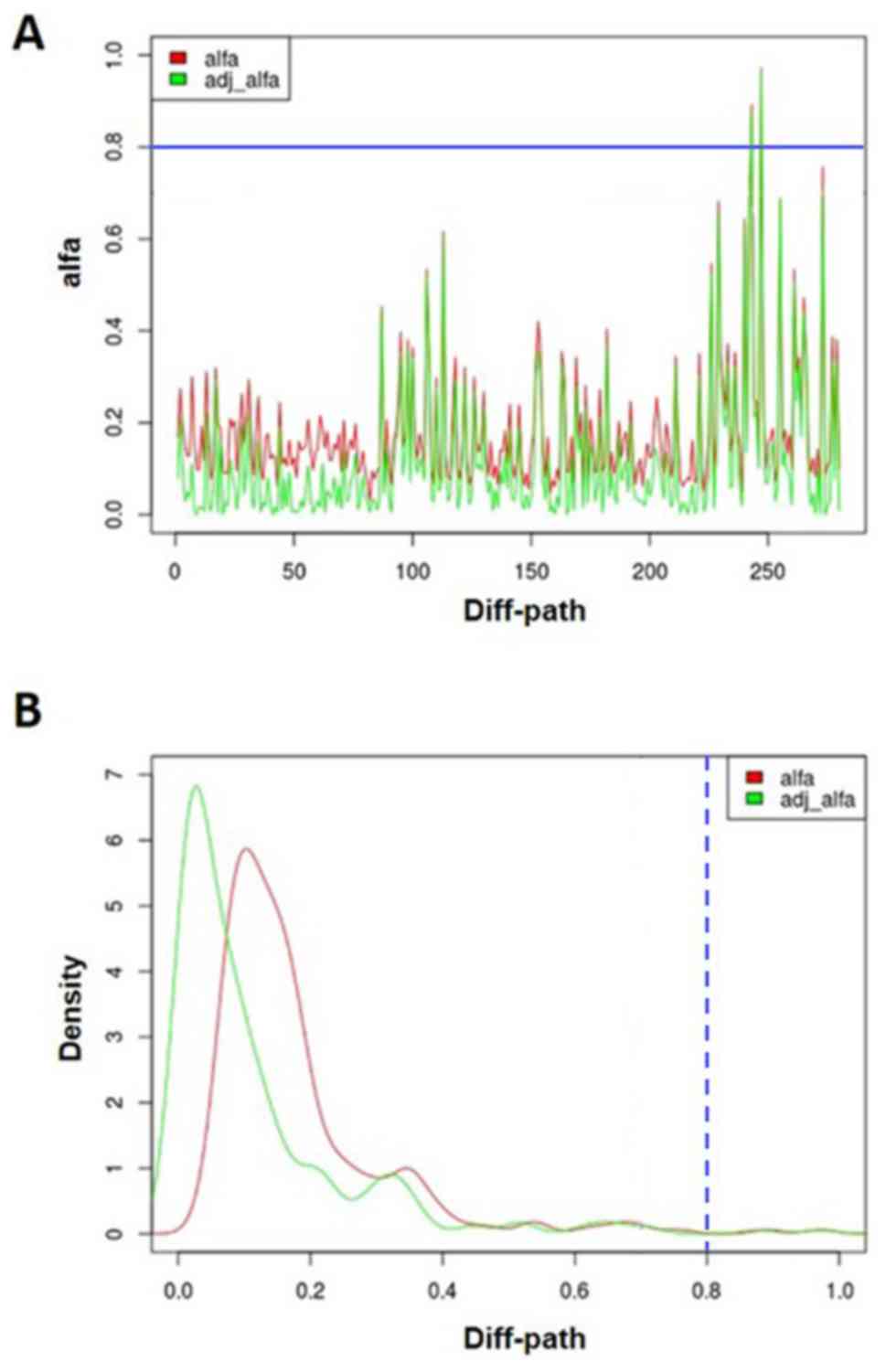
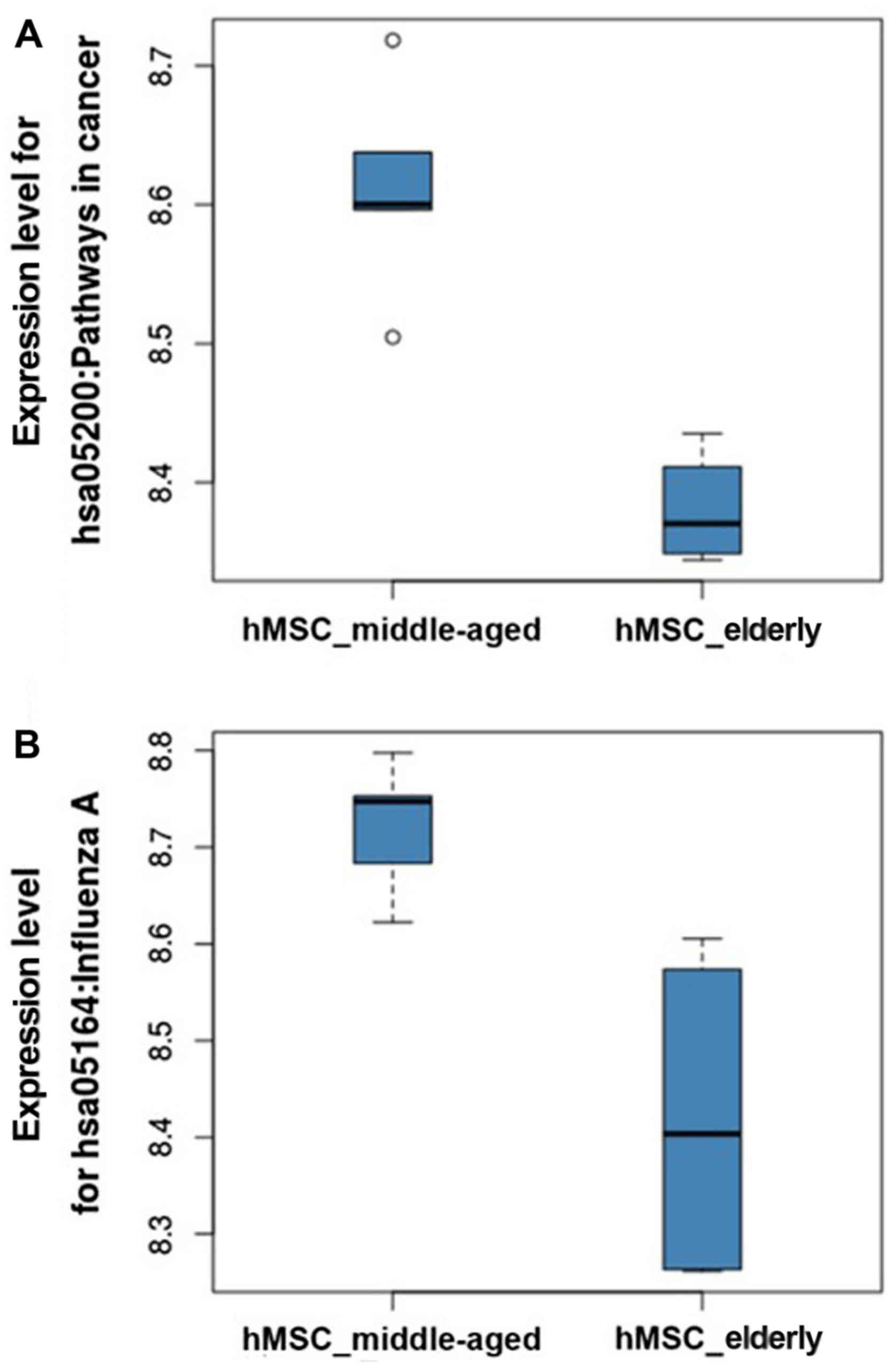
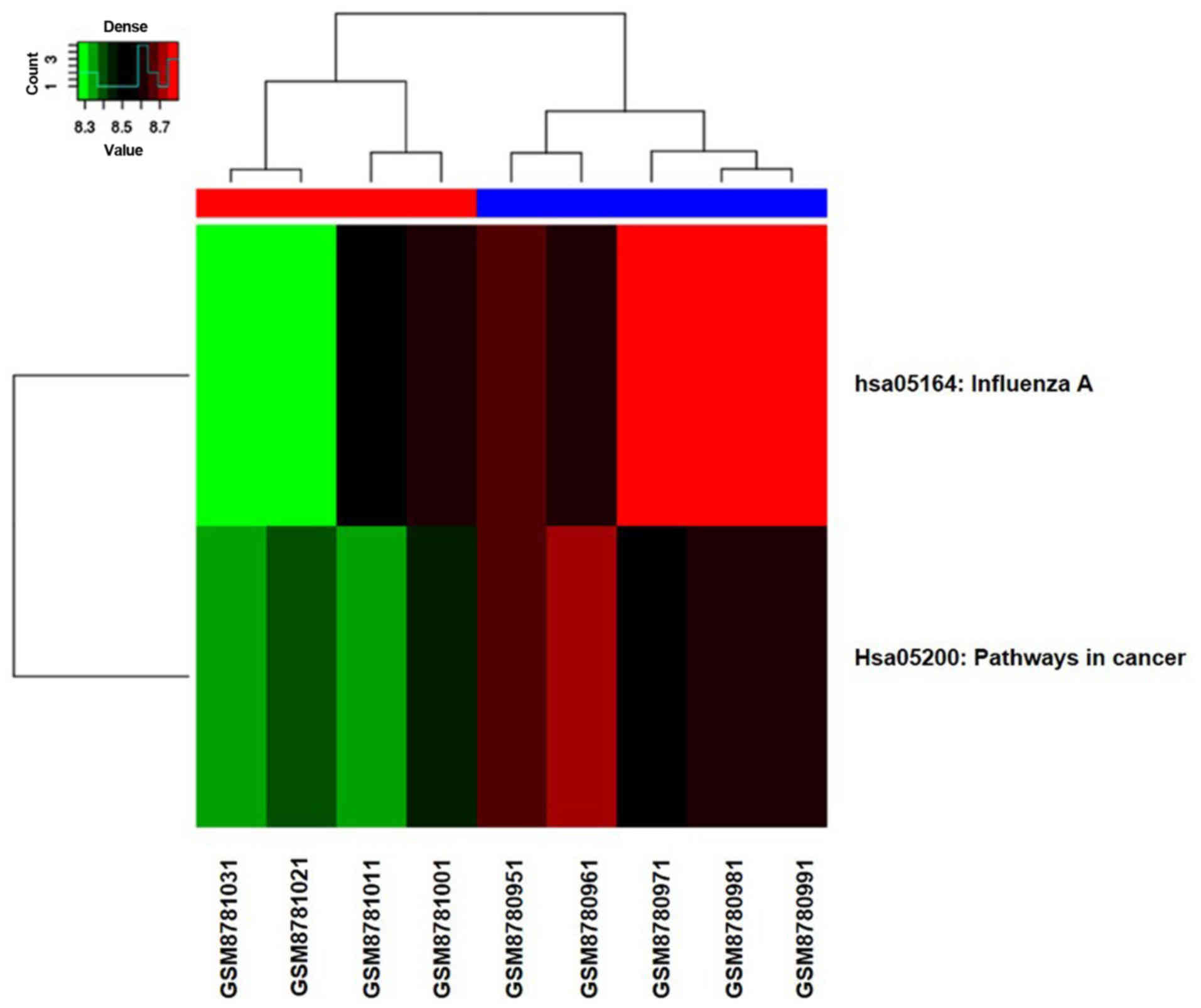
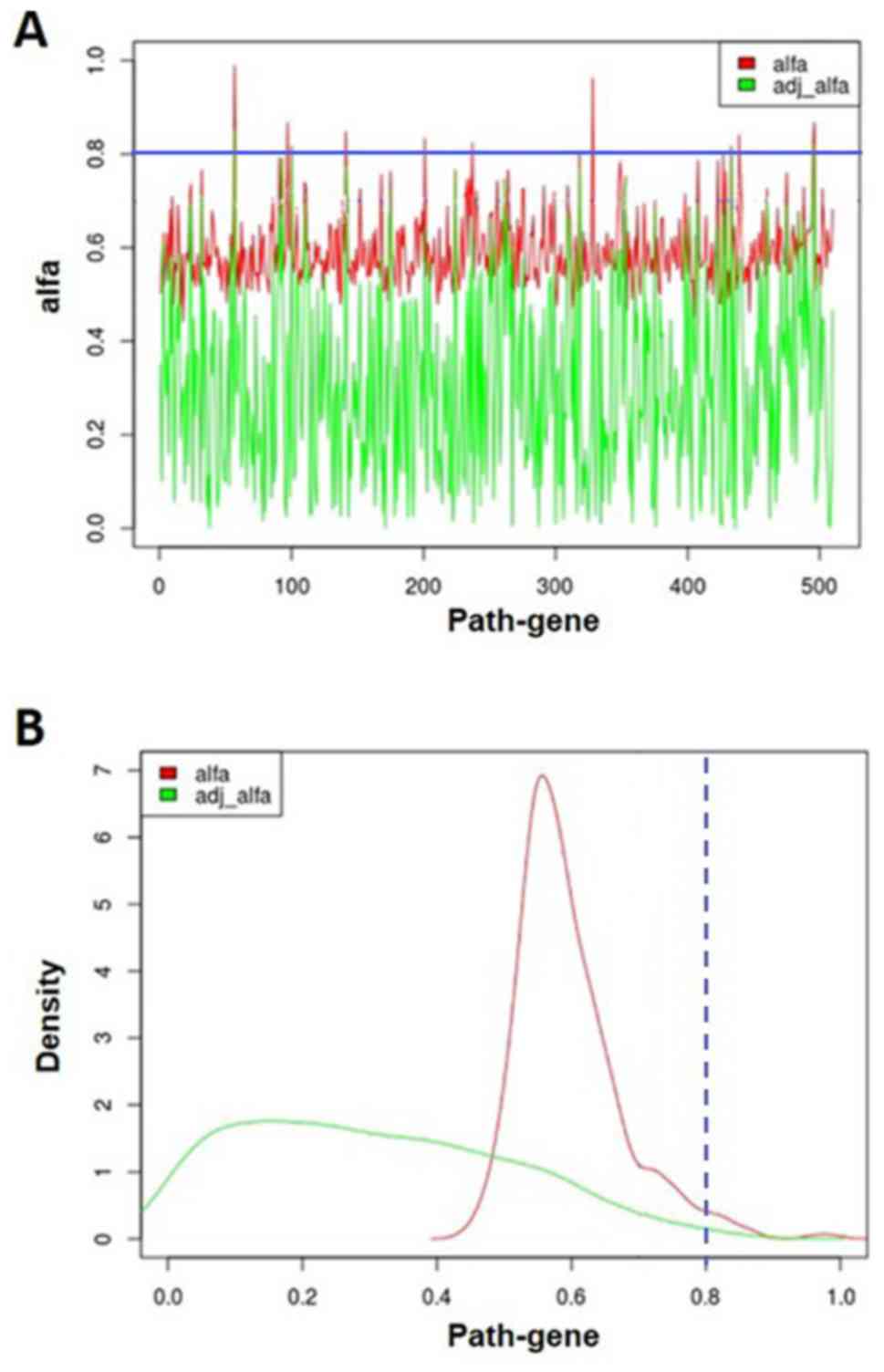
|
1
|
Muschitz C, Kocijan R, Haschka J, Pahr D,
Kaider A, Pietschmann P, Hans D, Muschitz GK, Fahrleitner-Pammer A
and Resch H: TBS reflects trabecular microarchitecture in
premenopausal women and men with idiopathic osteoporosis and
low-traumatic fractures. Bone. 79:259–266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harsløf T and Langdahl BL: New horizons in
osteoporosis therapies. Curr Opin Pharmacol. 28:38–42. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wright NC, Looker AC, Saag KG, Curtis JR,
Delzell ES, Randall S and Dawson-Hughes B: The recent prevalence of
