Introduction
Breast cancer is the most frequently diagnosed
cancer and the second leading cause of cancer-related mortality
among women worldwide (1). The
overall incidence of breast cancer worldwide has been increasing
since the late 1970s (2). Breast
cancer is the second most common cause of brain metastases among
solid malignancies and it is estimated to be present at the time of
diagnosis of breast cancer in 0.41% of patients, with 7.56% of
patients presenting with metastatic disease to any site (3). Although progress has been made in the
diagnosis and treatment of breast cancer, the survival rate of
breast cancer patients remains low (4). It is therefore important to develop
novel therapeutic targets for the treatment of breast cancer.
microRNAs (miRNAs or miRs) are a group of endogenous
non-coding RNAs 21–23 nucleotides in length, which are present in
eukaryotes (5). Increasing evidence
suggests that miRNAs can regulate the expression of a broad
spectrum of genes resulting in altered cellular processes including
cell proliferation, differentiation and apoptosis (6,7). In
addition, miRNAs can regulate progression and metastasis in several
types of cancer (8–14). Studies have demonstrated that miRNAs
can function as either oncogenes or tumor suppressor genes in
cancer (15,16). As a result, miRNAs are thought to be
promising biomarkers for diagnosis and therapeutic targets in
cancer (17–20).
SRC kinase signaling inhibitor 1 (SRCIN1), also
known as p140 Cas-associated protein, contains two regions of
highly charged amino acids, two proline-rich regions and two
coiled-coil domains (21–23). Previous studies have demonstrated
that SRCIN1 serves an important role in Src inactivation and it can
also act as a tumor suppressor gene in several types of cancer
(24,25). A previous study revealed that miR-150
promotes the proliferation and migration of lung cancer cells by
targeting SRCIN1 (26). However, the
role of miR-150 and its underlying mechanism in breast cancer
remains unknown.
Epithelial-mesenchymal transition (EMT) is a
biological process, which involves the transformation from a
polarized epithelium cell to a mesenchymal cell, which has enhanced
migratory capacity and invasiveness (27,28). EMT
presents tumor cells with specific stem cell-like characteristics,
which include reduced apoptosis and resistance to immunosuppression
and senescence (29). These
characteristics serve a role in development, however they are also
associated with tissue healing, organ fibrosis, cancer development
and other biological processes.
The aim of the present study was to investigate the
cellular function of miR-150 and its underlying mechanism in breast
cancer cells.
Materials and methods
Clinical samples
Triple-negative breast cancer tissue and adjacent
healthy tissue samples were collected during biopsies from 30
female (age range, 25–57 years) triple-negative breast cancer
patients from the First Affiliated Hospital of Soochow University
(Suzhou, China) from January 2015-June 2017. Patients did not
receive preoperative radiotherapy or chemotherapy. All tissue
samples were immediately flash-frozen in liquid nitrogen and stored
at −80°C until further use. The present study was approved by the
Ethics Committee of the First Affiliated Hospital of Soochow
University, and written informed consent was obtained from each
patient.
Cell culture
Human breast epithelial cell MCF10A, and breast
cancer cell lines (MCF7, MDA-MB-468, MDA-MB-231 and MDA-MB-157)
were purchased from the Shanghai Institute of Life Sciences Cell
Resource Centre (Shanghai, China). Cells were cultured in RPMI-1640
or DMEM medium (both Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 1%
streptomycin-penicillin mix solution (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany), and maintained at 37°C in a 5%
CO2-humidified incubator.
Cell transfection
MDA-MB-468 cells were seeded in 6-well plates at a
density of 4×105 cells/well. miR-150-5p mimic
(5′-UCUCCCAACCCUUGUACCAGUG-3′), mimic control
(5′-UUCUCCGAACGUGUCACGUTT-3′), miR-150-5p inhibitor
(5′-CACUGGUACAAGGGUUGGGAGA-3′) and inhibitor control
(5′-CAGUACUUUUGUGUAGUACAA-3′) were synthesized by Shanghai
GenePharma Co., Ltd. (Shanghai, China). MDA-MB-468 cells were
subsequently transfected with 100 nM miR-150-5p mimic, 100 nM mimic
control, 100 nM miR-150-5p inhibitor or 100 nM inhibitor control
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Following 48-h transfection, cells were collected and used for
subsequent experimentation. Transfection efficiency was detected
using reverse transcription-quantitative polymerase chain reaction
(RT-qPCR).
