YouAn Hospital
(Beijing, China). The study was conducted in accordance with the
principles of the Declaration of Helsinki and the standards of Good
Clinical Practice (as defined by the International Conference on
Harmonization) (10).
Liver histology
Specimens were collected from patients with HBV
through two methods: Ultrasound-guided percutaneous liver biopsy or
wedge biopsy during an open splenectomy and esophagogastric
devascularisation (hereafter referred to as surgery). Meta-analysis
of Histological Data in Viral Hepatitis (METAVIR) scoring was
applied to determine the severity of the fibrosis (11). F0 refers to no fibrosis, and F4
refers to liver cirrhosis. This procedure to determine which
METAVIR stage each specimen was in was performed by two
pathologists.
Noninvasive assessment of liver
fibrosis
The aspartate aminotransferase-to-platelet ratio
index (APRI) and fibrosis-4 (FIB-4) scores were derived from
routine blood markers, and calculated as follows: APRI score,
APRI=[aspartate transaminase (AST) × (upper limit of normal of
AST)] × 100/platelet count (PLT) (12); FIB-4 score, FIB-4=(age of patient ×
AST)/PLT × alanine aminotransferase (ALT1/2) (13). The liver stiffness measurement (LSM)
was calculated using FibroScan® 502 transient elasticity
imager (Echosens, Paris, France). For the LSM, patients were placed
in a supine position and the right arm was lifted to fully expose
the intercostal space. The 7th, 8th and 9th intercostal spaces of
the right axillary line were selected for the continuous detection
of LSM. Data were represented as the median value of ≥10 valid
shots. A reliable result was defined as ≥10 valid shots, a success
rate of ≥60% and an interquartile range <30% of the median LSM
value (14).
Serum ALT and AST activity levels
Blood specimens were collected using a BD Vacutainer
Plus (BD Biosciences, Franklin Lakes, NJ, USA) and separated from
serum by centrifugation at 3,000 × g for 5 min at room temperature.
The separated serum was aliquoted and stored at −80°C. Serum ALT
and AST activity levels were determined on the ADVIA 2400 Clinical
Chemistry System using ALT (cat. no. 74046) and AST (cat. no.
74045) reagents (all Siemens AG, Munich, Germany). These
contemporary methods for measurement of enzyme activity are all
standardized to International Federation of Clinical Chemistry
reference measurement procedures (15,16).
RNA isolation and quantification
In the present study, 7 candidate miRNAs were
selected: Homo sapiens (Hsa)-miR-122-5p, hsa-miR-21-5p,
hsa-miR-146a-5p, hsa-miR-29c-3p, hsa-miR-223, hsa-miR-22-3p and
hsa-miR-381-3p based on a previous study by our group (17). Human plasma samples were collected in
Biomedical Information Center at Beijing YouAn Hospital. Venous
blood samples were collected using a BD Vacutainer Plus and
centrifuged at 700 × g at room temperature for 15 min within 4 h of
collection. The plasma supernatant was immediately recovered and
stored at −80°C. RNA was isolated using QuantoBio Total RNA
Isolation Kit (Beijing QuantoBio Biotechnology Co., Ltd., Beijing,
China), then sample quantity and quality were evaluated with a
Nanodrop™ 8000 spectrophotometer (Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA). All samples successfully passed quality
control [RNA purity: (optical density) OD260/OD280=1.8–2.1; RNA
concentration: ≥1.0 ng/µl] for miRNA isolation and were screened
using a QuantoBio three-step quantitative polymerase chain reaction
(qPCR) method. Quanto EC1 and EC2 (Beijing QuantoBio Biotechnology
Co., Ltd.) were introduced as external controls during RNA
isolation, polyadenylation, cDNA synthesis and qPCR. Reverse
transcription was started using Escherichia coli polyA
polymerase (New England Biolabs, Ipswich, MA, USA) to add a polyA
tail at the 3′ end of RNAs. Then, with RT primer annealing, a
universal tag was attached to the 3′ end of cDNAs during the cDNA
synthesis using a M-MLV reverse transcriptase (Beijing QuantoBio
Biotechnology Co., Ltd.) for 1 h at 42°C. The whole polyA product
was added into the 40 µl RT solution, which included 500 U M-MLV
with 1X RT buffer, 0.5 mM dNTP and 20 µM RT primer (Table I). qPCR was performed with
SYBR® Premix Ex Taq™ II (TaKaRa) and the primers in
Table I. Universal Primers Mix,
which target the universal tag in RT primer, were the down stream
primers used in qPCR for every miRNA. Upstream primer were specific
to the miRNA. The thermocycling conditions were as follows: Initial
denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for
30 sec and 60°C for 1 min. The dissociation stage was as follows:
95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. The data were
normalized using the external controls Quanto EC1 and Quanto EC2
and analysed with a RealTime StatMiner (Integromics, Granada,
Spain). The relative expression level of miRNA was calculated using
the 2−ΔΔCq method (18).
 | Table I.Primers used in the quantitative
polymerase chain reaction analysis. |
Table I.
Primers used in the quantitative
polymerase chain reaction analysis.
| Name | Sequence
(5′-3′) | Target miRNA |
|---|
| RT |
AAGCAGTGGTATCAACGCAGAGTGCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT | Universal |
| FP1 |
CAAGGGCAAGCTCTCTG | hsa-miR-381-3P |
| FP2 |
GAGTTCTTCAGTGGCAAGC | hsa-miR-22-3p |
| FP3 |
GAGAACTGAATTCCATGGGT |
hsa-miR-146a-5p |
| FP4 |
GAACGCCATTATCACACT | hsa-miR-122-5p |
| FP5 |
GCTTATCAGACTGATGTTG | hsa-miR-21-5p |
| FP6 |
GCACCATTTGAAATCGGT | hsa-miR-29c-3p |
| FP7 |
TGTCAGTTTGTCAAATACC | hsa-miR-223 |
| FP-EC1 |
CAACCTCCTAGAAAGAGTA | cel-miR-67-3p |
| FP-EC2 |
TGAGCAACGCGAACAAATCA | cel-miR-356a |
| UPM-L |
CTCACACGACTCACGACAGGGCAAGCAGTGGTATCAACGCAGAGTG | Universal |
| UPM-S |
CTCACACGACTCACGACAGGGC | Universal |
HBV load measurements
Human blood samples were collected in Biomedical
Information Center at Beijing YouAn Hospital. Venous blood samples
were collected using a BD Vacutainer Plus and centrifuged at 1,000
× g at room temperature for 20 min within 24 h of collection. DNA
was isolated from plasma using the QIAamp DNA Mini kit (Qiagen,
Inc., Valencia, CA, USA) and stored at −20°C prior to use. The qPCR
reactions were performed using the CFX384 Touch Real-Time PCR
Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
according to the manufacturer's instructions. The 20 µl qPCR
mixture comprised of 250 nmol/l 2X SsoAdvanced Universal Probes
Supermix (Bio-Rad Laboratories, Inc.), 900 nmol/l HBV-specific
sense primers (5′-CTCTCTTTACGCGGTCTC-3′), HBV-specific antisense
primers (5′-GTCGTTGACATTGCTGAG-3′), 250 nmol/l HBV probe
(5′-CCGTCTGTGCCTTCTCATCTGC-3′) and 4 µl DNA sample. The
thermocycling conditions were as follows: Initial denaturation at
95°C for 10 min, followed by 45 cycles of denaturation at 94°C for
30 sec, annealing and extension step at 57°C for 60 sec. The final
extension step at 98°C for 10 min, then continued at 4°C
indefinitely. Cq values were generated and linear regression
analyses of the calibration curves were performed by CFX™ Manager
Software 3.1 (Bio-Rad Laboratories, Inc.). The data generated by
the qPCR was normalized according to the guidelines of the World
Health Organization International Standard for Hepatitis B Virus
DNA Nucleic Acid Amplification Techniques (19). HBV DNA levels are expressed in the
units, IU/ml.
