Introduction
Osteoporosis (OP), as a common metabolic bone
disease with high incidence, is caused by the abnormal activation
of osteoclasts and the downregulation of osteoblastic
proliferation, which accelerate with age (1,2). There
are >200 million women suffering from OP worldwide and 1/3 women
aged over 50 years will have an osteoporotic fracture in their
lifetime, which becomes an important disease factor threating
women's both physical and mental health as well as quality of life
(3). Studies have demonstrated that
OP is a polygenic disease affected by the combined effects of
multiple genes, each with a small effect (4). Previous studies of candidate genes and
genome-wide association on OP have been performed mainly with
case-control design, showing that >60 genes including vitamin D
receptor (VDR) (5), ESR1 (6) and LRP5 (7) are related to OP susceptibility.
However, a lack of replication of these associations is common.
Besides, these studies show inconsistencies due to the use of
different platforms, sample sources and analytic techniques. In
addition, studies on key genes and pivotal pathways in OP are
limited. Therefore, there is an urgent need to find new ways to
solve these problems by clarifying the pathological mechanisms of
OP.
Recently, microarray studies in peripheral blood
mononuclear cells of osteoporotic patients have identified many
genes and pathophysiological mechanisms for OP (8). The main mechanism of OP is the
imbalance between bone formation and resorption. Studies have
reported that it is important to analyze the related genes and
transcription factors (TFs) responsible for skeletal disease such
as OP (9). However, most of the
studies were focused on related genes individually with unilateral
analysis, leading to a lack of understanding of the comprehensive
interactions between genes and proteins. Despite some modern
methods, such as meta-analysis, the relevant information could be
difficult to obtain due to many important issues related to
cross-platform annotations and comparisons. To solve these
problems, we used Gibbs sampling. Gibbs sampling is often used as a
means of statistical inference. In statistics, Gibbs sampling is
generally chosen when direct sampling is difficult (10). Gibbs sampling is a Markov Chain Monte
Carlo (MCMC) algorithm that is used to obtain observation sequences
approximated from a specified multivariate probability distribution
(11). Specifically, each component
is sampled sequentially, and when a certain component is sampled,
other components are considered to be fixed. It translates
multidimensional sampling problems into sampling one-dimensional
distributions. When the Gibbs sampling algorithm is executed
multiple times, the resulting samples are subject to the
distribution of the real samples (12). Based on the probability distribution,
researchers can identify pivotal pathways and hub genes in complex
disease pathologies.
Thus, in this study, we used a new method Gibbs
sampling to predict hub genes and pivotal pathways of OP. We
converted 280 pathways based on gene crossings >5 into Markov
chain (MC). Gibbs sampling is then performed to obtain a new MC. At
the same time, MCMC algorithm was used to calculate the average
probability of each gene expression to obtain the hub genes
(expression probability >0.8) and pivotal pathways (expression
probability >0.8). These findings will help to understand the
molecular mechanism of OP and provide a reference for the
identification of potential molecular markers and treatments.
Materials and methods
Data sources and preprocessing
In the present study, to identify disturbed pathways
and genes associated with OP, the gene expression profile dataset
(no. GSE 35955) were obtained from Gene Expression Omnibus (GEO;
http://www.ncbi.nlm.nih.gov/geo/)
database. All data in GEO are freely available to the research
community. The dataset contains 9 samples, the first group (5
samples, name human mesenchymal stem cells (hMSC)_middle-aged) and
the second group (4 samples, name hMSC_elderly). Affymetrix Human
Genome U133 plus 2.0 (HG-U133_Plus_2) arrays were used in gene chip
hybridization.
Kyoto Encyclopedia of Genes and
Genomes (KEGG) metabolic pathway enrichment
For the discovery of disturbed pathways related to
OP, all human pathways were obtained from KEGG database (www.genome.jp/kegg/). In KEGG database, there are a
total of 287 pathways (covering 6,894 genes). In the present study,
gene symbols were enriched to KEGG pathways, from which we chose
pathways with gene interaction in pathways ≥5.
Construction of path initial
state
According to the enrichment of genes expressed in
each pathway, the average value of gene expression of each pathway
was calculated as the expression of the path under the condition of
state 1 hMSC_middle-aged and state 2 hMSC_elderly. State 1 acted as
the initial state of the path, and state 2 acted as a priori of the
path. According to MC theory, the initial transition probability is
obtained from the expression of state 1 and state 2 in the access
system, and state 3 derives from state 2. The state n is
sequentially deduced, and the state n is only related to state n-1.
The original state of the system has nothing to do with other
Markov processes.
Gibbs sampling
The specific algorithm of Gibbs sampling is as
follows. Suppose a k-dimensional vector of n samples was needed for
construction. First, a k-dimensional vector was initialized
randomly. Then the remaining element was extracted by fixing the
k-1 elements of the vector, and a given posteriori random number
was generated. After circulating this process k times to update the
entire vector, a new sample was generated. Repeating this whole
process n times, a new MC was yielded. Here, k = 280 and n =
10,000, which was 10,000 iterations for the MC of 280 KEGG
pathways. A convergence of all parameters in all chains was
obtained after 2,000 iterations.