RT-qPCR
Total RNA was extracted from cultured cell lines
(MCF10A, MCF7, MDA-MB-468, MDA-MB-231 and MDA-MB-157) and tissue
samples using TRIzol® reagent (Life Technologies; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
Total RNA was reverse transcribed into cDNA using the PrimeScript
RT Reagent kit (Takara Bio, Inc., Otsu, Japan), according to the
manufacturer's protocol. RNA concentration was detected by
NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Inc.).
qPCR was subsequently performed using the SYBR® RT-PCR
kit (Takara Bio, Inc.). The following primer sequences were used
for the qPCR: miR-150-5p forward, 5′-TCGGCGTCTCCCAACCCTTGTAC-3′ and
reverse, 5′-GTCGTATCCAGTGCAGGGTCCGAGGT-3′; SRCIN1 forward,
5′-AGCCCCGACAAAAGCAAAC-3′ and reverse,
5′-CCAAAGGAAGTCAATACAGGGATAG-3′; zonula occludens (ZO)-1 forward,
5′-CCTCTGATCATTCCACACAGTC-3′ and reverse,
5′-TAGACATGCGCTCTTCCTCTCT-3′; E-cadherin forward,
5′-CGAGAGCTACACGTTCACGG-3′ and reverse,
5′-GGGTGTCGAGGGAAAAATAGG-3′; N-cadherin forward,
5′-TTTGATGGAGGTCTCCTAACACC-3′ and reverse,
5′-ACGTTTAACACGTTGGAAATGTG-3′; vimentin forward,
5′-GACGCCATCAACACCGAGTT-3′ and reverse,
5′-CTTTGTCGTTGGTTAGCTGGT-3′; β-catenin forward,
5′-AACAGGGTCTGGGACATTAGTC-3′ and reverse,
5′-CGAAAGCCAATCAAACACAAAC-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; and GAPDH forward,
5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse,
5′-GTAGAGGCAGGGATGATGTTCT-3′. The following thermocycling
conditions were used for the qPCR: Initial denaturation at 95°C for
10 min; 35 cycles of 95°C for 15 sec and 55°C for 40 sec. Relative
expression levels of miR-150, SRCIN1 mRNA and other related genes
were quantified using the 2−ΔΔCq method (30) and normalized to the internal
reference genes U6 and GAPDH. All experiments were carried out in
triplicate.
Cell viability assay
Cell viability was analyzed using the cell counting
kit-8 (CCK-8; cat. no. C0038; Beyotime Institute of Biotechnology,
Haimen, China). Logarithmic phase MDA-MB-468 cells were seeded in
96-well plates at a density of 1×104 cells/well and
incubated at 37°C in a 5% CO2-humidified incubator for
12 h. Following incubation, 10 µl CCK-8 reagent was added to each
well and cells were incubated for a further 2 h at 37°C in a 5%
CO2-humidified incubator. Cell viability was determined
by measuring the absorbance at a wavelength of 490 nm using a
microplate reader.
Western blot analysis
Total protein was extracted from MDA-MB-468 cells
using radioimmunoprecipitation assay buffer (Thermo Fisher
Scientific, Inc.). Bicinchoninic protein assay kit (Pierce; Thermo
Fisher Scientific, Inc.) was used to measure the protein
concentration. Proteins (30 µg/lane) were separated via SDS-PAGE on
a 10% gel. The separated proteins were subsequently transferred
onto polyvinyl difluoride membranes (EMD Millipore, Billerica, MA,
USA) and blocked at room temperature for 2 h with 5% skimmed milk
in PBS containing 0.1% Tween® 20. The membranes were
incubated with primary antibodies against SRCIN1 (1:1,000; cat. no.
3757), E-cadherin (1:1,000; cat. no. 3195), ZO-1 (1:1,000; cat. no.
13663), vimentin (1:1,000; cat. no. 12826), N-cadherin (1:1,000;
cat. no. 13116), β-catenin (1:1,000; cat. no. 25362) or β-actin
(1:1,000; cat. no. 4970; all Cell Signaling Technology, Inc.,
Danvers, MA, USA) overnight at 4°C. Following primary incubation,
membranes were incubated with horseradish peroxidase-labeled
anti-rabbit IgG secondary antibody (1:1,000; cat. no. 7074; Cell
Signaling Technology, Inc.) for 2 h at room temperature. Proteins
bands were visualized using the enhanced chemiluminescence super
sensitive liquid (cat no. P1010; Applygen Technologies, Inc.,
Beijing, China) and imaged using the ChemiDoc XRS+ system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Protein expression was
quantified using Gel-Pro Analyzer software (version 6.3; Media
Cybernetics, Inc., Rockville, MD, USA).