Statistical analysis
All data were analysed using SPSS 19.0 statistical
software (IBM Corp., Armonk, NY, USA) and GraphPad Prism 5 software
(GraphPad Software, Inc., La Jolla, CA, USA). Data are expressed as
the mean ± standard deviation (SD), number (percentage) or median
(25–75% percentiles) when appropriate. Categorical data were
compared by χ2 or Fischer's exact test when appropriate.
Clinical data from the five independent fibrosis groups (F0-F4)
were normally distributed and analysed using a one-way analysis of
variance followed by a Least Significant Difference test. HBV load
were measured using qPCR and log-transformed for parametric
statistical tests. miRNA expression levels from independent samples
from two fibrosis groups (≥F2 vs. F0-F1; ≥F3 vs. F0-F2; F4 vs.
F0-F3) were compared by Mann-Whitney U-test, and multiple groups,
including the 5 fibrosis subgroups, were compared by Kruskal-Wallis
test and a Dunn-Bonferroni test for post-hoc comparisons. The
diagnostic accuracy of plasma miRNAs was examined using the area
under the curve (AUC) of corresponding receiver operating
characteristic (ROC) curves. Univariate and multivariate logistic
regression analyses were performed to identify predictor miRNAs
associated with the risk of ≥F2, ≥F3 or F4. The predicted
probability according to the panel was used as a diagnostic marker
to construct the ROC curve. Spearman correlation analysis was used
to examine the association between clinical parameters including
the LSM, FIB-4 and APRI scores, serum ALT and AST activity levels,
hepatitis B virus (HBV) expression, and miRNA expression. MedCalc
(version 12.7; MedCalc Software bvba, Ostend, Belgium) was used to
calculate and compare the AUCs and assess the diagnostic values of
the miRNAs of interest. P<0.05 was considered to indicate a
statistically significant difference.
Results
Description and clinicopathologic
features of patients
A total of 92 patients (26 females and 66 males),
who provided liver tissue and plasma specimens in Beijing YouAn
Hospital affiliated with Capital Medical University from June 2014
to October 2016, were enrolled in the present study. The mean age
of these patients was 40.58±11.43 years (range, 16–60 years). The
mean body mass index (BMI) was 22.87±3.04 kg/m2, and the
Model for end-stage liver disease (MELD) index was 5.25±4.40.
Patients were divided into five subgroups by liver histology
result: F0; F1; F2; F3; and F4. Among these patients, 11 patients
exhibited F0 stage, 16 patients exhibited F1 stage, 12 patients
exhibited F2 stage, 13 patients exhibited F3 stage, and 40 patients
exhibited F4 stage disease. The characteristics of the participants
are presented in Table II. LSM,
FIB-4, serum ALT, and HBV DNA levels were significantly different
among the subgroups (P<0.05). There was no significant
difference in the distribution of age, sex, MELD, Child-Pugh score
(20), APRI, BMI or AST among the
five groups (P>0.05).
 | Table II.Characteristics of participants by
fibrosis stage. |
Table II.
Characteristics of participants by
fibrosis stage.
|
| Fibrosis stage |
|
|---|
|
|
|
|
|---|
|
Characteristics | F0 | F1 | F2 | F3 | F4 | P-value |
|---|
| No. of cases
(%) | 11 (12) | 16 (17.4) | 12 (13) | 13 (14.1) | 40 (43.5) |
|
| Sex |
|
|
|
|
| 0.481 |
| Male, n
(%) | 9 (9.8) | 13 (14.1) | 8 (8.7) | 7 (7.6) | 29 (31.5) |
|
| Female,
n (%) | 2 (2.2) | 3 (3.3) | 4 (4.3) | 6 (6.5) | 11 (12.0) |
|
| Age, years | 34.7±12.8 | 34.8±13.0 | 40.2±10.1 | 38.0±10.2 |
45.5±9.3a–d | 0.192 |
| Child-Pugh
score | 5.3±0.7 | 5.6±1.4 | 5.5±1.0 | 5.8±0.6 |
6.1±1.2a | 0.228 |
| MELD | 3.5±5.0 | 4.5±5.6 | 3.7±2.9 | 4.4±3.5 |
6.8±4.0a,c | 0.055 |
| BMI | 22.8±4.0 | 24.0±2.4 | 23.8±3.8 | 22.2±3.9 | 22.7±2.7 | 0.783 |
| LSM, KPa | 5.9±2.0 | 7.8±4.4 | 9.4±3.5 | 14.0±9.1 |
22.8±14.1a–d | <0.0001 |
| HBV DNA, log
IU/ml | 5.2±2.5 | 4.9±2.4 | 3.9±2.0 |
4.5±2.2a |
3.0±1.6a–d | 0.003 |
| ALT, U/l | 32.1±16.8 |
185.7±222.0a,d,e | 105.7±145.3 | 83.62±89.0 | 37.7±34.3 | 0.001 |
| AST, U/l | 26.6±13.6 | 101.1±107.5 | 50.3±46.6 |
158.1±379.2a,e | 42.9±35.9 | 0.121 |
| FIB-4 | 0.8±0.4 | 1.8±1.6 | 3.0±4.6 | 3.0±2.7 |
6.1±6.2a–d | 0.002 |
| APRI |
0.3±0.1d | 1.6±1.6 | 1.0±0.9 | 3.4±7.6 | 1.9±1.9 | 0.192 |
| Method of obtaining
liver tissue samples |
|
|
|
|
| <0.0001 |
| Liver
biopsy, n | 11 | 15 | 10 | 9 | 4 |
|
| OSED,
n | 0 | 1 | 2 | 4 | 36 |
|
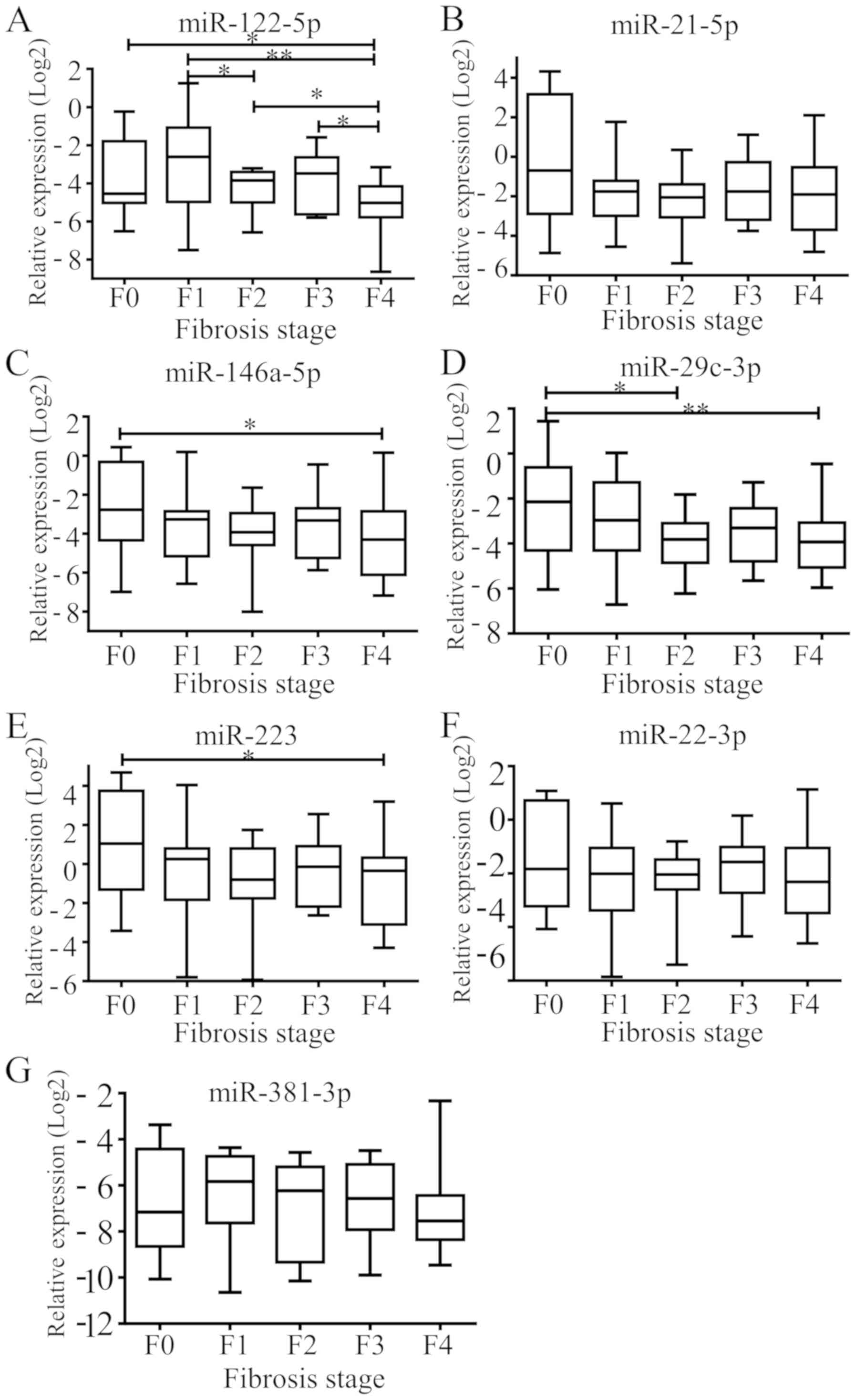
Relative expression of the 7 selected
miRNAs at different fibrosis stages
Plasma miRNAs were analysed using the Cq value by
qPCR. The threshold levels were set as follows: miRNA, Cq <35;
detection rate >75%. Then, these values were transformed using
the 2−ΔΔCq method (18).
