Introduction
Preterm is a situation in which the pregnant women
have a delivery during pregnancy between 28–37 weeks. Compared with
normal infants, preterm infants have an immature immune system
which lead to higher morbidity (1).
Therefore, preterm is a significant cause for the death of
perinatal infants (2). However,
present research is still not able to explain clearly the mechanism
of the occurrence of preterm, including the theory of cervical
ripening, neurotransmitter (3),
immune, mechanical hypothesis (4)
and endocrine control (5). Due to
the lack of the detailed molecular characterizations and
understanding of gene-regulatory mechanisms, the therapy developing
and avoiding adverse effects are difficult. Therefore, making sure
the genetic mechanism on the critical aspects of embryonic
development is important to predict the occurrence of preterm and
ensure smooth delivery (6).
MicroRNA (miRNA) is a small, non-coding,
single-stranded RNA, which can participate in the
post-transcriptional regulation of gene expression of either mRNA
degradation or inhibiting translation, thereby controlling protein
synthesis or inhibition (7). miRNAs
contain approximately 19–24 nucleotides, and their regulating
function on mRNA is accomplished by base-pairing to the
3′-untranslated region (UTR) of target mRNAs (8). However, the same miRNA in the mechanism
of different diseases has different target genes and biological
functions (9). As early as 1997,
scholars found cell-free fetal mRNAs which existed in the blood of
pregnant women. The abnormal increase of these cell-free fetal
mRNAs was confirmed to have a correlation with preterm (10). Therefore, we believe that the
abnormal expression of miRNA-regulated mRNAs have a close contact
with the occurrence of preterm. Recently, it was identified that
the miRNA-200 family is highly upregulated in myometrium both of
mice and humans, and the coordinately downregulated targets are
ZEB1 and ZEB2 (11).
ZEB1 and ZEB2 could inhibit the
contraction-associated gene expression, including the expression of
oxytocin receptor and connexin 43. It was believed that if the
expression of ZEB1 and ZEB2 is decreased, the
transcription of contraction-associated genes leading to delivery
is increased. Pineles et al used the real-time quantitative
reverse transcription-polymerase chain reaction to compare the
different expression of 157 miRNAs in the normal placenta and the
premature placenta (12). The
differential expression of miRNA-210 and miRNA-182 in the two
groups was found, and the specific biology of miRNA-182 also was
analyzed. It suggests that miRNA-200 and miRNA-182 may have an
important significance for the inchoate diagnosis and prevention of
preterm. However, the existing reports, do not entirely predict the
mRNAs which are regulated by the relevant miRNAs. In order to
clarify the mechanism of preterm and understanding the regulatory
role of miRNAs in premature delivery, we selected miRNA-200 and
miRNA-182 to predict the target genes.
Because every miRNA has numerous target mRNAs, the
accurate prediction on target mRNAs is difficult to characterize.
The common solution for this problem is to first predict the target
mRNAs by the bioinformatics tools, and then to verify these miRNA:
mRNA interactions by experiment (13). Presently, the common method is the
TargetScan algorithm which predicts mRNAs by the principle of
sequence complementarity. In order to improve the accuracy of the
prediction, we predict the target genes by the targetscore
algorithm which is developed on the basis of TargetScan algorithm.
The mathematical model of targetscore algorithm is the variational
Bayesian-Gaussian mixture model (VB-GMM) which was modified on the
basis of prediction model revised by Nam in 2014 (14). Unlike the TargetScan method, it is
not limited to predict conserved genes, so it has a higher accuracy
which could improve the efficiency of experimental verification
later and reduce the error rate.
In this study, we used the targetscore method to
predict the target genes of miRNA-200 and miRNA-182 which affect
preterm. Furthermore, we used the |logFC| and targetscore value of
each gene to determine the ultimate prediction results. The target
genes of miRNA-200 are KIAA0754, POGZ, ACKR3, N4BP2, RAMP3,
GS1-259H13.2, SPX, MAST4, KRBA2, PFN2, RNF145, SYDE2, RIN2, ZNF665,
ACOXL, NMRK1, TSNAX, ZNF274, DENND4B, and SGK494. The
target genes of miRNA-182 are KIAA1549, AMOTL2, NUMB, NRN1,
TARBP1, SAMD5, KLHL5, FAM107A, TSPAN14, DACT2, EP300, RASGRP1,
CA10, ATM, OTOGL, KIF3B, SYDE2, MAST4, TSNAX, and
LCOR.