Transwell assay
Cell invasion and migration was examined using the
Transwell assay using Transwell inserts (Corning, Inc., Corning,
NY, USA) with or without Matrigel® (BD Biosciences, San
Jose, CA, USA). Following transfection, MDA-MB-468 cells
(1×104 cells/well) in 100 µl serum-free medium added to
the upper chamber of the Transwell insert, whereas 500 µl medium
supplemented with 10% FBS was added to the lower chamber and the
plates were incubated at 37°C for 48 h. The invasive and migratory
cells were fixed with methanol for 20 min, and then stained with
0.1% crystal violet for 20 min at 37°C. The total number of
migratory and invasive cells on the underside of the membrane was
counted using five randomly selected visual fields under an
inverted light microscope at a magnification of ×200.
Dual-luciferase reporter assay
TargetScan 7.2 (http://www.targetscan.org/vert_72/) was used to
analyze the potential targets of miR-150-5p, and it was determined
that the binding site of miR-150-5p corresponds to that of SRCIN1.
To confirm the direct binding between miR-150-5p and SRCIN1, the
wild type (WT-SRCIN1) and mutant (MUT-SRCIN1) 3′UTR of SRCIN1 was
cloned into the pmiR-RB-Report™ luciferase reporter vector plasmids
(Guangzhou RiboBio Co., Ltd., Guangzhou, China). Cells
(5×104 cells/well) were seeded into 24-well plates, and
co-transfected with 100 nM miR-150-5p mimic or 100 nM mimic control
and the WT-SRCIN1 (40 ng) or MUT-SRCIN1 (40 ng) plasmids using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h. Following 48-h transfection, relative
luciferase activities were detected using a Dual-Luciferase
Reporter assay system (Promega Corporation, Madison, WI, USA),
according to the manufacturer's protocol. Firefly luciferase
activity was normalized to Renilla luciferase activity.
Statistical analysis
Data are presented as the mean ± standard deviation.
All statistical analyses were performed using SPSS software