Kruskal-Wallis and a Dunn-Bonferroni test for post-hoc comparisons
was used to compare 5 subgroups (Fig.
1). It was identified that hsa-miR-122 exhibited a
significantly different expression between F4 and F3, F2, F1 and F0
stages (P=0.012, 0.045, 0.001 and 0.028, respectively; Fig. 1A) and between F2 and F1 stages
(P=0.032; Fig. 1A). hsa-miR-21-5p
expression was not identified to be significant between groups
(Fig. 1B). hsa-miR-146a-5p exhibited
significantly different expression levels between F4 and F0 stages
(P=0.037; Fig. 1C), and
hsa-miR-29c-3p exhibited significantly different expression levels
between F0 and F2 stages (P=0.034), and between F0 and F4 stages
(P=0.007; Fig. 1D). hsa-miR-223
exhibited significantly different expression between F4 and F0
stages (P=0.017; Fig. 1E).
hsa-miR-22-3p and hsa-miR-381-3p expression was not identified to
be significant between groups (Fig. 1F
and G).
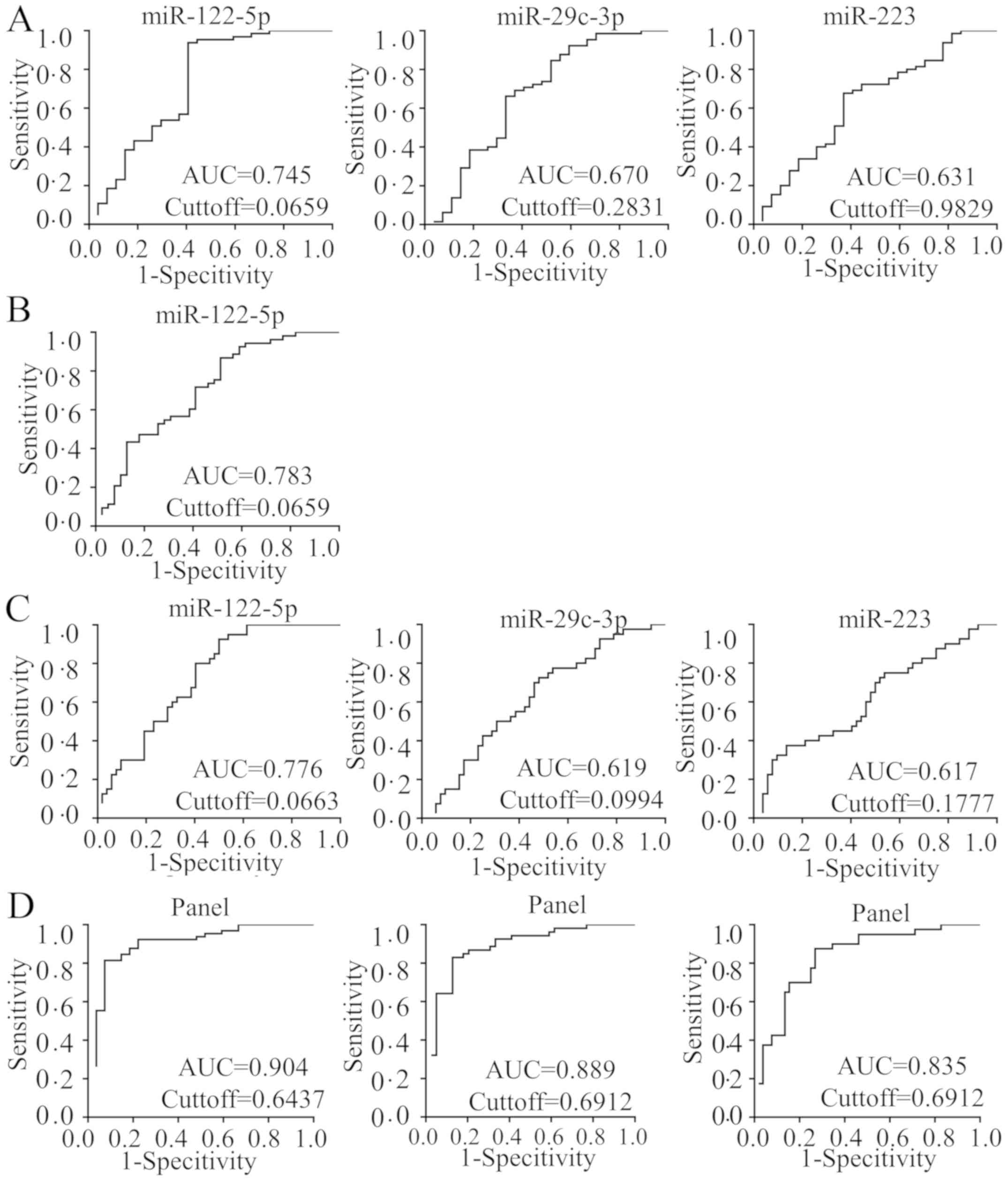
Diagnostic potential of individual
plasma miRNAs
To evaluate whether the 7 selected plasma miRNAs
were able to be used as potential diagnostic markers for liver
fibrosis, ROC curves were generated to evaluate their performance.
The ROC curves revealed that several plasma miRNAs significantly
identified ≥F2, ≥F3 and F4 stages (P<0.05; Fig. 2). hsa-miR-122-5p, hsa-miR-223 and
hsa-miR-29c-3p identified patients with ≥F2 stage with AUC=0.745
(0.643–0.830), 0.631 (0.524–0.730) and 0.670 (0.564–0.765),
respectively. hsa-miR-122-5p identified patients with ≥F3 stage
disease with AUC=0.783 (0.685–0.862). hsa-miR-122-5p, hsa-miR-223
and hsa-miR-29c-3p were able to significantly identify patients
with cirrhosis (F4 stage) with AUC=0.776 (0.677–0.856), 0.617
(0.510–0.717), and 0.619 (0.512–0.719), respectively. The 95%
confidence intervals (CI), sensitivities, specificities, positive
predictive values (PPV) and negative predictive values (NPV) for
the miRNAs that identified patients with ≥F2, ≥F3, and F4 stage
disease are summarized in Table
III.
 | Table III.Performance of individual plasma
miRNAs. |
Table III.