Materials and methods
miRNA expression profile data
We obtained the miRNA expression profile GSE18809
which was stored in the Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo/) by
Stephen and other scholars. The GSE18809 dataset based on GPL570
contained 10 samples, including 5 placentas of term and 5 placentas
of preterm spontaneous delivery samples. The placentas were all
obtained from Prince of Wales Hospital in Hong Kong (China). Each
placental biopsy was collected, rinsed in phosphate-buffered
saline, and submerged into RNA later (Ambion: Thermo Fisher
Scientific, Inc., Waltham, MA, USA) as soon as possible after
delivery.
Preprocessing of genes
The miRNA expression profile is RNA. We preprocessed
the expression profile data and obtained the relative expression
quantity which is more advantageous to follow-up gene differences
in the calculation. The relative expression quantity cannot include
the missing value, the nearly unvaried value, the maximum value and
the minimum value. Therefore, the gene expression we obtained does
not contain these values. We selected the database hgu133plus2 for
gene preprocessing. The new gene expression set consists of the
expression values and the sample annotation information. In the
first step of preprocessing, we sorted out the expression values of
all genes. If there are several probes which represent the same
gene, we calculated the average of these probes as the expression
value of this gene. Then, we modified the sample annotation
information and renamed the groups as ‘preterm’ and ‘control’.
Finally, we reprocessed the gene expression values and changed the
row name of the expression matrix to GENESYMBOL.
The calculation of targetscore
We use the targetscore algorithm to predict the
target gene for miRNAs. The targetscore method is improved based on
the TargetScan method through the algorithms which can improve
match prediction accuracy. It passed by genome-wide comparison of
differential gene expression to predict the target genes without
starting to rely on evolutionary conservation (15). We deduced the maximum likelihood by
the VB-GMM, and obtained the posterior probability of each gene. In
other words, the posterior probability is calculated by the prior
probability and the likelihood function.
First of all, we used the limma package to calculate
the logFC value of all genes. The limma package provides a
framework for analyzing gene expression experiments from beginning
to end in a flexible and statistically rigorous way (16). It is often used to detect differently
expressed genes. In this study, we do not need to detect
differentially expressed genes, but we had to calculate the logFC
values of each gene by limma package. Therefore, we calculated the
logFC values for each gene with the data of the miRNA expression
profile. And the calculated logFC value is known as the priori
value in target gene prediction. The next step is to calculate the
posterior value with the equation. This algorithm is available at
www.genelibs.com/gb. TargetScan context
score (TSCS) and the probabilities of conserved targeting (PCT)
data of the desired gene are acquired in the TargetScan data
website. Based on the TSCS value, the PCT value and the
experimental logFC value, we inferred the maximum likelihood value
by the VB-GMM. In other words, the posterior value of each gene is
calculated with the prior probability and likelihood function. The
variational Bayesian algorithm is a process of iterating to
optimize the maximum likelihood estimate. It is based on
variational principle to find the edge distribution of minimality
Kullback-Leibler distance to approximate the joint distribution,
and use the mean field to reduce the complexity of the joint
estimation. It is the reason that we regarded the targetscore
method as an extension of the Expectation Maximization (EM)
algorithm in some ways. The VB-GMM model follows the standard
Bayesian GMM reported by Bishop with only tiny modifications. We
initialized the model parameters based on priors and randomly
sample κ data points μ, then we evaluated the Equation 1 using the
model parameters and update the model parameters using Equation 1
at the ith iteration.
p(Znk|χn,θ)≡∑[Znk]=ρnk∑j=1kρnj
Finally, we calculated the targetscore value and
predicted the target gene for the miRNA according to the
targetscore value of each gene. The targetscore is defined as an
integrative probabilistic score of a target gene of a miRNA. The
targetscore value is calculated by the Equation 2, and the target
genes of chosen miRNAs are evaluated quantitatively according to
the value.