(version 17.0; SPSS, Inc., Chicago, IL, USA). The statistical
significance of differences between two groups was analyzed using
both paired and unpaired Student's t-test. One-way analysis of
variance followed by Tukey's post hoc test was used to analyze
differences among multiple groups. All experiments were repeated
three times. P<0.05 was considered to indicate a statistically
significant difference.
Results
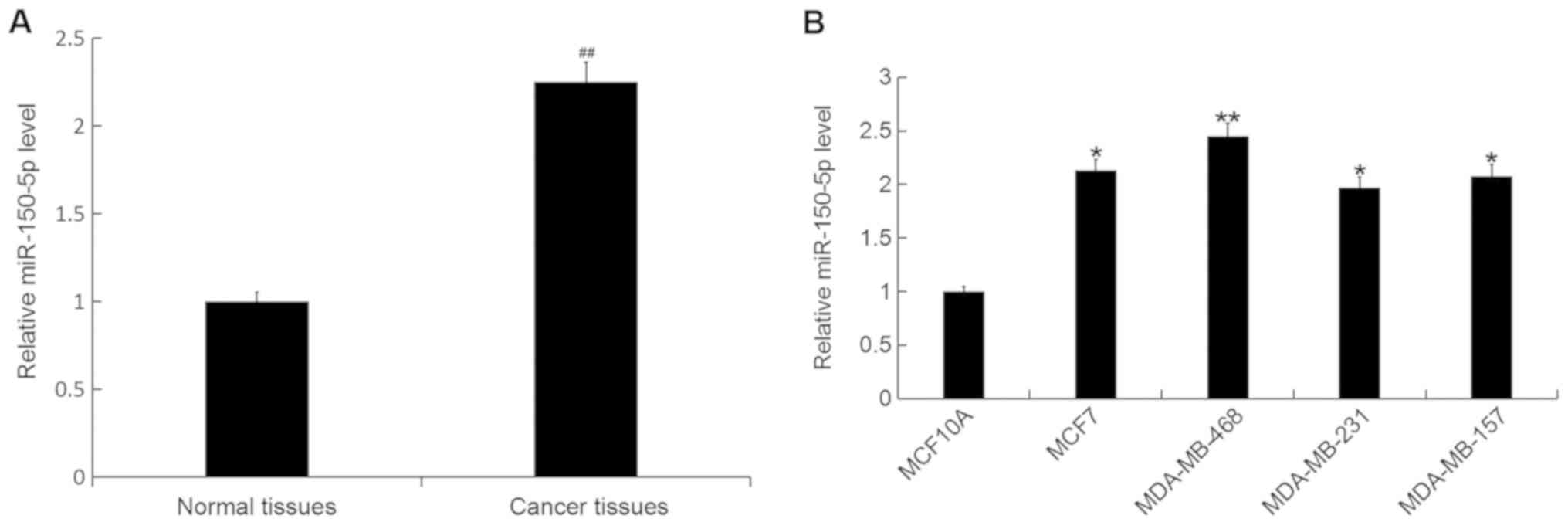
miR-150-5p expression in breast
cancer
The expression level of miR-150-5p was detected by
RT-qPCR in breast cancer tissue samples and cell lines. The
expression level of miR-150 was significantly increased in breast
cancer tissue compared with adjacent healthy tissue samples
(Fig. 1A). In addition, the
expression level of miR-150-5p was significantly increased in all
breast cancer cell lines (MCF7, MDA-MB-468, MDA-MB-231 and
MDA-MB-157) compared with the normal human breast epithelial cell
line MCF10A (Fig. 1B). The greatest
increase was observed in the MDA-MB-468 cell line, and therefore
these cells were selected for all subsequent experimentation.
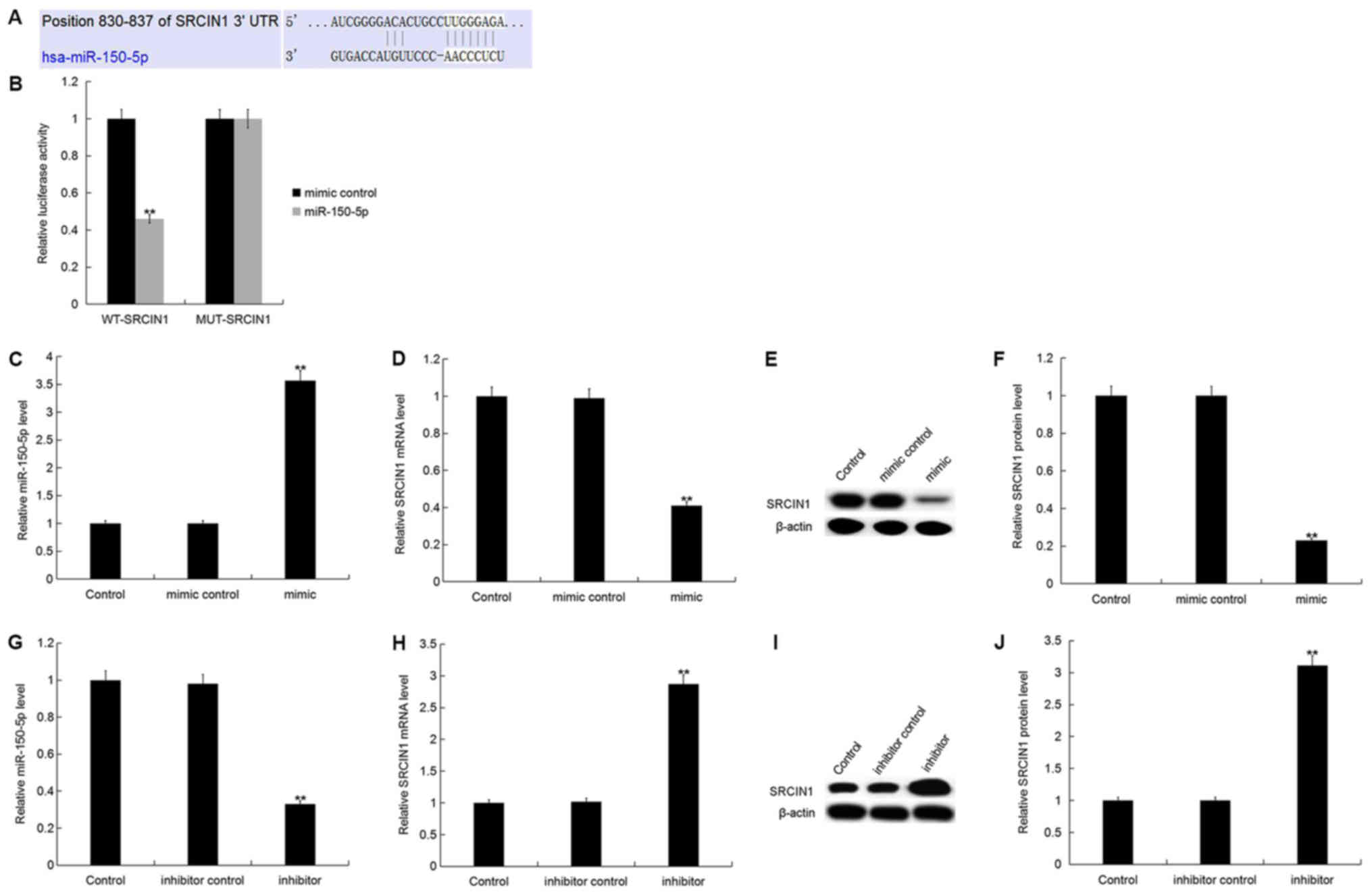
SRCIN1 is a direct target of
miR-150-5p
SRCIN1, an inhibitor of Src activity and downstream
signaling (23), was identified as a
putative target gene of miR-150-5p. TargetScan was used to predict
the miR-150-5p binding site in the 3′UTR of SRCIN1 (Fig. 2A). Luciferase reporter assays were
used to validate the direct interaction between miR-150-5p and
SRCIN1. The current study demonstrated that miR-150-5p
overexpression significantly decreased SRCIN1-WT luciferase
activity compared with SRCIN1-MUT, which had no marked effect on
luciferase activity (Fig. 2B). The
results suggest that SRCIN1 is a direct target gene of
miR-150-5p.
To investigate whether miR-150-5p regulates
endogenous SRCIN1 expression in breast cancer, SRCIN1 expression