Performance of individual plasma
miRNAs.
| Disease stage vs.
control | miRNA | Area under the
curve | 95% Confidence
interval | Cut-off value | Sensitivity, % | Specificity, % | Positive predictive
value, % | Negative predictive
value, % |
|---|
| ≥F2 | miR-122-5p | 0.745 | 0.643–0.830 | 0.066 | 72.3 | 74.1 | 87.0 | 52.6 |
|
| miR-223 | 0.631 | 0.524–0.730 | 0.983 | 67.7 | 63.0 | 81.5 | 44.7 |
|
| miR-29c-3p | 0.670 | 0.564–0.765 | 0.283 | 92.3 | 40.7 | 78.9 | 68.8 |
| ≥F3 | miR-122-5p | 0.783 | 0.685–0.862 | 0.066 | 79.2 | 69.2 | 77.8 | 71.1 |
| F4 | miR-122-5p | 0.776 | 0.677–0.856 | 0.066 | 90.0 | 61.5 | 64.3 | 88.9 |
|
| miR-223 | 0.617 | 0.510–0.717 | 0.178 | 37.5 | 86.5 | 68.2 | 64.3 |
|
| miR-29c-3p | 0.619 | 0.512–0.719 | 0.099 | 72.5 | 51.9 | 53.7 | 71.1 |
Establishment of a predictive and
non-invasive diagnostic panel with selected plasma miRNAs
A logistic regression model was applied to predict
the probability of diagnosing different fibrosis statuses using
miRNA diagnostic markers. The panel, which included these 7 miRNAs,
was identified to be a significant predictor for the risk of ≥F3
diagnosis and was used to construct the ROC curve. The predicted
probability of diagnosing ≥F3 stage disease was calculated from the
logit model, and the ROC curve was drawn using the following
equation: Logit (P)=0.912–5.696 × hsa-miR-122-5p+0.060 ×
hsa-miR-21-5p+6.444 × hsa-miR-146a-5p-6.234 × hsa-miR-29c-3p-1.955
× hsa-miR-223+10.789 × hsa-miR-22-3p-45.954 × hsa-miR-381-3p.
Diagnostic performance of the
predictive panel
The ROC analysis demonstrated that the complete
panel exhibited increased accuracy in discriminating between severe
fibrosis (≥F3) and patients with no/mild/moderate (F0-F2) fibrosis
compared with individual miRNAs, with AUC=0.889 (95% CI,
0.806–0.945; P<0.0001), sensitivity=83.02%, specificity=87.18%,
PPV=89.8% and NPV=79.1%. In addition, it also exhibited improved
performance in discriminating between patients with significant
fibrosis (≥F2) from patients with no/mild fibrosis (F0-F1) compared
with individual miRNAs, with AUC=0.904 (95% CI, 0.825–0.956;
P<0.0001), sensitivity=81.54%, specificity=92.59%, PPV=96.4% and
NPV=67.6%. The model was also able to significantly identify
patients with F4 disease from patients at other fibrosis stages
with AUC=0.835 (95% CI, 0.743–0.904; P<0.0001),
sensitivity=87.50%; specificity=73.08%, PPV=71.4%, and NPV
(certainty to exclude F4)=88.4% (Fig.
2D).
Correlation of plasma miRNA expression
with fibrosis, LSM, FIB-4, APRI and serum biochemical values
It was identified that LSM, FIB-4 and APRI exhibited
positive correlations with fibrosis stage (r=0.701, P<0.001;
r=0.661, P<0.001; and r=0.416, P<0.001, respectively;
Fig. 3A), and hsa-miR-122-5p,
hsa-miR-146a, hsa-miR-29c and hsa-miR-223 were identified to be
positively correlated with fibrosis stage (r=0.409, P<0.001;
r=0.212, P=0.043; r=0.252, P=0.015; and r=0.232, P=0.026,
respectively) (Fig. 3B). A total of
6 miRNAs (hsa-miR-122-5p, hsa-miR-146a-5p, hsa-miR-29c-3p,
hsa-miR-223, hsa-miR-22-3p and hsa-miR-381-3p) exhibited a positive
correlation with LSM (r=0.350, P=0.003; r=0.290, P=0.014; r=0.342,
P=0.004; r=0.274, P=0.021; r=0.260, P=0.028; and r=0.304, P=0.010,
respectively; Fig. 3C). A total of 6
miRNAs (hsa-miR-122-5p, hsa-miR-21-5p, hsa-miR-146a-5p,
hsa-miR-29c-3p, hsa-miR-223 and hsa-miR-381-3p) were positively
correlated with FIB-4 (r=0.419, P<0.001; r=0.272, P=0.009;
r=0.376, P<0.001; r=0.360, P<0.001; r=0.409, P<0.001; and
r=0.238, P=0.023, respectively; Fig.
4A) and 5 miRNAs (hsa-miR-21-5p, hsa-miR-146a-5p,
hsa-miR-29c-3p, hsa-miR-223 and hsa-miR-22-3p) were positively
correlated with APRI (r=0.277, P=0.008; r=0.363, P<0.001;
r=0.225, P=0.031; r=0.414, P<0.001; and r=0.212, P=0.043,
respectively; Fig. 4B). A total of 3
miRNAs (hsa-miR-122-5p, hsa-miR-29c-3p and hsa-miR-381-3p) were
negatively correlated with HBV load (r=−0.514, P=0.000; r=−0.332,
P=0.001; and r=−0.437, P<0.001, respectively; Fig. 4C). hsa-miR-122-5p (r=−0.458,
P<0.001) and hsa-miR-381-3p (r=−0.242, P=0.021) were negatively
correlated with ALT activity (Fig.
4D) and AST levels (hsa-miR-122-5p, r=−0.328, P=0.001; and
hsa-miR-381-3p, r=−0.239, P=0.022; Fig.
4E).