Analysis of different paths
According to the posterior value of access to MC and
the formula of probability alfa.pi, the probability alfa of each
pathway was obtained.
alfa.pi=∑i=200010000pi10000-2000+1
Pi represents the posterior value
of the path in the ith sample. To improve the
reliability of the results, changes in gene expression were taken
into account to adjust the probability of Gibbs sampling. According
to the gene expression values between two conditions, the pathway
expression differences between hMSC_middle-aged and hMSC_elderly
were calculated by using t-test. Rank was obtained by sorting
P-value. Combined with the probability alfa, the correction
coefficients (Rvalue) was calculated. The adjusted
probability of pathway (alfa-adj) was calculated as follows:
Rvalue=1-rankin
n represents the number of pathways, ranki
represents the rank of path i. The values of adj-alfa >0.8 were
screened out as differential pathways.
Screening hub genes
After obtaining the disturbed pathways, the hub
genes from disturbed pathways were screened by Gibbs sampling. The
genes in disturbed pathways were converted to MCs. Then, the
probability distributions of genes in disturbed pathways were
estimated by conducting a posteriori inference. Similarly, gene
expression variation was considered to adjust the probability.
Based on the gene expression values between two conditions, the
gene expression differences between hMSC_middle-aged and
hMSC_elderly were calculated by using t-test. Then, according to
the gene expression differences, the probability was adjusted. This
process was similar to the method of identifying interfering paths
by Gibbs sampling. Finally, the hub genes were identified in the
criterion of adj-alfa >0.8.
Results
Preparatory work
The expression profiles of 20,514 genes were
obtained after preprocessing. The human pathways were obtained from
KEGG database. There are 287 pathways and 6,894 genes in database
for OP. Then, using KEGG enrichment analysis, a total of 280
pathways with gene intersections ≥5 were obtained for further
analysis.
Pivotal pathways
Based on the gene expression data and KEGG pathways,
the probabilities of these pathways were evaluated by using MCMC
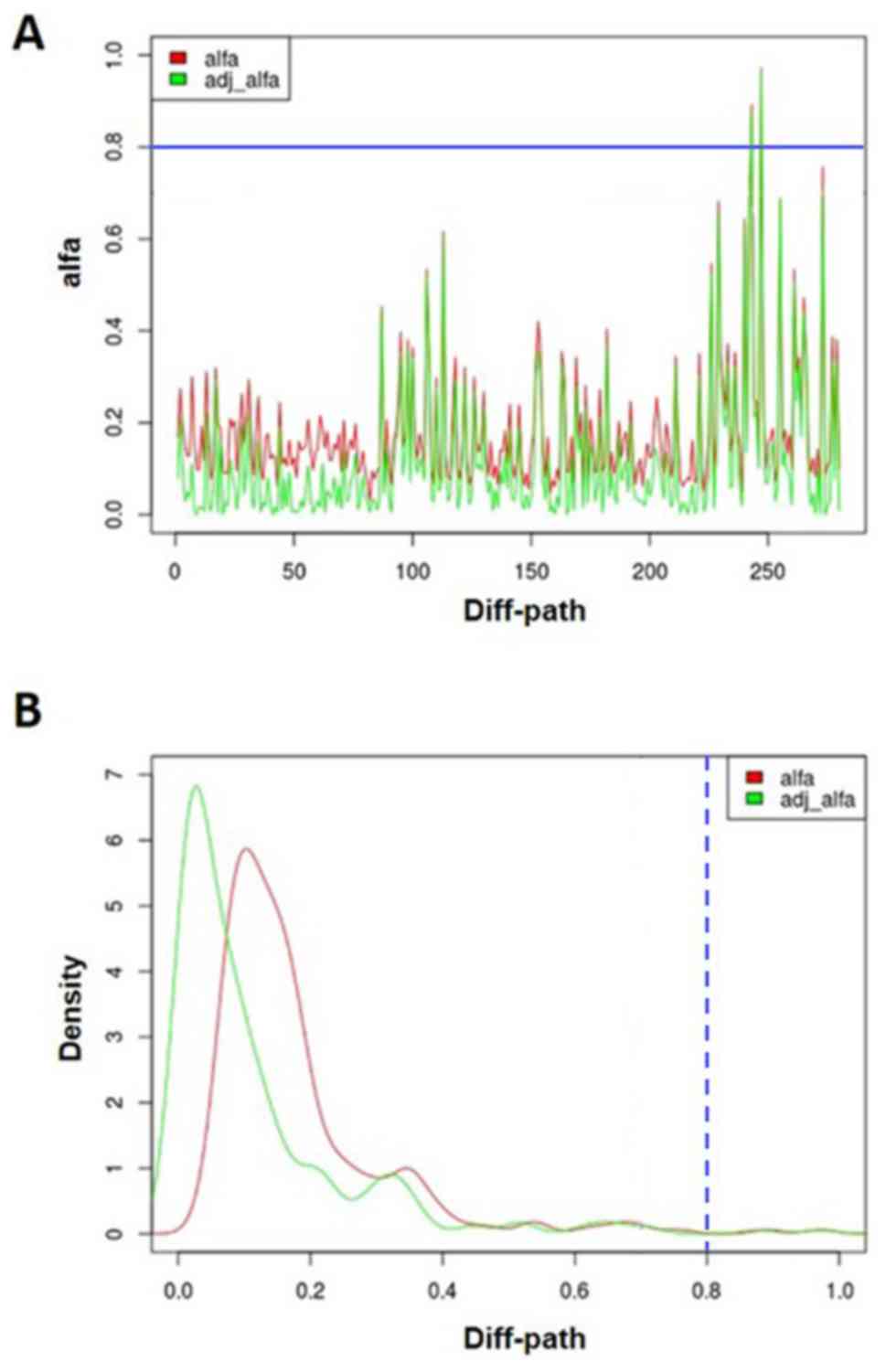
algorithm and Gibbs sampling. Fig. 1
shows the probability distribution of all KEGG pathways. In this
study, we obtained two disturbed pathways under the criterion of
alfa >0.8, including pathways in cancer (alfa = 0.973) and
influenza A (alfa = 0.892). Then the alfa was adjusted by gene
expression variation between hMSC_middle-aged and hMSC_elderly. The
adjusted alfa of pivotal pathways (pathways in cancer and influenza
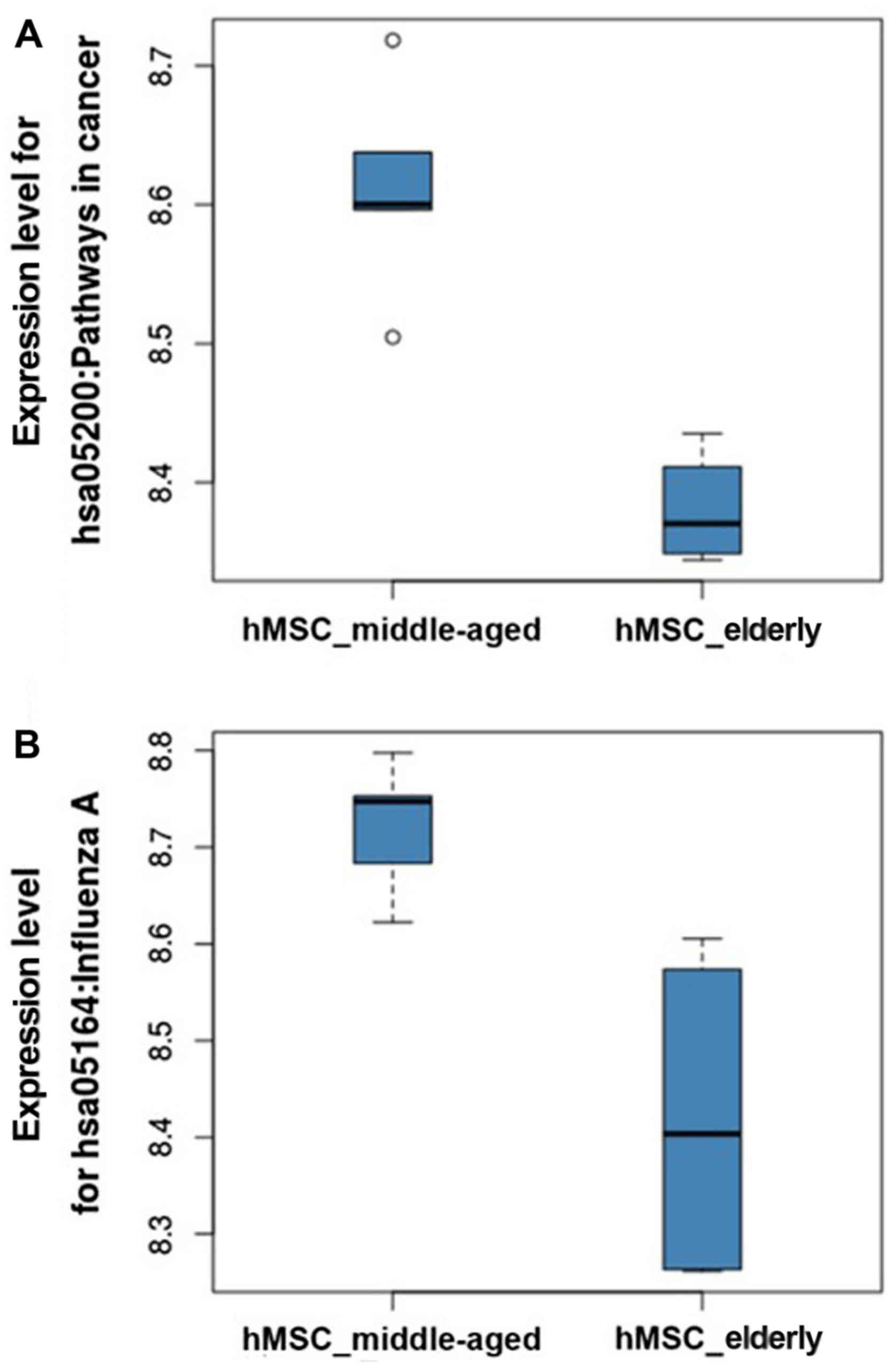
A) were 0.970 and 0.876, respectively. The expression levels of
pathways in cancer and influenza A in hMSC_middle-aged and
hMSC_elderly are shown in Fig. 2.