was analyzed in MDA-MB-468 cells following 48-h transfection with
miR-150-5p mimic, mimic control, miR-150-5p inhibitor or inhibitor
control. The relative miR-150-5p expression level was significantly
increased, whilst the SRCIN1 mRNA expression level was
significantly decreased in MDA-MB-468 cells following transfection
with miR-150-5p mimic compared with the control (Fig. 2C and D). In addition, the protein
expression level of SRCIN1 was significantly decreased in
MDA-MB-468 cells following transfection with miR-150-5p mimic
compared with the control (Fig. 2E and
F).
Furthermore, the relative miR-150-5p expression
level was significantly decreased, whilst the SRCIN1 mRNA
expression level was significantly increased in MDA-MB-468 cells
following transfection with miR-150-5p inhibitor compared with the
control (Fig. 2G and H). In
addition, the protein expression level of SRCIN1 was significantly
increased in MDA-MB-468 cells following transfection with
miR-150-5p inhibitor compared with the control (Fig. 2I and J).
SRCIN1 expression in breast
cancer
To further investigate the association between
miR-150-5p and SRCIN1, the expression level of SRCIN1 was detected
by RT-qPCR in breast cancer tissue samples and cell lines. The mRNA
expression level of SRCIN1 was significantly decreased in breast
cancer tissue compared with adjacent healthy tissue samples
(Fig. 3A). In addition, the mRNA
expression level of SRCIN1 was significantly decreased in all
breast cancer cell lines (MCF7, MDA-MB-468, MDA-MB-231 and
MDA-MB-157) compared with the normal human breast epithelial cell
line MCF10A (Fig. 3B). Furthermore,
the protein expression level of SRCIN1 was significantly decreased
in each breast cancer cell line compared with MCF10A (Fig. 3C and D). The greatest decrease was
observed in the MDA-MB-468 cell line, and therefore these were
selected for all subsequent experimentation.