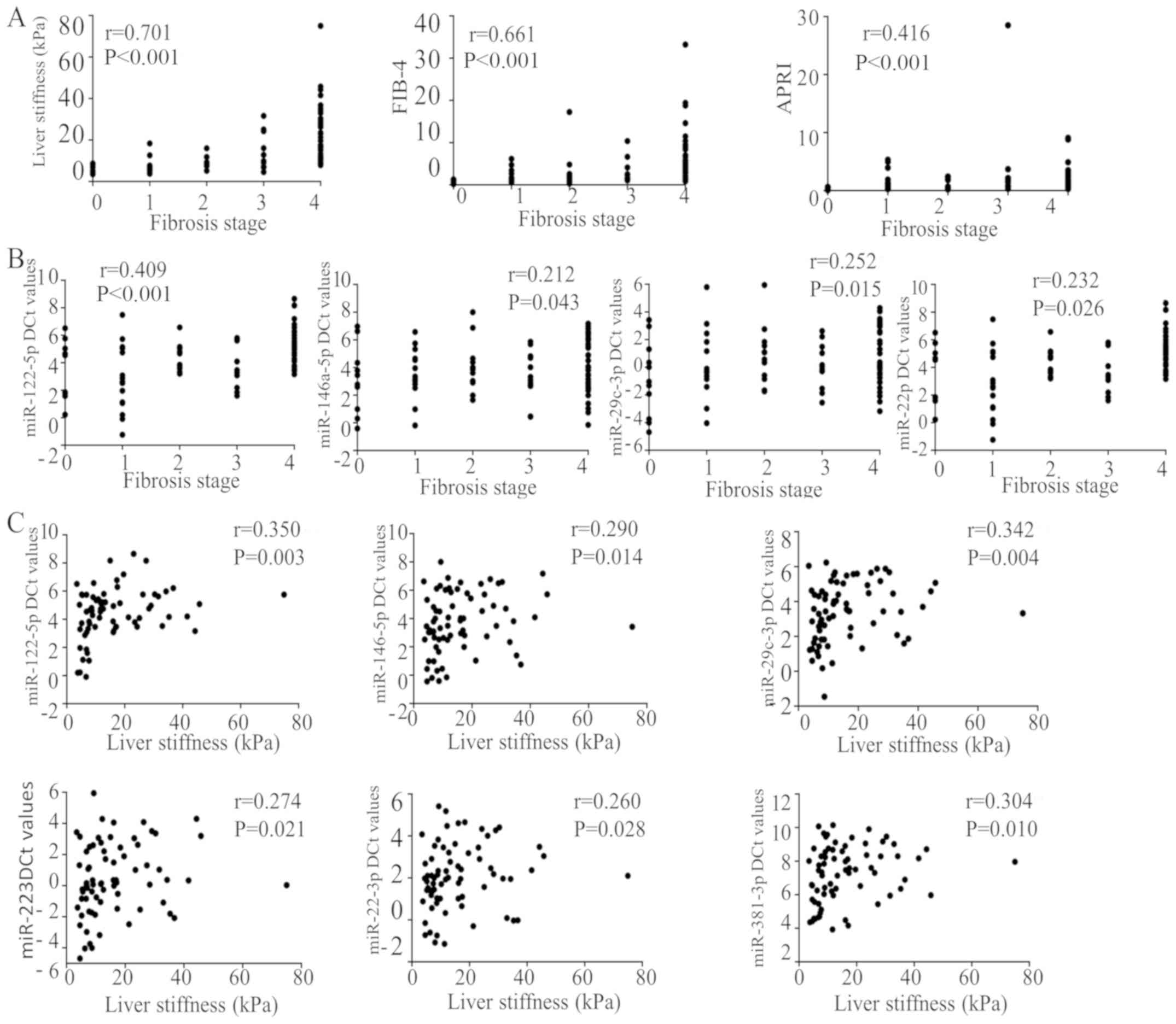 | Figure 3.Correlations between plasma miRNA
expression and LSM and fibrosis stage. All miRs are human (i.e.,
hsa-miR). (A) LSM, FIB-4 and APRI exhibited positive correlations
with fibrosis stage. (B) miR-122-5p, miR-146a miR-29c and miR-223
were identified to be positively correlated with fibrosis stage.
(C) A total of 6 miRNAs (miR-122-5p, miR-146a-5p, miR-29c-3p,
miR-223, miR-22-3p and miR-381-3p) demonstrated positive
correlations with LSM. miR, microRNA; FIB-4, fibrosis-4; APRI,
aspartate transaminase to platelet ratio index; LSM, Liver
stiffness measurement. |
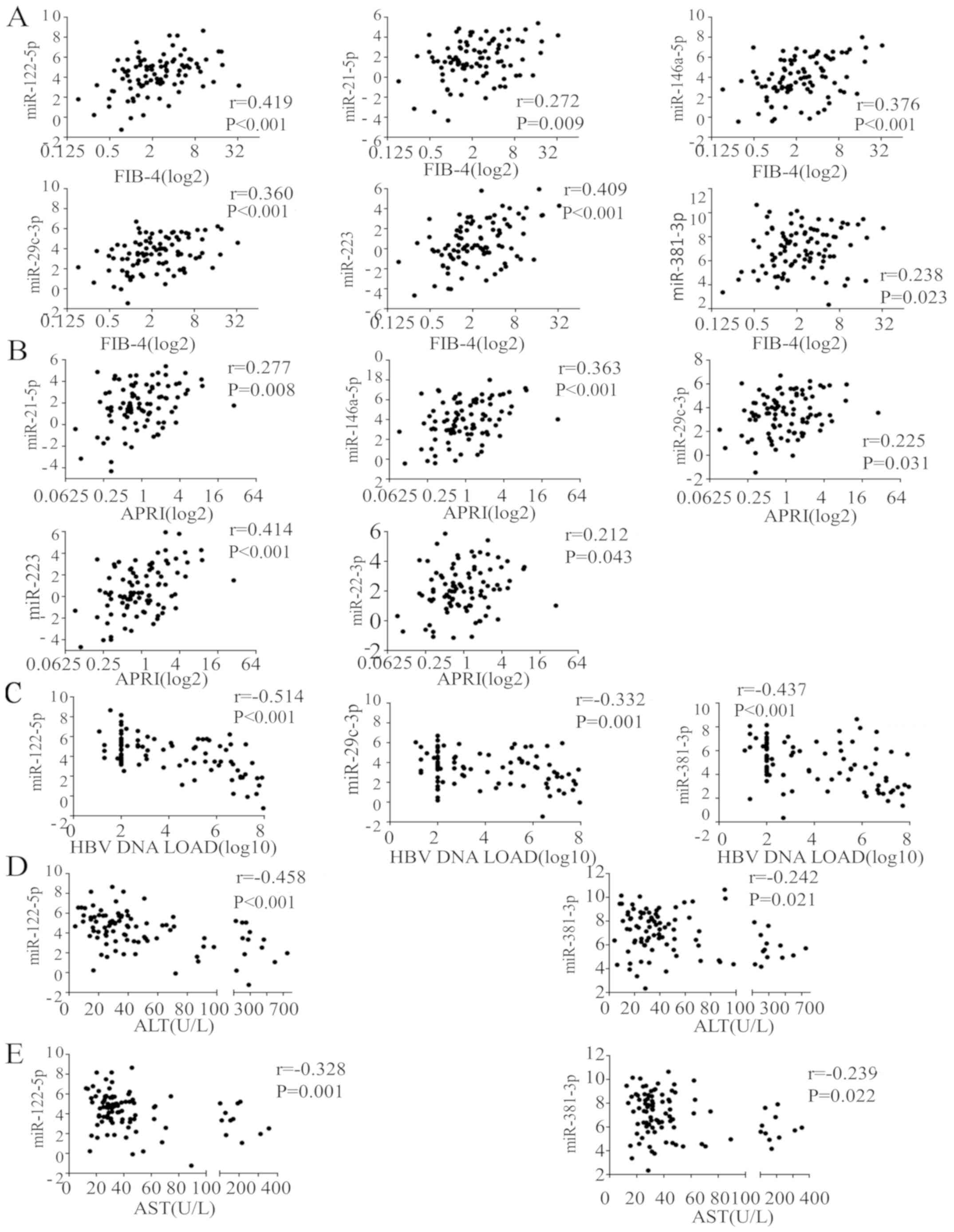 | Figure 4.Correlations between plasma miRNA
expression levels and hepatic function and viral load. All miRs are
human (i.e., hsa-miR). (A) A total of 6 miRNAs (miR-122-5p,
miR-21-5p, miR-146a-5p, miR-29c-3p, miR-223 and miR-381-3p) were
positively correlated with FIB-4. (B) A total of 5 miRNAs
(miR-21-5p, miR-146a-5p, miR-29c-3p, miR-223 and miR-22-3p) were
positively correlated with APRI. (C) A total of 3 miRNAs
(miR-122-5p, miR-29c-3p, and miR-381-3p) were negatively correlated
with HBV DNA load levels. miR-122-5p and miR-381-3p were negatively
correlated with (D) ALT and (E) AST activity levels. miR, microRNA;
FIB-4, fibrosis-4; HBV, hepatitis B virus; ALT, alanine
aminotransferase; AST, aspartate transaminase; APRI, AST to
platelet ratio index. |
Discussion
The pathophysiological mechanism of hepatic fibrosis
is a disruption between the production and dissolution of the
extracellular matrix triggered by chronic and inflammatory injury
(21). There are several other
factors that cause liver disease, including viruses, bacteria,
parasites, other infectious factors and non-infectious factors,
including alcohol, drugs, autoimmune disease, metabolic syndromes,
genetic defects and congenital dysplasia. HBV is the most common
cause of liver fibrosis in China (1). A previous study hypothesized that the
clinical pathway of the majority of HBV-associated HCC follows four
stages: Healthy; hepatitis; cirrhosis; and HCC (22). With the gradual improved
understanding of liver fibrosis and the prevalence of treatment
options, we hypothesized that the hepatic fibrosis stage should be
included in this classification of clinicopathological progression.