Targetscore=11+ex×p¯
In Equation 2, the × means the logFC value of the
genes which calculate by limma package, and the p means the
posterior value inferred.
Results
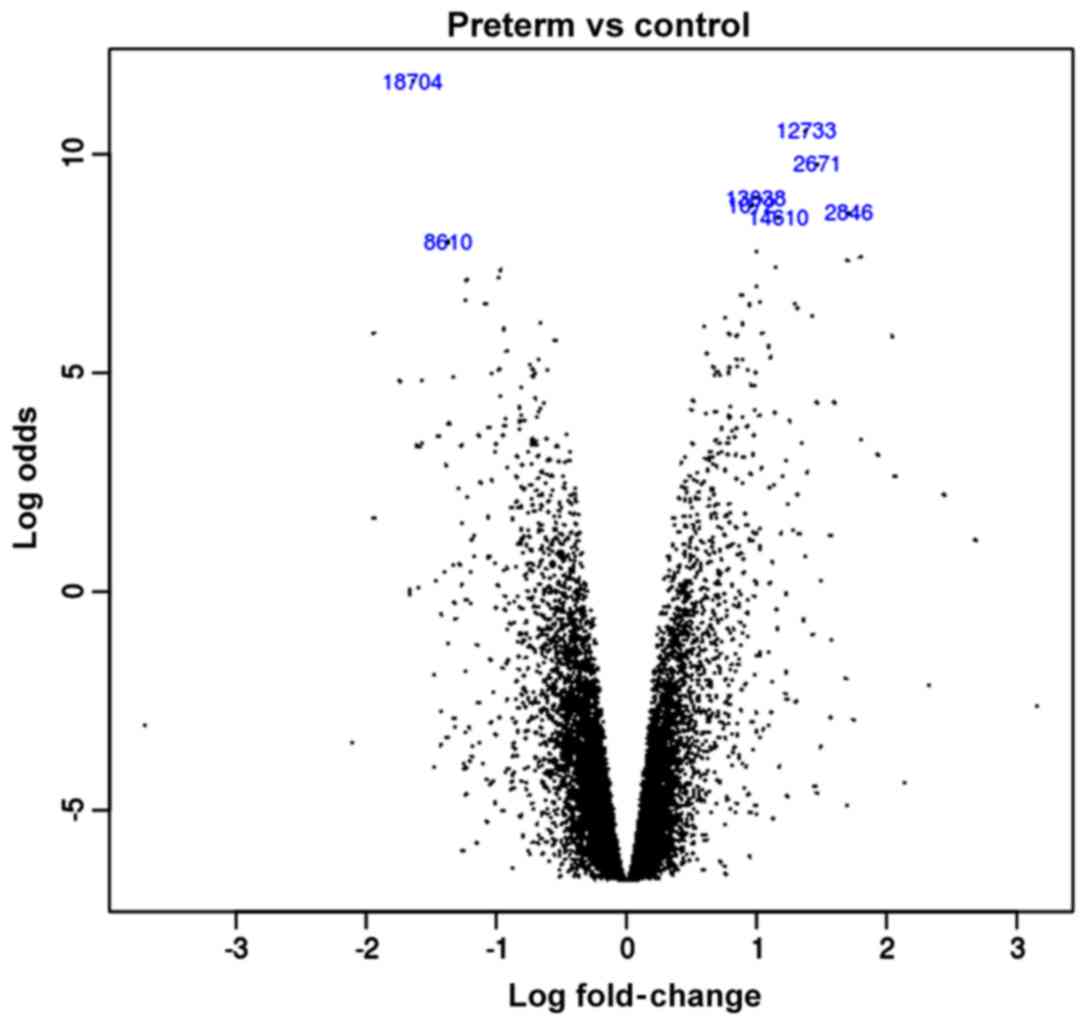
The volcano plot of the logFC
In this study, we extracted RNA from 5 cases of
preterm and 5 cases of normal placenta for analysis. Through the
mapping between the probe and the gene we obtained a total of
20,514 genes. We used the limma package to calculate the discrepant
expression of the genes between the preterm placenta experimental
group and the normal placenta control group. Then we obtained the
logFC value of each gene according to the data with linear fit. The
acquired logFC value is regarded as a priori value in our study and
all data are integrated into the volcano plot (Fig. 1). In all different genes, there are
9,783 downregulated genes and 10,731 upregulated genes.