The increased expression levels were easily found in
hMSC_middle-aged relative to hMSC_elderly in both pathways. The
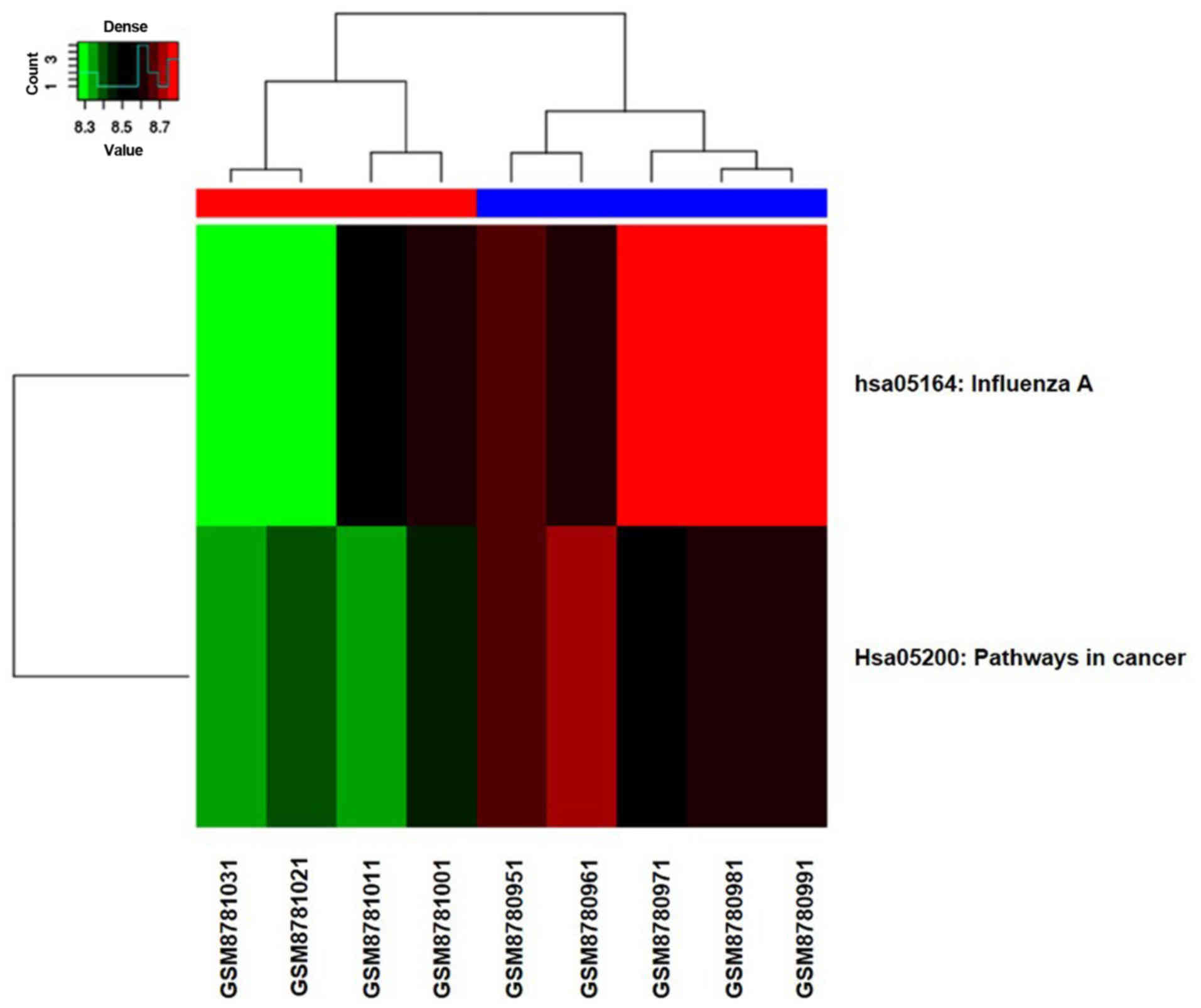
heatmap of two pathways is shown in Fig.
3, from which we can see the distribution of the two critical
pathways in 9 samples.
Hub genes
To further identify the hub genes in disturbed
pathways, the probability distributions of genes in disturbed
pathways were estimated by Gibbs sampling. We analyzed the gene
composition of two disturbed pathways, and found that pathways in
cancer contained 390 genes and influenza A contained 158 genes.