The advantage is that the natural course of HBV infection may be
more accurately studied, but it does require sufficient
pathological examination or LSM, APRI and FIB-4 analysis. It was
demonstrated that altered miRNA patterns following chronic liver
disease markedly affect the progression of fibrosis by potentially
targeting the expression of extracellular matrix proteins and the
synthesis of mediators of profibrogenic pathways (23). The pathological changes in liver
fibrosis, which may be caused by different pathogens, may have a
combination of commonalities and individual characteristics.
Therefore, HBV-associated liver fibrosis patients were
intentionally enrolled in the present study to remove the potential
interference of other factors. Cirrhosis is the most advanced stage
of fibrosis, which may develop into HCC. Therefore, the diagnosis
and staging of HBV-associated liver fibrosis in patients is
important and may guide the appropriate timing of antiviral
therapy. In the present study, it was identified that the miRNA
expression data of several patients with clinically-diagnosed CHB
and cirrhosis were not consistent with their pathological results,
therefore patients who were diagnosed with liver cirrhosis did not
have pathological results the verified the diagnosis. As the liver
has a strong compensatory capacity (24); consequently certain patients with
liver cirrhosis may have mild or asymptomatic clinical
manifestations. Concomitantly, specific cases exhibited
clinically-diagnosed cirrhosis with minor pathological changes. In
the present study, the pathological results of 7 cases (16.2%) (1
case of F1, 2 cases of F2, and 4 cases of F3) did not reach an
early diagnosis of liver cirrhosis (F4). The reason may be that the
wedge biopsies during surgery were not representative of the actual
clinical state. Although liver biopsy remains the ‘gold standard’
for the diagnosis of liver fibrosis, it has limitations; its
invasive nature and sampling errors (i.e., the most severely
fibrotic may not be obtained) have not yet been overcome.
Therefore, the present study aimed to investigate the potential of
circulating miRNAs as a method of diagnosis.
Circulating miRNAs, as diagnostic markers for
different stages of hepatic fibrosis, have been extensively studied
in previous years (6,25–27).
miRNAs are derived from blood or body fluids, making them easily
accessible as potential biomarkers for the evaluation of hepatic
fibrosis. Lee et al (28)
identified the first miRNA, termed lin-4, from Caenorhabditis
elegans in 1993. Previous evidence has suggested that there is
a complex association between miRNAs and HBV genes (29–31). The
mechanism of HBV infection in regulating miRNAs may be through
either RNA interference or the inhibition of translational
initiation and elongation (23,32–34). In
addition, deregulated expression of miRNAs may be present for a
long period prior to the onset of disease (35).
In the present study, it was identified that
hsa-miR-122, has-miR-146a-5p, hsa-miR-29c-3p and hsa-miR-223
exhibited significantly different expression levels between the F4
and F0 stage, which suggests that these 4 miRNAs may participate in
the progression of liver cirrhosis from CHB infection, and that
they may be used as diagnostic biomarkers. Furthermore, the
expression levels of hsa-miR-122 were significantly different
between F4 and F3, F2 and F1 disease stages (P=0.012, 0.045 and
0.001, respectively), demonstrating that it may be used as a
precise fibrosis staging biomarker. These data are consistent with
certain studies investigating HCV-associated diseases (36,37),
non-alcoholic fatty liver disease (NAFLD) (38,39) and
HCC. hsa-miR-122 is specifically and most abundantly expressed in
the liver, accounting for >70% of the total miRNAs in
hepatocytes (40), which may reflect
the fact that the accumulation of fibrosis may be not only
associated with the expression level of hsa-miR-122, but also with
the duration of HBV infection. The downregulation of miR-122
between F2 from F1 (P=0.032), which had been previously supported
by studies involving HCV and NAFLD (36,37,39),
maybe a sensitive signal of hepatic injury. The mechanism may
involve the positive regulation of the accumulation of cholesterol
and triglycerides and the metabolism of fatty acids (39). hsa-miR-122 has been identified to
suppress the proliferation of hematopoietic stem cells (HSCs),
resulting in the decreased maturation of collagen by downregulating
the expression of prolyl 4-hydroxylase subunit alpha 1, a key
enzyme in collagen maturation (41).
In addition, the expression level of plasma hsa-miR-122 may predict
the survival of patients with liver cirrhosis (42). Following liver injury and fibrosis,
levels of plasma hsa-miR-122 were markedly decreased depending on
the severity of fibrosis (43).
These data indicate that hsa-miR-122 has other unknown functions in
the viral life cycle. Previous data have revealed that hsa-miR-146a
promotes viral replication and deregulates the metabolic pathways
associated with liver disease (32).
It has also been demonstrated that hsa-miR-146a is downregulated in
liver fibrotic tissues and suppresses the activation and
proliferation of HSCs in the progression of fibrosis (44,45). The
mechanism regulates transforming growth factor-β signalling by
targeting mothers against decapentaplegic homolog 4 (46). hsa-miR-29 is also involved in the
pathogenesis of fibrotic diseases, including liver cirrhosis: It
was identified that hsa-miR-29 levels were significantly decreased
in advanced liver cirrhosis compared with healthy controls
(33). In addition, a low expression
level of hsa-miR-29 was identified to be correlated with the
severity of hepatic fibrosis (23).
The deregulation of hsa-miR-223 was described in HBV-positive HCC
and HBV/HCV-positive cirrhosis (47). In the present study, the expression
level of hsa-miR-223 was detected to be downregulated in early
hepatic cirrhosis (F4) compared with CHB infection (F0). We
hypothesized that deregulated levels of hsa-miR-223 are present
throughout the entire process of HBV infection, and are associated
with infection, inflammation and cancer, which is supported by
previous studies (48,49).
There have been a number of studies examining
potential biomarkers for CHB infection, liver cirrhosis and HCC
(17,50). However, miRNAs for liver fibrosis
staging have not been well-studied. In the METAVIR fibrosis-scoring
system, F0 indicates no fibrosis, and F4 indicates liver cirrhosis.
Within this system, there are several different classifications of
liver fibrosis stages. For example, F0/1 represents no significant
liver fibrosis, F2/3 represents progressive fibrosis and F4
represents cirrhosis. The present study explored the diagnostic
performance of 7 fibrosis-associated miRNAs in identifying no/mild
fibrosis (F0-F1), significant fibrosis (≥F2), severe fibrosis (≥F3)
and cirrhosis (F4) based on the METAVIR fibrosis-scoring system.
hsa-miR-122-5p, hsa-miR-223 and hsa-miR-29c-3p identified ≥F2
(AUC=0.745, 0.631 and 0.670, respectively) and F4 (AUC=0.776, 0.617
and 0.619, respectively). hsa-miR-223 and hsa-miR-29c-3p exhibited
good predictive potential in the diagnosis of significant fibrosis
or cirrhosis. In addition, hsa-miR-122-5p was also able to identify
patients with ≥F3 disease with AUC=0.783. As a staging biomarker,
the authors of the current study hypothesis that hsa-miR-122-5p may
participate in the whole process of liver fibrosis and may be used
to determine fibrosis status.
Although ROC analysis revealed that
differentially-expressed plasma miRNAs were able to significantly
identify ≥F2, ≥F3, and F4 disease (P<0.05), the performance of
individual plasma miRNAs was not as satisfactory (AUC=0.617–0.783).