osteoporosis and low bone mass in the United States based on bone
mineral density at the femoral neck or lumbar spine. J Bone Miner
Res. 29:2520–2526. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang D, Ge Z, Ma X, Zhi L, Zhang Y, Wu X,
Yao S and Ma W: Genetic association study identified a 20 kb
regulatory element in WLS associated with osteoporosis and bone
mineral density in Han Chinese. Sci Rep. 7:136682017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mohammadi Z, Fayyazbakhsh F, Ebrahimi M,
Amoli MM, Khashayar P, Dini M, Zadeh RN, Keshtkar A and Barikani
HR: Association between vitamin D receptor gene polymorphisms (Fok1
and Bsm1) and osteoporosis: A systematic review. J Diabetes Metab
Disord. 13:982014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiao P, Chen Y, Jiang H, Liu YZ, Pan F,
Yang TL, Tang ZH, Larsen JA, Lappe JM, Recker RR, et al: In vivo
genome-wide expression study on human circulating B cells suggests
a novel ESR1 and MAPK3 network for postmenopausal osteoporosis. J
Bone Miner Res. 23:644–654. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Astiazarán MC, Cervantes-Sodi M,
Rebolledo-Enríquez E, Chacón-Camacho O, Villegas V and Zenteno JC:
Novel homozygous LRP5 mutations in Mexican patients with
osteoporosis-pseudoglioma syndrome. Genet Test Mol Biomarkers.
21:742–746. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li JJ, Wang BQ, Fei Q, Yang Y and Li D:
Identification of candidate genes in osteoporosis by integrated
microarray analysis. Bone Joint Res. 5:594–601. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xie W, Ji L, Zhao T and Gao P:
Identification of transcriptional factors and key genes in primary
osteoporosis by DNA microarray. Med Sci Monit. 21:1333–1344. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pei C, Wang SL, Fang J and Zhang W: GSMC:
Combining parallel Gibbs sampling with maximal cliques for hunting
DNA motif. J Comput Biol. 24:1243–1253. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nan N, Chen Q, Wang Y, Zhai X, Yang CC,
Cao B and Chong T: Screening disrupted molecular functions and
pathways associated with clear cell renal cell carcinoma using
Gibbs sampling. Comput Biol Chem. 70:15–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen P, Guo LH, Guo YK, Qu ZJ, Gao Y and
Qiu H: Identification of disturbed pathways in heart failure based
on Gibbs sampling and pathway enrichment analysis. Genet Mol Res.
15:gmr79562016.
|
|
13
|
Yang K, Wang X, Zhang H, Wang Z, Nan G, Li
Y, Zhang F, Mohammed MK, Haydon RC, Luu HH, et al: The evolving
roles of canonical WNT signaling in stem cells and tumorigenesis:
Implications in targeted cancer therapies. Lab Invest. 96:116–136.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Park JI: Growth arrest signaling of the
Raf/MEK/ERK pathway in cancer. Front Biol (Beijing). 9:95–103.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gasparri ML, Bardhi E, Ruscito I, Papadia
A, Farooqi AA, Marchetti C, Bogani G, Ceccacci I, Mueller MD and
Benedetti Panici P: PI3K/AKT/mTOR pathway in ovarian cancer
treatment: Are we on the right track? Geburtshilfe Frauenheilkd.