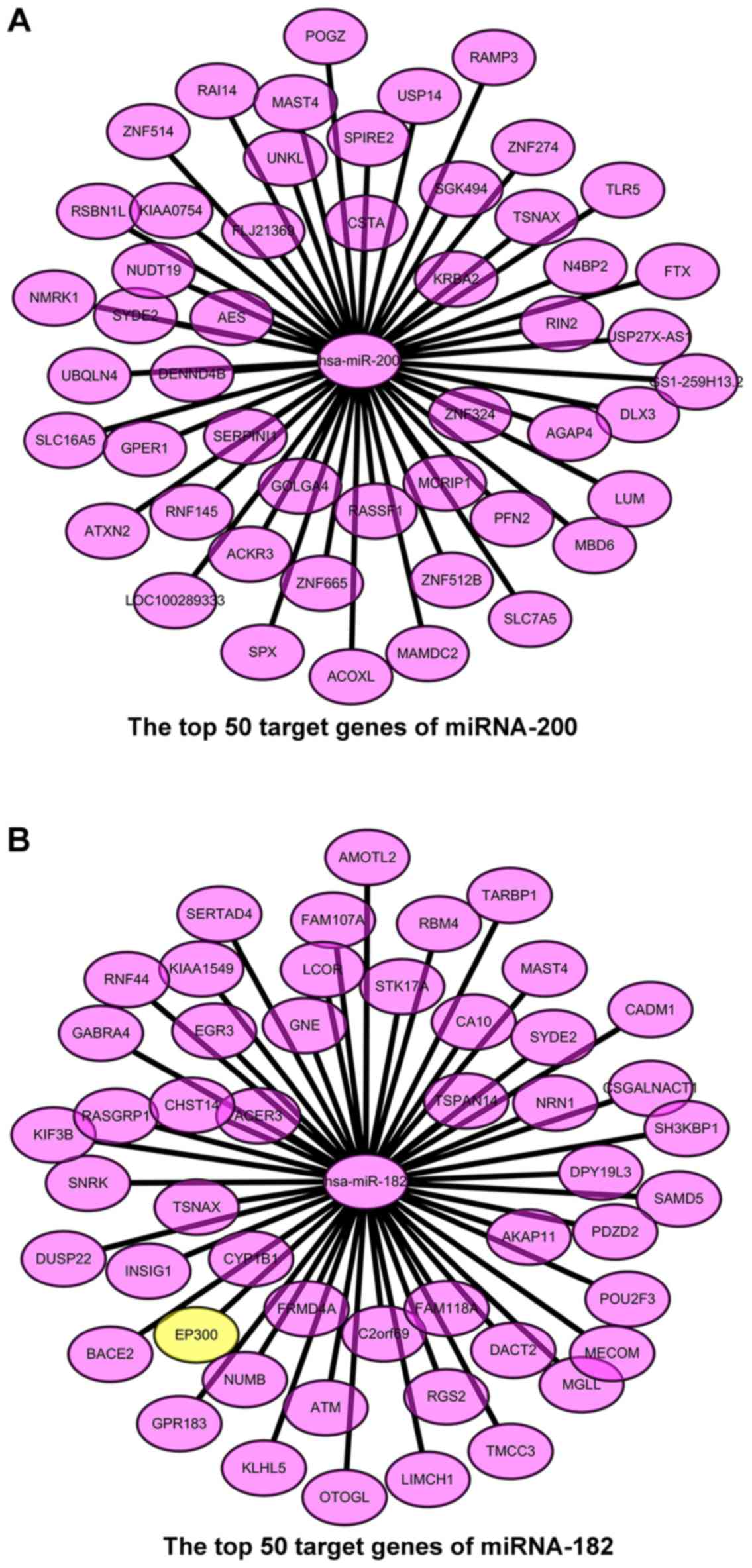
Preliminary screening of the target
genes with the targetscore
We predicted the target genes of the chosen miRNAs,
and the targetscore = 0.62 was considered as the preliminary
screening level. After preliminary screening, we selected 140
target genes of miRNA-200 and 260 target genes of miRNA-182. The
target genes of miRNA-200 are all non-validated, while the
CLOCK, EP300, F0X01, and F0X03 are validated target
genes of miRNA-182. In order to increase the accuracy of target
gene prediction, we made further target gene screening with the
higher targetscore values and drew the top 50 target genes into the
cytoscape chart (Fig. 2).