Multivariate logistic analysis revealed that the panel combining
the 7 miRNAs exhibited a high diagnostic accuracy for ≥F3
(AUC=0.889) and F4 (AUC=0.904) disease stages. However, the panel
had modest results in identifying ≥F2 disease (AUC=0.835). The
miRNA panel significantly improved the diagnostic efficacy of
fibrosis compared with the diagnostic efficacy of a single miRNA,
which may provide a strategy for using miRNAs to study the staging
of HBV-associated liver fibrosis. In fact, a large number of
studies have used miRNA panels for the diagnosis of HCC (22,50).
Additional studies are required to confirm this panel and validate
its significance in liver fibrosis of other aetiologies.
In the present study, the correlation between plasma
miRNAs levels and disease progression in patients with CHB
infection was also investigated. Plasma hsa-miR-122-5p,
hsa-miR-146a, hsa-miR-29c and hsa-miR-223 levels were identified to
be positively correlated with fibrosis stage, suggesting that these
4 miRNAs were involved in the progression of HBV-associated liver
fibrosis. hsa-miR-122-5p and hsa-miR-381-3p were negatively
correlated with ALT, AST and HBV viral DNA load. The data are
presented as ΔCq values and not as ΔΔCq values. These results
indicated that increased levels of hsa-miR-122-5p and
hsa-miR-381-3p are accompanied by HBV replication and high activity
levels of ALT and AST in cultured cells. Plasma hsa-miR-122 levels
were increased with the severity of liver injury, indicating a good
correlation with aminotransferase levels and necroinflammation
(51,52). A correlation between hsa-miR-21
expression and viral load, fibrosis and plasma liver transaminase
levels, as described in the literature (36), was not identified in the present
study. In the present study, it was not determined whether
hsa-miR-21 induction was a causative factor in fibrosis. However, a
previous study revealed that increased hsa-miR-21 levels were
associated with fibroblast dysfunction and fibrosis outcome
(53). In a number of other organs,
hsa-miR-21 is one of the most important miRNAs that is abundantly
expressed following fibrosis induction (54–56).
In the present study, three non-invasive parameters,
LSM, APRI and FIB-4, were thoroughly examined. Although they were
identified to be positively associated with liver fibrosis stage,
they lacked inter-stage specificity and were unable to detect the
early stages of fibrosis. The majority of the miRNAs included in
the present study demonstrated significant positive correlations
with these non-invasive techniques, indicating that there is an
established association between miRNAs and these non-invasive
examination markers.
In conclusion, the present study revealed that
plasma miRNAs are potential circulating markers for fibrosis and
early cirrhosis diagnosis. An optimized miRNAs panel may improve
diagnostic accuracy and decrease the necessity for clinical liver
punctures.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Grand
Program on Key Infectious Diseases (grant no. 2015ZX10004801), the
National Major Scientific and Technological Special Project during
the Thirteenth Five-year Plan Period Beijing (grant no.
2017ZX10203205-006-003), and the Beijing Health System High-level
Health Technology Talents Training Program (grant no.
2013-3-074).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
D-DL and NL conceived and designed the study. T-ZW,
B-XJ and X-YS performed the experiments. T-ZW wrote the paper.
T-ZW, D-DL and NL reviewed and edited the manuscript. All authors
read and approved the manuscript and agree to be accountable for
all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
Written informed consent was obtained from each
participant, and the study was approved by the Ethics Review
Committee of Beijing YouAn Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Qin G, Shao JG, Zhu YC, Xu AD, Yao JH,
Wang XL, Qian YK, Wang HY, Shen Y, Lu P and Wang LJ:
Population-representative incidence of acute-on-chronic liver
failure: A prospective cross-sectional study. J Clin Gastroenterol.
50:670–675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Perz JF, Armstrong GL, Farrington LA,
Hutin YJ and Bell BP: The contributions of hepatitis B virus and
hepatitis C virus infections to cirrhosis and primary liver cancer
worldwide. J Hepatol. 45:529–538. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou WC, Zhang QB and Qiao L: Pathogenesis
of liver cirrhosis. World J Gastroenterol. 20:7312–7324. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sebastiani G: Non-invasive assessment of
liver fibrosis in chronic liver diseases: Implementation in
clinical practice and decisional algorithms. World J Gastroenterol.
15:2190–2203. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen SL, Zheng MH, Shi KQ, Yang T and Chen
YP: A new strategy for treatment of liver fibrosis: Letting
MicroRNAs do the job. BioDrugs. 27:25–34. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang
Y, Li J, Bian Z, Liang X, Cai X, et al: Exogenous plant MIR168a
specifically targets mammalian LDLRAP1: Evidence of cross-kingdom
regulation by microRNA. Cell Res. 22:107–126. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mentz RJ, Hernandez AF, Berdan LG, Rorick
T, O'Brien EC, Ibarra JC, Curtis LH and Peterson ED: Good clinical
practice guidance and pragmatic clinical trials: Balancing the best
of both worlds. Circulation. 133:872–880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Poynard T, Bedossa P and Opolon P: Natural
history of liver fibrosis progression in patients with chronic
hepatitis C. The OBSVIRC METAVIR, CLINIVIR, andDOSVIRC groups.
Lancet. 349:825–832. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trang T, Petersen JR and Snyder N:
Non-invasive markers of hepatic fibrosis in patients co-infected
with HCV and HIV: Comparison of the APRI and FIB-4 index. Clin Chim
Acta. 397:51–54. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vallet-Pichard A, Mallet V, Nalpas B,
Verkarre V, Nalpas A, Dhalluin-Venier V, Fontaine H and Pol S:
FIB-4: An inexpensive and accurate marker of fibrosis in HCV
infection. Comparison with liver biopsy and fibrotest. Hepatology.
46:32–36. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Castera L, Forns X and Alberti A:
Non-invasive evaluation of liver fibrosis using transient
elastography. J Hepatol. 48:835–847. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schumann G, Bonora R, Ceriotti F, Férard
G, Ferrero CA, Franck PF, Gella FJ, Hoelzel W, Jørgensen PJ, Kanno
T, et al: IFCC primary reference procedures for the measurement of
catalytic activity concentrations of enzymes at 37 degrees C.
International Federation of Clinical Chemistry and Laboratory
Medicine. Part 5. Reference procedure for the measurement of
catalytic concentration of aspartate aminotransferase. Clin Chem
Lab Med. 40:725–733. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schumann G, Bonora R, Ceriotti F, Férard
G, Ferrero CA, Franck PF, Gella FJ, Hoelzel W, Jørgensen PJ, Kanno
T, et al: IFCC primary reference procedures for the measurement of
catalytic activity concentrations of enzymes at 37 degrees C.
International Federation of Clinical Chemistry and Laboratory
Medicine. Part 4. Reference procedure for the measurement of
catalytic concentration of alanine aminotransferase. Clin Chem Lab
Med. 40:718–724. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jin BX, Zhang YH, Jin WJ, Sun XY, Qiao GF,
Wei YY, Sun LB, Zhang WH and Li N: MicroRNA panels as disease
biomarkers distinguishing hepatitis B virus infection caused
hepatitis and liver cirrhosis. Sci Rep. 5:150262015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Saldanha J, Gerlich W, Lelie N, Dawson P,
Heermann K and Heath A: WHO Collaborative Study Group: An
international collaborative study to establish a World Health
Organization international standard for hepatitis B virus DNA
nucleic acid amplification techniques. Vox Sang. 80:63–71. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu AX, Baron AD, Malfertheiner P, Kudo M,
Kawazoe S, Pezet D, Weissinger F, Brandi G, Barone CA, Okusaka T,
et al: Ramucirumab as second-line treatment in patients with
advanced hepatocellular carcinoma: Analysis of REACH trial results
by child-pugh score. JAMA Oncol. Sep 22–2016.(Epub ahead of
print).
|
|
21
|
Lee UE and Friedman SL: Mechanisms of
hepatic fibrogenesis. Best Pract Res Clin Gastroenterol.