77:1095–1103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo G, Xu B and Huang Y: Icariside II
promotes the osteogenic differentiation of canine bone marrow
mesenchymal stem cells via the PI3K/AKT/mTOR/S6K1 signaling
pathways. Am J Transl Res. 9:2077–2087. 2017.PubMed/NCBI
|
|
17
|
Zhang JH, Xin J, Fan LX and Yin H:
Intervention effects of Zuoguiwan containing serum on osteoblast
through ERK1/2 and Wnt/β-catenin signaling pathway in models with
kidney-Yang-deficiency, kidney-Yin-deficiency osteoporosis
syndromes. Zhongguo Zhong Yao Za Zhi. 42:3983–3989. 2017.(In
Chinese). PubMed/NCBI
|
|
18
|
Marjuki H, Gornitzky A, Marathe BM,
Ilyushina NA, Aldridge JR, Desai G, Webby RJ and Webster RG:
Influenza A virus-induced early activation of ERK and PI3K mediates
V-ATPase-dependent intracellular pH change required for fusion.
Cell Microbiol. 13:587–601. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dam S, Kracht M, Pleschka S and Schmitz
ML: The influenza A virus genotype determines the antiviral
function of NF-κB. J Virol. 90:7980–7990. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou RP, Lin SJ, Wan WB, Zuo HL, Yao FF,
Ruan HB, Xu J, Song W, Zhou YC, Wen SY, et al: Chlorogenic acid
prevents osteoporosis by Shp2/PI3K/Akt pathway in ovariectomized
rats. PLoS One. 11:e01667512016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang X, Chen L and Peng W: Protective
effects of resveratrol on osteoporosis via activation of the
SIRT1-NF-κB signaling pathway in rats. Exp Ther Med. 14:5032–5038.
2017.PubMed/NCBI
|
|
22
|
Yang B, Miao S, Zhang LN, Sun HB, Xu ZN
and Han CS: Correlation of CCNA1 promoter methylation with
malignant tumors: A meta-analysis introduction. Biomed Res Int.
2015:1340272015.PubMed/NCBI
|
|
23
|
Miftakhova R, Hedblom A, Batkiewicz L,
Anagnosaki L, Zhang Y, Sjölander A, Wingren AG, Wolgemuth DJ and
Persson JL: Cyclin A1 regulates the interactions between mouse
haematopoietic stem and progenitor cells and their niches. Cell
Cycle. 14:1948–1960. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu S, Sharma R, Nie D, Jiao H, Im HJ, Lai
Y, Zhao Z, Zhu K, Fan J, Chen D, et al: ADAR1 ablation decreases
bone mass by impairing osteoblast function in mice. Gene.
513:101–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang C, Ma R, Xu Y, Li N, Li Z, Yue J, Li
H, Guo Y and Qi D: Wnt2 promotes non-small cell lung cancer
progression by activating WNT/β-catenin pathway. Am J Cancer Res.
5:1032–1046. 2015.PubMed/NCBI
|
|
26
|
Jiang H, Li Q, He C, Li F, Sheng H, Shen
X, Zhang X, Zhu S, Chen H, Chen X, et al: Activation of the Wnt
pathway through Wnt2 promotes metastasis in pancreatic cancer. Am J
Cancer Res. 4:537–544. 2014.PubMed/NCBI
|
|
27
|
Tan SH, Senarath-Yapa K, Chung MT,
Longaker MT, Wu JY and Nusse R: Wnts produced by Osterix-expressing
osteolineage cells regulate their proliferation and
differentiation. Proc Natl Acad Sci USA. 111:E5262–E5271. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Monroe DG, McGee-Lawrence ME, Oursler MJ
and Westendorf JJ: Update on Wnt signaling in bone cell biology and
bone disease. Gene. 492:1–18. 2012. View Article : Google Scholar : PubMed/NCBI
|