Re-screening of the target genes
In order to make the results more accurate, we
re-screened the predicted target genes. We used the |logFC| and
targetscore value as the quantitative standard of re-screening. In
all |logFC| value >0.6 of predicted genes, we selected 20 target
genes which were the most connected with miRNAs as the final
screening results. Since the targetscore value is calculated by
TargetScan CS, TargetScan PCT, and logFC value, the relevance
between the target gene and the miRNA is measured by the
targetscore value. Thus, we re-screened the predictions of
miRNA-200 and miRNA-182 by the |logFC| values and targetscore
values. The target genes of miRNA-200 are KIAA0754, POGZ, ACKR3,
N4BP2, RAMP3, GS1-259H13.2, SPX, MAST4, KRBA2, PFN2, RNF145, SYDE2,
RIN2, ZNF665, ACOXL, NMRK1, TSNAX, ZNF274, DENND4B, and
SGK494. The target genes of miRNA-182 are KIAA1549,
AMOTL2, NUMB, NRN1, TARBP1, SAMD5, KLHL5, FAM107A, TSPAN14, DACT2,
EP300, RASGRP1, CA10, ATM, OTOGL, KIF3B, SYDE2, MAST4, TSNAX,
and LCOR (Table I). In these
genes, only EP300 is a validated target gene, so we
reorganized the reference information into a table (Table II).
 | Table I.The miRNA-200 and miRNA-182 target
genes. |
Table I.
The miRNA-200 and miRNA-182 target
genes.
|
| miRNA-200 | miRNA-182 |
|---|
|
|
|
|
|---|
| External gene ID | LogFC | Targetscore | External gene ID | LogFC | Targetscore |
|---|
| KIAA0754 | −0.65328 | 0.622305 | KIAA1549 | −0.800179644 | 0.657947267 |
| POGZ | −0.65319 | 0.622305 | AMOTL2 | −0.774416547 | 0.657750851 |
| ACKR3 | −0.65379 | 0.622305 | NUMB | −0.810013576 | 0.657727 |
| N4BP2 | −0.65401 | 0.622305 | NRN1 | −0.770132168 | 0.657618084 |
| RAMP3 | −0.6541 | 0.622305 | TARBP1 | −0.767998978 | 0.657542041 |
|
GS1-259H13.2 | −0.65426 | 0.622305 | SAMD5 | −0.744871627 | 0.656319805 |
| SPX | −0.65236 | 0.622304 | KLHL5 | −0.839652128 | 0.655947693 |
| MAST4 | −0.65219 | 0.622304 | FAM107A | −0.848040295 | 0.655109486 |
| KRBA2 | −0.65135 | 0.622302 | TSPAN14 | −0.728294334 | 0.655035392 |
| PFN2 | −0.65558 | 0.622301 | DACT2 | −0.725113964 | 0.654753677 |
| RNF145 | −0.65111 | 0.622301 | EP300 | −0.720548552 | 0.654330512 |
| SYDE2 | −0.65576 | 0.6223 | RASGRP1 | −0.855821334 | 0.654202657 |
| RIN2 | −0.65084 | 0.6223 | CA10 | −0.863356689 | 0.65318794 |
| ZNF665 | −0.65602 | 0.622299 | ATM | −0.705431839 | 0.652779173 |
| ACOXL | −0.65035 | 0.622297 | OTOGL | −0.697877318 | 0.65192387 |
| NMRK1 | −0.6501 | 0.622296 | KIF3B | −0.6717802 | 0.648598376 |
| TSNAX | −0.64971 | 0.622293 | SYDE2 | −0.655760282 | 0.646305057 |
| ZNF274 | −0.6495 | 0.622292 | MAST4 | −0.652193502 | 0.645771108 |
| DENND4B | −0.649 | 0.622288 | TSNAX | −0.649707123 | 0.645394125 |
| SGK494 | −0.64895 | 0.622288 | LCOR | −0.635573098 | 0.643180143 |
 | Table II.The reference information of
validated gene. |
Table II.