After removing genes absent from expression profile and repetitive
genes, a total of 510 genes were remained for Gibbs sampling.
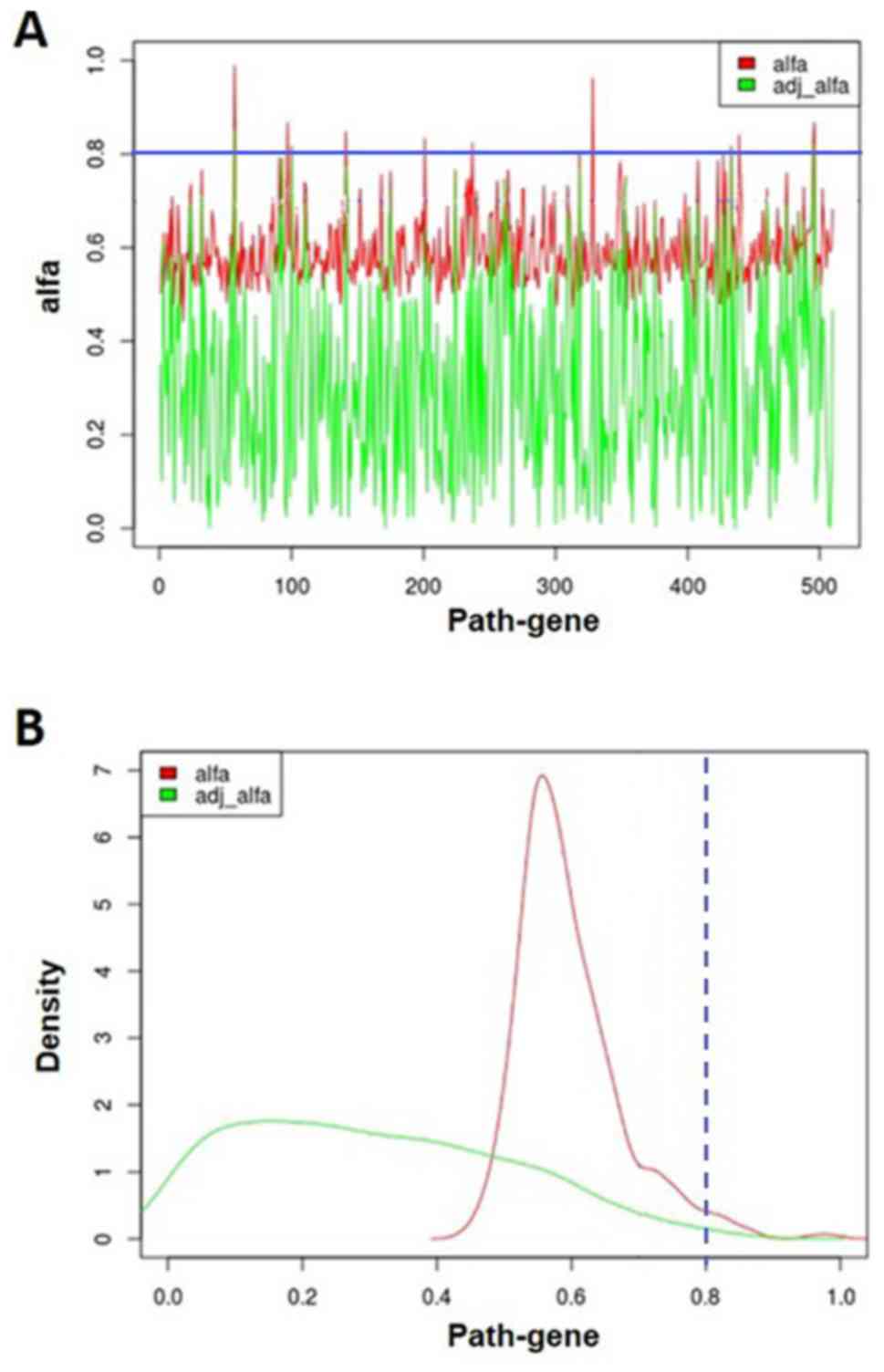
Fig. 4 shows the probability
distribution of genes in disturbed pathways. Under the criterion of
alfa >0.8, we identified two key genes associated with OP,
including cyclin A1 (CCNA1) (alfa = 0.989) and WNT2 (alfa = 0.867).
Then the probability was adjusted by gene expression variation. The
key genes CCNA1 and WNT2 had adjusted probabilities of 0.851 and
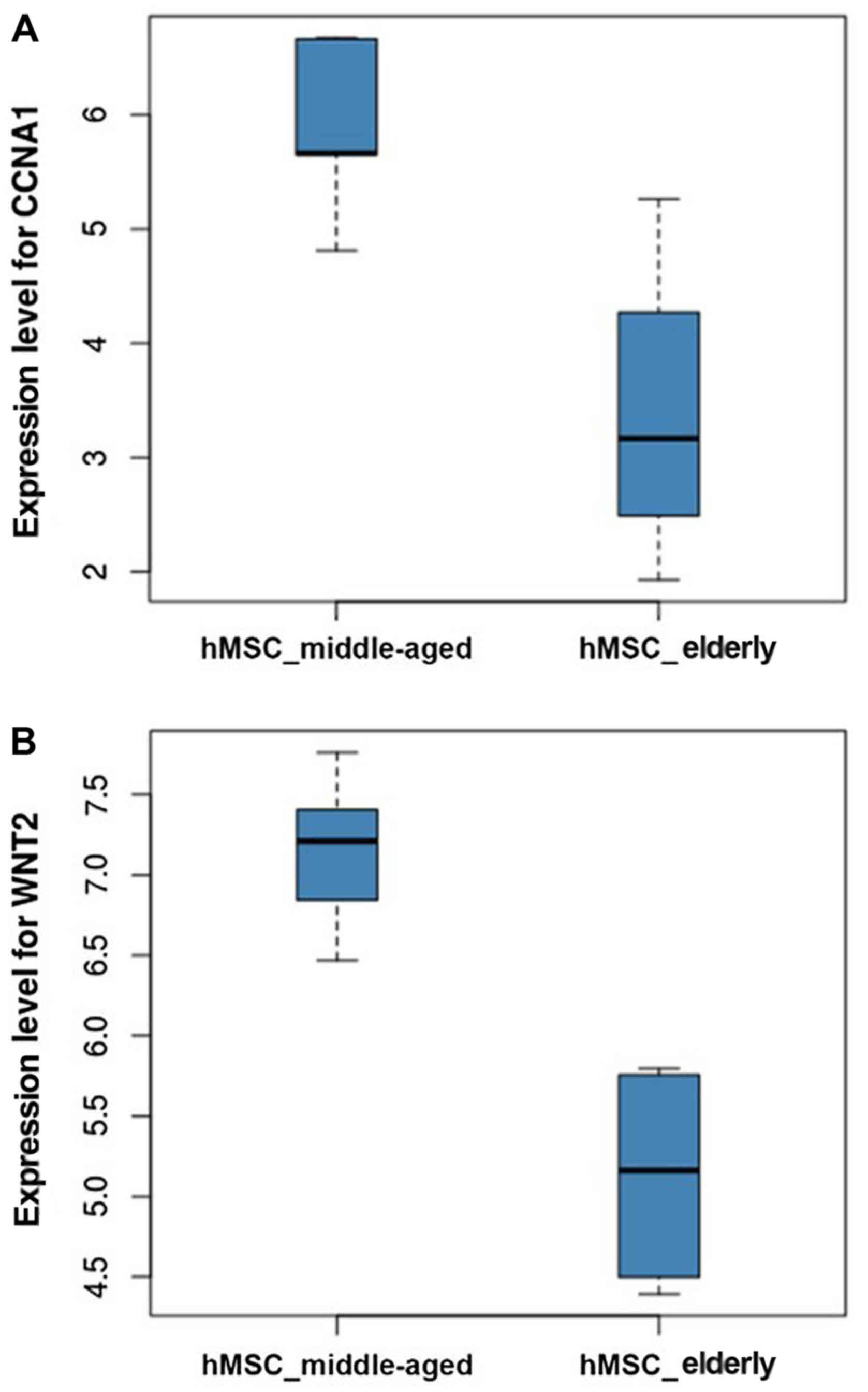