25:195–206. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X,
Zhou X and Gan J: A serum microRNA panel as potential biomarkers
for hepatocellular carcinoma related with hepatitis B virus. PLoS
One. 9:e1079862014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Noetel A, Kwiecinski M, Elfimova N, Huang
J and Odenthal M: microRNA are central players in anti- and
profibrotic gene regulation during liver fibrosis. Front Physiol.
3:492012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Itoh T and Miyajima A: Liver regeneration
by stem/progenitor cells. Hepatology. 59:1617–1626. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Appourchaux K, Dokmak S, Resche-Rigon M,
Treton X, Lapalus M, Gattolliat CH, Porchet E, Martinot-Peignoux M,
Boyer N, Vidaud M, et al: MicroRNA-based diagnostic tools for
advanced fibrosis and cirrhosis in patients with chronic hepatitis
B and C. Sci Rep. 6:349352016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Roderburg C and Luedde T: Circulating
microRNAs as markers of liver inflammation, fibrosis and cancer. J
Hepatol. 61:1434–1437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sharma S, Khalili K and Nguyen GC:
Non-invasive diagnosis of advanced fibrosis and cirrhosis. World J
Gastroenterol. 20:16820–16830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dai X, Zhang W, Zhang H, Sun S, Yu H, Guo
Y, Kou Z, Zhao G, Du L, Jiang S, et al: Modulation of HBV
replication by microRNA-15b through targeting hepatocyte nuclear
factor 1α. Nucleic Acids Res. 42:6578–6590. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fan HX and Tang H: Complex interactions
between microRNAs and hepatitis B/C viruses. World J Gastroenterol.
20:13477–13492. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu FL, Jin WB, Li JH and Guo AG: Targets
for human encoded microRNAs in HBV genes. Virus Genes. 42:157–161.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bandiera S, Pernot S, El Saghire H, Durand
SC, Thumann C, Crouchet E, Ye T, Fofana I, Oudot MA, Barths J, et
al: Hepatitis C virus-induced upregulation of MicroRNA miR-146a-5p
in hepatocytes promotes viral infection and deregulates metabolic
pathways associated with liver disease pathogenesis. J Virol.
90:6387–6400. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Roderburg C, Urban GW, Bettermann K, Vucur
M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi
M, et al: Micro-RNA profiling reveals a role for miR-29 in human
and murine liver fibrosis. Hepatology. 53:209–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou Y, Sun L, Wang X, Zhou L, Li J, Liu
M, Wang F, Peng J, Gui X, Zhao H, et al: Heroin use promotes HCV
infection and dysregulates HCV-related circulating microRNAs. J
Neuroimmune Pharmacol. 10:102–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ciesla M, Skrzypek K, Kozakowska M, Loboda
A, Jozkowicz A and Dulak J: MicroRNAs as biomarkers of disease
onset. Anal Bioanal Chem. 401:2051–2061. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marquez RT, Bandyopadhyay S, Wendlandt EB,
Keck K, Hoffer BA, Icardi MS, Christensen RN, Schmidt WN and
McCaffrey AP: Correlation between microRNA expression levels and
clinical parameters associated with chronic hepatitis C viral
infection in humans. Lab Invest. 90:1727–1736. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Morita K, Taketomi A, Shirabe K, Umeda K,
Kayashima H, Ninomiya M, Uchiyama H, Soejima Y and Maehara Y:
Clinical significance and potential of hepatic microRNA-122
expression in hepatitis C. Liver Int. 31:474–484. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kerr TA, Korenblat KM and Davidson NO:
MicroRNAs and liver disease. Transl Res. 157:241–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lakner AM, Bonkovsky HL and Schrum LW:
microRNAs: Fad or future of liver disease. World J Gastroenterol.
17:2536–2542. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Burns DM, D'Ambrogio A, Nottrott S and
Richter JD: CPEB and two poly(A) polymerases control miR-122
stability and p53 mRNA translation. Nature. 473:105–108. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li J, Ghazwani M, Zhang Y, Lu J, Li J, Fan
J, Gandhi CR and Li S: miR-122 regulates collagen production via
targeting hepatic stellate cells and suppressing P4HA1 expression.
J Hepatol. 58:522–528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Waidmann O, Köberle V, Brunner F, Zeuzem
S, Piiper A and Kronenberger B: Serum microRNA-122 predicts
survival in patients with liver cirrhosis. PLoS One. 7:e456522012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheung O, Puri P, Eicken C, Contos MJ,
Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA and Sanyal AJ:
Nonalcoholic steatohepatitis is associated with altered hepatic
MicroRNA expression. Hepatology. 48:1810–1820. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Du J, Niu X, Wang Y, Kong L, Wang R, Zhang
Y, Zhao S and Nan Y: MiR-146a-5p suppresses activation and
proliferation of hepatic stellate cells in nonalcoholic fibrosing
steatohepatitis through directly targeting Wnt1 and Wnt5a. Sci Rep.
5:161632015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
He Y, Huang C, Sun X, Long XR, Lv XW and
Li J: MicroRNA-146a modulates TGF-beta1-induced hepatic stellate
cell proliferation by targeting SMAD4. Cell Signal. 24:1923–1930.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Pu W, Shang Y, Shao Q and Yuan X: miR-146a
promotes cell migration and invasion in melanoma by directly
targeting SMAD4. Oncol Lett. 15:7111–7117. 2018.PubMed/NCBI
|
|
47
|
Oksuz Z, Serin MS, Kaplan E, Dogen A,
Tezcan S, Aslan G, Emekdas G, Sezgin O, Altintas E and Tiftik EN:
Serum microRNAs; miR-30c-5p, miR-223-3p, miR-302c-3p and miR-17-5p
could be used as novel non-invasive biomarkers for HCV-positive
cirrhosis and hepatocellular carcinoma. Mol Biol Rep. 42:713–720.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Haneklaus M, Gerlic M, O'Neill LA and
Masters SL: miR-223: Infection, inflammation and cancer. J Intern
Med. 274:215–226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wan L, Yuan X, Liu M and Xue B:
miRNA-223-3p regulates NLRP3 to promote apoptosis and inhibit
proliferation of hep3B cells. Exp Ther Med. 15:2429–2435.
2018.PubMed/NCBI
|
|
50
|
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z,
Wang JF, Zhang Z, Lu S, Huang X, et al: Plasma microRNA panel to
diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin
Oncol. 29:4781–4788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Waidmann O, Bihrer V, Pleli T, Farnik H,
Berger A, Zeuzem S, Kronenberger B and Piiper A: Serum microRNA-122
levels in different groups of patients with chronic hepatitis B
virus infection. J Viral Hepat. 19:e58–e65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Arataki K, Hayes CN, Akamatsu S, Akiyama
R, Abe H, Tsuge M, Miki D, Ochi H, Hiraga N, Imamura M, et al:
Circulating microRNA-22 correlates with microRNA-122 and represents
viral replication and liver injury in patients with chronic
hepatitis B. J Med Virol. 85:789–798. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou XL, Xu H, Liu ZB, Wu QC, Zhu RR and
Liu JC: miR-21 promotes cardiac fibroblast-to-myofibroblast
transformation and myocardial fibrosis by targeting Jagged1. J Cell
Mol Med. May 28–2018.(Epub ahead of print).
|
|
54
|
Liu G, Friggeri A, Yang Y, Milosevic J,
Ding Q, Thannickal VJ, Kaminski N and Abraham E: miR-21 mediates
fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J
Exp Med. 207:1589–1597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu H and Fan GC: Role of microRNAs in the
reperfused myocardium towards post-infarct remodelling. Cardiovasc
Res. 94:284–292. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Thum T, Gross C, Fiedler J, Fischer T,
Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, et
al: MicroRNA-21 contributes to myocardial disease by stimulating
MAP kinase signalling in fibroblasts. Nature. 456:980–984. 2008.
View Article : Google Scholar : PubMed/NCBI
|