The reference information of
validated gene.
| Target gene | Target gene (Entrez
ID) | Experiments | Support type | References
(PMID) |
|---|
| EP300 | 2033 | Microarray | Functional MTI
(weak) | 19569050 |
Discussion
miRNAs have important functions because the
post-transcriptional level regulation is accomplished by them
binding to the 3′-UTR of mRNAs. Some studies have shown that many
miRNAs are dysregulated in preterm placentas, and the occurrence of
preterm has significant correlations with the different expression
levels of these miRNAs (17). The
disordered miRNAs include miRNA-200 and miRNA-182. The relationship
between miRNAs and their target genes is dynamic in different
conditions. Therefore, the functions of miRNAs are different in
each context. For the occurrence of preterm, it is considered that
miRNAs have an effect on gene regulation of uterine
quiescence/contractility during pregnancy and delivery (18). Based on these studies, Williams et
al confirmed that miRNA-200, which is highly upregulated in
myometrium of both human and mouse, is an important factor to
control progesterone (P4) and progesterone receptor (PR) expression
(11). Furthermore, the induction of
miR-200 family expression led to the decline of PR function and the
increase of contraction-associated gene transcription. For this
reason, the occurrence of preterm is closely associated with the
abnormal expression of miRNA-200. The scholars have found that some
specific miRNAs were abnormally upregulated in placentas of
preeclampsia and small-for-gestational age neonates, including
miRNA-182. The relevance between preeclampsia and upregulated
miRNA-182 expression indicates the possibility of a functional role
for miRNA-182 in preeclampsia or preterm (12). However, these studies did not
disclose the exact target genes which are regulated by miRNA-200
and miRNA-182, so that it cannot completely clarify the mechanism
of preterm. The potential target gene identification is fundamental
for the explanation of the biological role of individual miRNAs
in vivo. Therefore, we used the targetscore algorithm to
reveal the target genes of miRNAs in the preterm to provide
evidence for variations in miRNA expression in obstetric
disease.
As the accurate prediction of target mRNAs is
difficult to characterize, an increasing number of new
bioinformatics tools have been developed to improve the accuracy of
prediction. TargetScan is the first algorithm used to predict miRNA
vertebrate targets. The first edition of TargetScan algorithm was
developed in 2003 by Bartel's group (19). In order to improve match prediction
accuracy scholars utilized more databases and genomes to develop
new versions of this algorithm, such as TargetScanS, TargetScan
PCT, TSCS (20–22). In our study, we chose the targetscore
algorithm. This algorithm calculates the score for each of the
context first and then weights the score by the AIR of a specific
site in each cell type, and obtains the cell-type-specific score
for each site ultimately. Compared with other versions of the
TargetScan algorithms, the targetscore algorithm is more closely
related to preterm, and the predicted target genes of miRNA are
more specific.