0.821, respectively. As illustrated in Fig. 5, gene expression analysis indicated
that both CCNA1 and WNT2 showed increased expression levels in
hMSC_middle-aged samples compared with hMSC_elderly samples. The
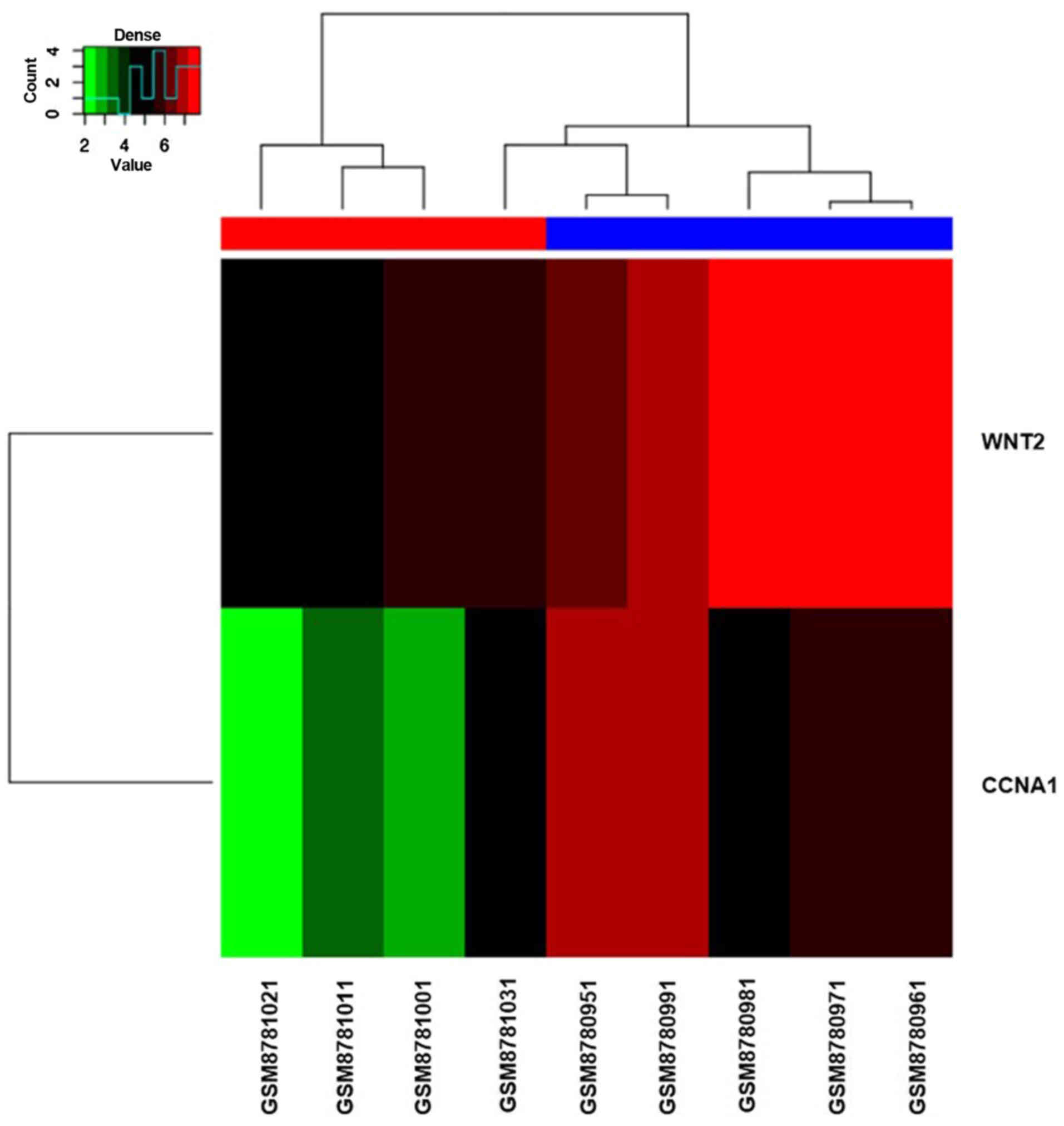
heatmap of two hub genes is shown in Fig. 6, from which we can see the
distribution of the two hub genes in 9 samples.
Discussion
In this study, two pivotal pathways associated with
OP, the pathways in cancer and influenza A, were identified through
Gibbs sampling. Moreover, two hub genes CCNA1 and WNT2 involved in
OP were also identified by using Gibbs sampling.