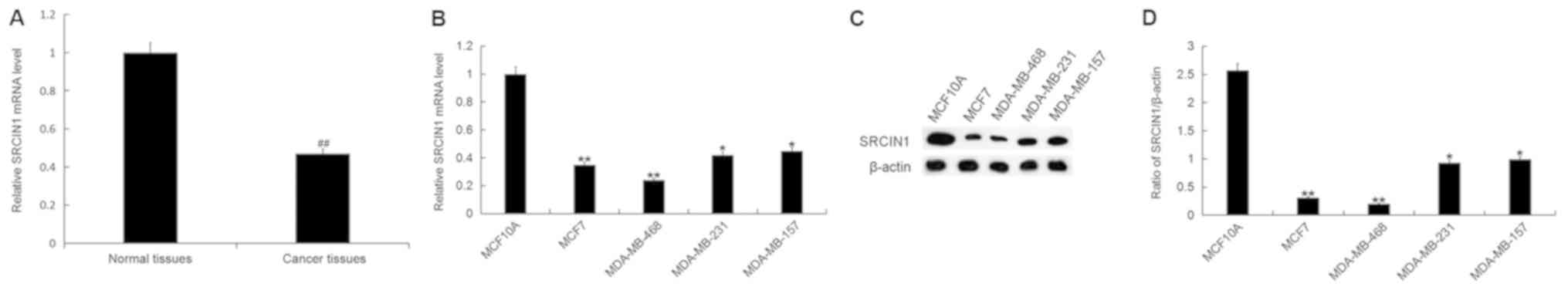 | Figure 3.SRCIN1 expression in breast cancer
tissue and cells. (A) The relative SRCIN1 mRNA expression level was
determined by RT-qPCR in breast cancer tissue and adjacent healthy
tissue samples from patients with breast cancer. (B) The relative
SRCIN1 mRNA expression level was determined by RT-qPCR in breast
cancer cell lines MCF7, MDA-MB-468, MDA-MB-231 and MDA-MB-157, and
the normal human breast epithelial cell line MCF10A. (C) The
relative protein expression level of SRCIN1 was determined by
western blot analysis in breast cancer cell lines MCF7, MDA-MB-468,
MDA-MB-231 and MDA-MB-157, and the normal human breast epithelial
cell line MCF10A. (D) Quantification of SRCIN1 protein expression.
Data are presented as the mean ± standard deviation.
##P<0.01 vs. Normal tissues; *P<0.05, **P<0.01
vs. the MCF10A cell line. SRCIN1, SRC kinase signaling inhibitor 1;
miR, microRNA; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction. |
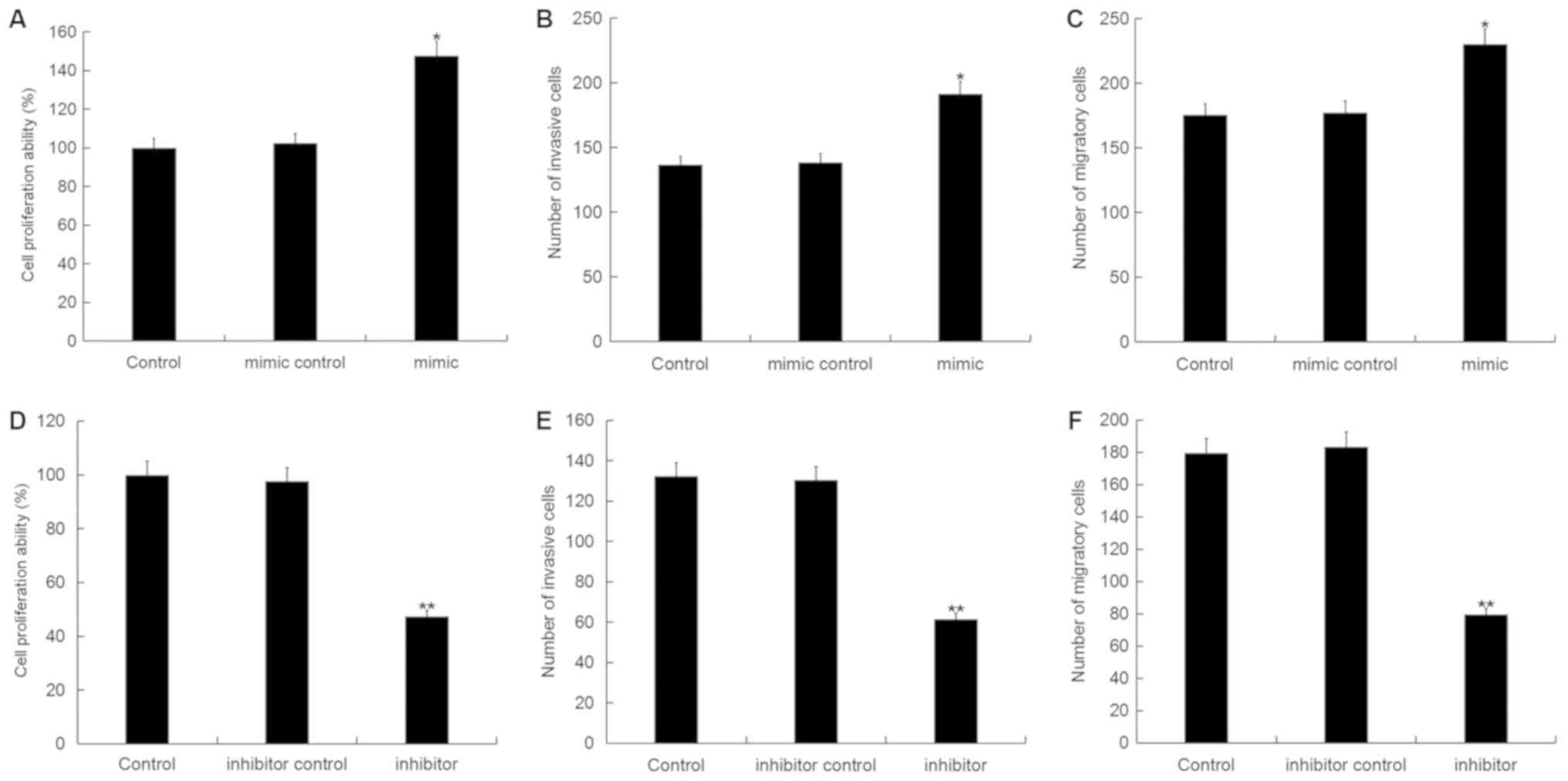
Effect of miR-150-5p on breast cancer
cell viability, migration and invasion
To investigate the role of miR-150-5p in breast
cancer, the CCK-8 and Transwell assays were used to examine the
effect of miR-150-5p on the cell viability, invasion and migration
ability of breast cancer cells following 48-h transfection with