miRNAs achieve their biological function by the
regulation of downstream target genes, and it is estimated that
>30% of human genes are potential targets for miRNAs. In order
to ascertain the mechanism of action of miRNAs in preterm, it is
crucial to predict their target genes. By predicting the target
genes, it is helpful to understand the expression patterns in
placenta of distinct pathologies and provide the theoretical basis
for the prevention and drug development of preterm. We calculated
the targetscore values with the TSCS, PCT and logFC values. For
this purpose, the prediction results are closer to the disease and
more accurate. After prediction, we found that there were 9,783
downregulated genes and 10,731 upregulated genes in the
differentially expressed genes. At targetscore = 0.62, there are
140 miRNA-200 target genes and 260 miRNA-182 target genes were
selected. In order to further confirm the predicted results
accurately, we used |logFC| and targetscore values to select the
target genes which are more closely related to miRNA in occurrence
of preterm. The prediction results of miRNA-200 are KIAA0754,
POGZ, ACKR3, N4BP2, RAMP3, GS1-259H13.2, SPX, MAST4, KRBA2, PFN2,
RNF145, SYDE2, RIN2, ZNF665, ACOXL, NMRK1, TSNAX, ZNF274,
DENND4B, and SGK494. The target genes of miRNA-182 are
KIAA1549, AMOTL2, NUMB, NRN1, TARBP1, SAMD5, KLHL5, FAM107A,
TSPAN14, DACT2, EP300, RASGRP1, CA10, ATM, OTOGL, KIF3B, SYDE2,
MAST4, TSNAX, and LCOR. However, in all results,
EP300 is the only gene which is validated by the other
scholars (23). It means that the
study of preterm birth mechanism is still a great shortage, and a
lot of genes are still not experimentally verified. This suggests
that in the future study of mechanism we can regard the other
non-validated target genes as the research goal and to explore the
influence of miRNA-200 and miRNA-182 on the preterm control though
their differential expression. We carefully read the literature
which described the only validated target gene EP300. Mees
et al analyzed the expression of the mRNAs which were
regulated by the specific expressed miRNAs during development and
metastasis of pancreatic ductal adenocarcinoma (23). By comparing the gene expression
profiles of the biological groups with different metastatic
potential, they identified EP300 as the most important
target mRNA of miRNA-182 and other significantly upregulated miRNAs
in an alignment of the epigenetic and genetic expression profiling.
This conclusion to some extent shows that the predicted target
genes in our study are credible.
The targetscore algorithm model which is
specifically designed for miRNA-overexpression experiments to
interrogate targets of a particular miRNA in a specific cell
condition is different from the overexpression model of miRNA
coupled with expression profiling of mRNA by either microarray or
RNA-seq. In our study, we respectively selected 20 genes as the
target genes which are regulated by miRNA-200 and miRNA-182 to be
affected preterm based on the |logFC| and targetscore values of
each gene. In the results, EP300 is one of the validated
target genes regulated by miRNA-182. This suggests that our
predictions are credible. It can effectively control the functional
validation of prediction genes and regulate the drug design in the
future, while it will have an important significance to prevent the
preterm early.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KL performed the experiments and analyzed the data,
and was also a major contributor in writing the manuscript. JH and
YY made a substantial contribution to the conception and design of
the study. KL, JH and YY performed the statistical analysis. DL was
involved in the conception and design of the study. All authors
read and approved the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gray C, McCowan LM, Patel R, Taylor RS and
Vickers MH: Maternal plasma miRNAs as biomarkers during
mid-pregnancy to predict later spontaneous preterm birth: A pilot
study. Sci Rep. 7:8152017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Peixoto AB, da Cunha Caldas TMR, Tahan LA,
Petrini CG, Martins WP, Costa FDS and Araujo Júnior E: Second
trimester cervical length measurement for prediction spontaneous
preterm birth in an unselected risk population. Obstet Gynecol Sci.
60:329–335. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kalikstad B, Kultima HG, Andersstuen TK,
Klungland A and Isaksson A: Gene expression profiles in preterm
infants on continuous long-term oxygen therapy suggest reduced
oxidative stress-dependent signaling during hypoxia. Mol Med Rep.
15:1513–1526. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Y, Song Y, Huang J, Qu M, Zhang Y,
Geng J, Zhang Z, Liu J and Yang GY: Increased circulating exosomal
miRNA-223 is associated with acute ischemic stroke. Front Neurol.
8:572017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dutta S, Singh S, Bhattacharya A,
Venkataseshan S and Kumar P: Relation of thyroid hormone levels
with fluid-resistant shock among preterm septicemic neonates.