There are many pathways in cancer, such as WNT
(13), Raf/MEK/ERK (14) and PI3K/Akt/mTOR (15). Studies have been performed to
illustrate the association between pathways and OP, and confirmed
that several pathways were involved in the progress of OP. Luo
et al suggested that icariside II promoted osteogenic
differentiation of canine BMSCs through the PI3K/AKT/mTOR signaling
pathway (16). Zhang et al
reported that Zuoguiwan could regulate the differentiation and
proliferation of bone cells by WNT/β-catenin signaling pathway
(17). Influenza A pathway involved
some specific pathways, such as PI3K (18) and NF-κB (19). Marjuki et al indicated that
influenza A virus induced early activation of PI3K pathway
(18). Dam et al described
that the role of NF-κB pathway in influenza A virus propagation and
adaptation was antiviral (19).
Studies have suggested that chlorogenic acid promoted proliferation
of osteoblastic differentiation of BMSCs through the
PI3K/Akt/cyclin D1 pathway (20).
Studies have shown that resveratrol plays a protective effect on OP
via the activation of NF-κB signaling pathway in rats (21). These findings confirmed that both
pathways were related to OP and played a significant role in OP,
which is similar to our results.
In addition, we identified two hub genes CCNA1 and
WNT2 involved in OP by using Gibbs sampling. CCNA1, as a member of
the highly conserved cyclin family, is manifested by a significant
periodicity in protein abundance through the cell division cycle
and functions as an active subunit of the enzyme complex in
conjunction with cyclin-dependent kinases (CDKs) (22). The mammalian cell cycle regulator
CCNA1 is specifically expressed in bone marrow cells. CCNA1
expression has also been detected in osteoblast cell lines
(23). Yu et al reported that
shRNA knockdown of the adenosine deaminase acting on RNA 1 (ADAR1)
gene in MC-4 preosteoblasts reduced CCNA1 expression and cell
growth (24). WNT proteins are a
secreted glycoprotein family that signals through the paracrine
pattern frizzled (Fz) receptor (25). WNT2 is an important member of the Wnt
family (26). Wnt signaling is a
critical regulator of bone development. It has been found that WNTs
produced by Osterix-expressing OP regulated their proliferation and
differentiation (27). In recent
years, the WNT signaling pathway has been the focus of research in
bone biology laboratories because of their importance in the
therapeutic potential of skeletal development and maintenance of
bone mass (28). Based on these
reports, CCNA1 and WNT2, as hub genes, are promising for predicting
and diagnosis of OP.
In conclusion, we identified two pivotal pathways
(pathways in cancer and influenza A) and two hub genes (CCNA1 and
Wnt2) in OP utilizing efficient Gibbs sampling. These results
contributed to the understanding of underlying pathogenesis and
might be potential biomarkers for early therapy of OP. However,
further support from animal experiments or clinical investigations
on the effectiveness of the identified pathways and hub genes are
still needed.
Acknowledgements
This study was completed under the careful guidance
and with strong support of director Fang-Ming Liu and Professor
Cheng-Yuan Wu.
Funding
This study was supported by 2017 Shandong Science
and Technology Association Assists Local Innovation-Driven
Development Project-Medical Class-Health Science and Technology
Association Science and Technology Project (ZLZXBJ20170031).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YT designed the study and contributed substantially
to its revision. LL was involved in data collection, performed the
statistical analysis, prepared the figures and drafted the
manuscript. Both authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
OP
|
osteoporosis
|
|
GEO
|
Gene Expression Omnibus
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
MC
|
Markov chains
|
|
hMSC
|
human mesenchymal stem cells
|
|
MCMC
|
Markov chain Monte Carlo
|
|
VDR
|
vitamin D receptor
|
|
TFs
|
transcription factors
|
References
|
1
|
Muschitz C, Kocijan R, Haschka J, Pahr D,
Kaider A, Pietschmann P, Hans D, Muschitz GK, Fahrleitner-Pammer A
and Resch H: TBS reflects trabecular microarchitecture in
premenopausal women and men with idiopathic osteoporosis and
low-traumatic fractures. Bone. 79:259–266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harsløf T and Langdahl BL: New horizons in
osteoporosis therapies. Curr Opin Pharmacol. 28:38–42. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wright NC, Looker AC, Saag KG, Curtis JR,
Delzell ES, Randall S and Dawson-Hughes B: The recent prevalence of
osteoporosis and low bone mass in the United States based on bone
mineral density at the femoral neck or lumbar spine. J Bone Miner
Res. 29:2520–2526. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang D, Ge Z, Ma X, Zhi L, Zhang Y, Wu X,
Yao S and Ma W: Genetic association study identified a 20 kb
regulatory element in WLS associated with osteoporosis and bone
mineral density in Han Chinese. Sci Rep. 7:136682017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mohammadi Z, Fayyazbakhsh F, Ebrahimi M,
Amoli MM, Khashayar P, Dini M, Zadeh RN, Keshtkar A and Barikani
HR: Association between vitamin D receptor gene polymorphisms (Fok1
and Bsm1) and osteoporosis: A systematic review. J Diabetes Metab
Disord. 13:982014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiao P, Chen Y, Jiang H, Liu YZ, Pan F,
Yang TL, Tang ZH, Larsen JA, Lappe JM, Recker RR, et al: In vivo
genome-wide expression study on human circulating B cells suggests
a novel ESR1 and MAPK3 network for postmenopausal osteoporosis. J
Bone Miner Res. 23:644–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Astiazarán MC, Cervantes-Sodi M,
Rebolledo-Enríquez E, Chacón-Camacho O, Villegas V and Zenteno JC:
Novel homozygous LRP5 mutations in Mexican patients with
osteoporosis-pseudoglioma syndrome. Genet Test Mol Biomarkers.