miR-150-5p mimic, mimic control, miR-150-5p inhibitor or inhibitor
control. Cell viability, invasion and migration were significantly
increased in MDA-MB-468 cells following transfection with
miR-150-5p mimic compared with the control (Fig. 4A-C). By contrast, the cell viability,
invasion and migration were significantly decreased in MDA-MB-468
cells following transfection with miR-150-5p inhibitor compared
with the control (Fig. 4D-F). These
results suggest that miR-150-5p up-regulation can promote cell
invasion and migration, whilst miR-150-5p down-regulation can
reduce cell invasion and migration.
Effect of miR-150-5p on EMT in breast
cancer cells
To investigate the effect of miR-150-5p on EMT in
breast cancer, the expression of specific EMT markers including
epithelial cell markers (E-cadherin, ZO-1) and interstitial cell
markers (N-cadherin, vimentin, β-catenin), were analyzed in
MDA-MB-468 cells following transfection with miR-150-5p mimic,
mimic control, miR-150-5p inhibitor or inhibitor control. The
protein expression level of N-cadherin, vimentin and β-catenin were
significantly increased, whilst E-cadherin and ZO-1 protein
expression were significantly decreased in MDA-MB-468 cells
following transfection with miR-150-5p mimic compared with the
control (Fig. 5A-F). Additionally,
similar results were obtained following mRNA expression analysis
(Fig. 5G-K).