Indian Pediatr. 54:121–124. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bukowski R, Sadovsky Y, Goodarzi H, Zhang
H, Biggio JR, Varner M, Parry S, Xiao F, Esplin SM, Andrews W, et
al: Onset of human preterm and term birth is related to unique
inflammatory transcriptome profiles at the maternal fetal
interface. PeerJ. 5:e36852017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Anton L, DeVine A, Sierra LJ, Brown AG and
Elovitz MA: miR-143 and miR-145 disrupt the cervical epithelial
barrier through dysregulation of cell adhesion, apoptosis and
proliferation. Sci Rep. 7:30202017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sanders AP, Burris HH, Just AC, Motta V,
Svensson K, Mercado-Garcia A, Pantic I, Schwartz J, Tellez-Rojo MM,
Wright RO, et al: microRNA expression in the cervix during
pregnancy is associated with length of gestation. Epigenetics.
10:221–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brunquell J, Snyder A, Cheng F and
Westerheide SD: HSF-1 is a regulator of miRNA expression in
Caenorhabditis elegans. PLoS One. 12:e01834452017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mayor-Lynn K, Toloubeydokhti T, Cruz AC
and Chegini N: Expression profile of microRNAs and mRNAs in human
placentas from pregnancies complicated by preeclampsia and preterm
labor. Reprod Sci. 18:46–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Williams KC, Renthal NE, Condon JC, Gerard
RD and Mendelson CR: MicroRNA-200a serves a key role in the decline
of progesterone receptor function leading to term and preterm
labor. Proc Natl Acad Sci USA. 109:7529–7534. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pineles BL, Romero R, Montenegro D, Tarca
AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P,
et al: Distinct subsets of microRNAs are expressed differentially
in the human placentas of patients with preeclampsia. Am J Obstet
Gynecol. 196:261.e1–261.e6. 2007. View Article : Google Scholar
|
|
13
|
L'Yi S, Jung D, Oh M, Kim B, Freishtat RJ,
Giri M, Hoffman E and Seo J: miRTarVis+: Web-based interactive
visual analytics tool for microRNA target predictions. Methods.
124:78–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nam JW, Rissland O, Koppstein D,
Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A and
Bartel DP: Global analyses of the effect of different cellular
contexts on microRNA targeting. Molecular Cell. 53:1031–1043. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Y, Goldenberg A, Wong KC and Zhang Z: A
probabilistic approach to explore human miRNA targetome by
integrating miRNA-overexpression data and sequence information.
Bioinformatics. 30:621–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Winger EE, Reed JL and Ji X: Early first
trimester peripheral blood cell microRNA predicts risk of preterm
delivery in pregnant women: Proof of concept. PLoS One.
12:e01801242017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu Q, Chen Y, Dai J, Wang B, Liu M, Wang
Y, Tao J and Li H: Methylenetetrahydrofolate reductase
polymorphisms at 3′-untranslated region are associated with
susceptibility to preterm birth. Transl Pediatr. 4:57–62.
2015.PubMed/NCBI
|
|
19
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Salehi Z and Akrami H: Target genes
prediction and functional analysis of microRNAs differentially
expressed in gastric cancer stem cells MKN-45. J Cancer Res Ther.
13:477–483. 2017.PubMed/NCBI
|
|
21
|
Zhao B and Xue B: Consensus datasets of
mouse miRNA-mRNA interactions from multiple online resources. Data
Brief. 14:143–147. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shi Y, Yang F, Wei S and Xu G:
Identification of key genes affecting results of hyperthermia in
osteosarcoma based on integrative ChIP-Seq/TargetScan analysis. Med
Sci Monit. 23:2042–2048. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mees ST, Mardin WA, Wendel C, Baeumer N,
Willscher E, Senninger N, Schleicher C, Colombo-Benkmann M and
Haier J: EP300 - a miRNA-regulated metastasis suppressor gene in
ductal adenocarcinomas of the pancreas. Int J Cancer. 126:114–124.
2010. View Article : Google Scholar : PubMed/NCBI
|