21:742–746. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li JJ, Wang BQ, Fei Q, Yang Y and Li D:
Identification of candidate genes in osteoporosis by integrated
microarray analysis. Bone Joint Res. 5:594–601. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie W, Ji L, Zhao T and Gao P:
Identification of transcriptional factors and key genes in primary
osteoporosis by DNA microarray. Med Sci Monit. 21:1333–1344. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pei C, Wang SL, Fang J and Zhang W: GSMC:
Combining parallel Gibbs sampling with maximal cliques for hunting
DNA motif. J Comput Biol. 24:1243–1253. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nan N, Chen Q, Wang Y, Zhai X, Yang CC,
Cao B and Chong T: Screening disrupted molecular functions and
pathways associated with clear cell renal cell carcinoma using
Gibbs sampling. Comput Biol Chem. 70:15–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen P, Guo LH, Guo YK, Qu ZJ, Gao Y and
Qiu H: Identification of disturbed pathways in heart failure based
on Gibbs sampling and pathway enrichment analysis. Genet Mol Res.
15:gmr79562016.
|
|
13
|
Yang K, Wang X, Zhang H, Wang Z, Nan G, Li
Y, Zhang F, Mohammed MK, Haydon RC, Luu HH, et al: The evolving
roles of canonical WNT signaling in stem cells and tumorigenesis:
Implications in targeted cancer therapies. Lab Invest. 96:116–136.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park JI: Growth arrest signaling of the
Raf/MEK/ERK pathway in cancer. Front Biol (Beijing). 9:95–103.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gasparri ML, Bardhi E, Ruscito I, Papadia
A, Farooqi AA, Marchetti C, Bogani G, Ceccacci I, Mueller MD and
Benedetti Panici P: PI3K/AKT/mTOR pathway in ovarian cancer
treatment: Are we on the right track? Geburtshilfe Frauenheilkd.
77:1095–1103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo G, Xu B and Huang Y: Icariside II
promotes the osteogenic differentiation of canine bone marrow
mesenchymal stem cells via the PI3K/AKT/mTOR/S6K1 signaling
pathways. Am J Transl Res. 9:2077–2087. 2017.PubMed/NCBI
|
|
17
|
Zhang JH, Xin J, Fan LX and Yin H:
Intervention effects of Zuoguiwan containing serum on osteoblast
through ERK1/2 and Wnt/β-catenin signaling pathway in models with
kidney-Yang-deficiency, kidney-Yin-deficiency osteoporosis
syndromes. Zhongguo Zhong Yao Za Zhi. 42:3983–3989. 2017.(In
Chinese). PubMed/NCBI
|
|
18
|
Marjuki H, Gornitzky A, Marathe BM,
Ilyushina NA, Aldridge JR, Desai G, Webby RJ and Webster RG:
Influenza A virus-induced early activation of ERK and PI3K mediates
V-ATPase-dependent intracellular pH change required for fusion.
Cell Microbiol. 13:587–601. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dam S, Kracht M, Pleschka S and Schmitz
ML: The influenza A virus genotype determines the antiviral
function of NF-κB. J Virol. 90:7980–7990. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou RP, Lin SJ, Wan WB, Zuo HL, Yao FF,
Ruan HB, Xu J, Song W, Zhou YC, Wen SY, et al: Chlorogenic acid
prevents osteoporosis by Shp2/PI3K/Akt pathway in ovariectomized
rats. PLoS One. 11:e01667512016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Chen L and Peng W: Protective
effects of resveratrol on osteoporosis via activation of the
SIRT1-NF-κB signaling pathway in rats. Exp Ther Med. 14:5032–5038.
2017.PubMed/NCBI
|
|
22
|
Yang B, Miao S, Zhang LN, Sun HB, Xu ZN
and Han CS: Correlation of CCNA1 promoter methylation with
malignant tumors: A meta-analysis introduction. Biomed Res Int.
2015:1340272015.PubMed/NCBI
|
|
23
|
Miftakhova R, Hedblom A, Batkiewicz L,
Anagnosaki L, Zhang Y, Sjölander A, Wingren AG, Wolgemuth DJ and
Persson JL: Cyclin A1 regulates the interactions between mouse
haematopoietic stem and progenitor cells and their niches. Cell
Cycle. 14:1948–1960. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu S, Sharma R, Nie D, Jiao H, Im HJ, Lai
Y, Zhao Z, Zhu K, Fan J, Chen D, et al: ADAR1 ablation decreases
bone mass by impairing osteoblast function in mice. Gene.
513:101–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang C, Ma R, Xu Y, Li N, Li Z, Yue J, Li
H, Guo Y and Qi D: Wnt2 promotes non-small cell lung cancer
progression by activating WNT/β-catenin pathway. Am J Cancer Res.
5:1032–1046. 2015.PubMed/NCBI
|
|
26
|
Jiang H, Li Q, He C, Li F, Sheng H, Shen
X, Zhang X, Zhu S, Chen H, Chen X, et al: Activation of the Wnt
pathway through Wnt2 promotes metastasis in pancreatic cancer. Am J
Cancer Res. 4:537–544. 2014.PubMed/NCBI
|
|
27
|
Tan SH, Senarath-Yapa K, Chung MT,
Longaker MT, Wu JY and Nusse R: Wnts produced by Osterix-expressing
osteolineage cells regulate their proliferation and
differentiation. Proc Natl Acad Sci USA. 111:E5262–E5271. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Monroe DG, McGee-Lawrence ME, Oursler MJ
and Westendorf JJ: Update on Wnt signaling in bone cell biology and
bone disease. Gene. 492:1–18. 2012. View Article : Google Scholar : PubMed/NCBI
|