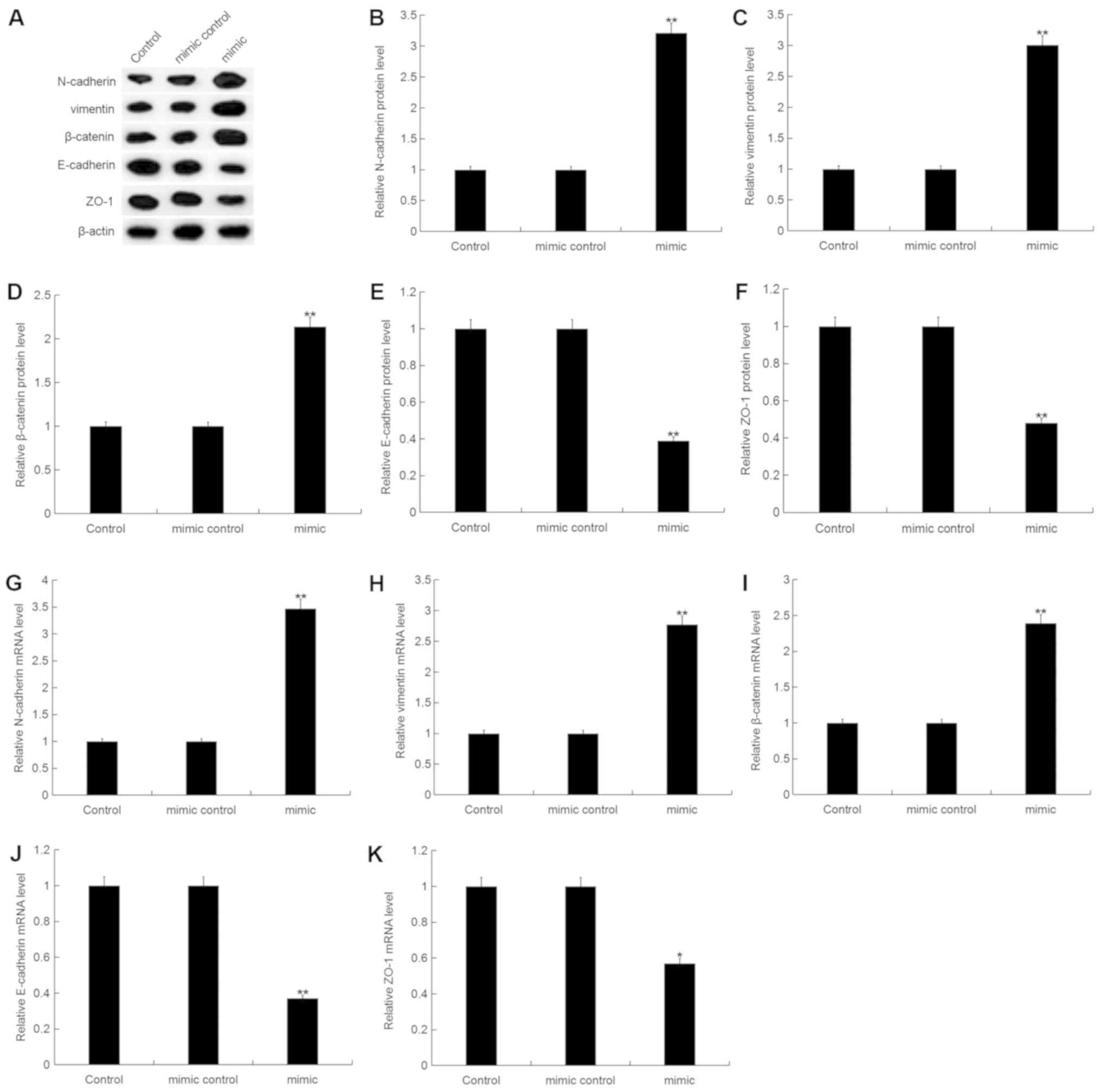 | Figure 5.Effect of miR-150-5p on
epithelial-mesenchymal transition in breast cancer cells. (A) The
relative protein expression level of N-cadherin, vimentin,
β-catenin, E-cadherin and ZO-1 were determined by western blot
analysis in MDA-MB-468 cells following 48-h transfection with
miR-150-5p mimic and mimic control. Quantification of (B)
N-cadherin, (C) vimentin, (D) β-catenin, (E) E-cadherin and (F)
ZO-1 protein expression. The relative (G) N-cadherin, (H) vimentin,
(I) β-catenin, (J) E-cadherin and (K) ZO-1 mRNA expression level
was determined by RT-qPCR in MDA-MB-468 cells following 48-h
transfection with miR-150-5p mimic and mimic control. Data are
presented as the mean ± standard deviation. *P<0.05 and
**P<0.01 vs. Control. miR, microRNA; ZO-1, zonula occludens-1;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction. |
Furthermore, the protein expression level of
N-cadherin, vimentin and β-catenin were significantly decreased,
whilst E-cadherin and ZO-1 protein expression were significantly
increased in MDA-MB-468 cells following transfection with
miR-150-5p inhibitor compared with the control (Fig. 6A-F). Additionally, similar results
were obtained following mRNA expression analysis (Fig. 6G-K).
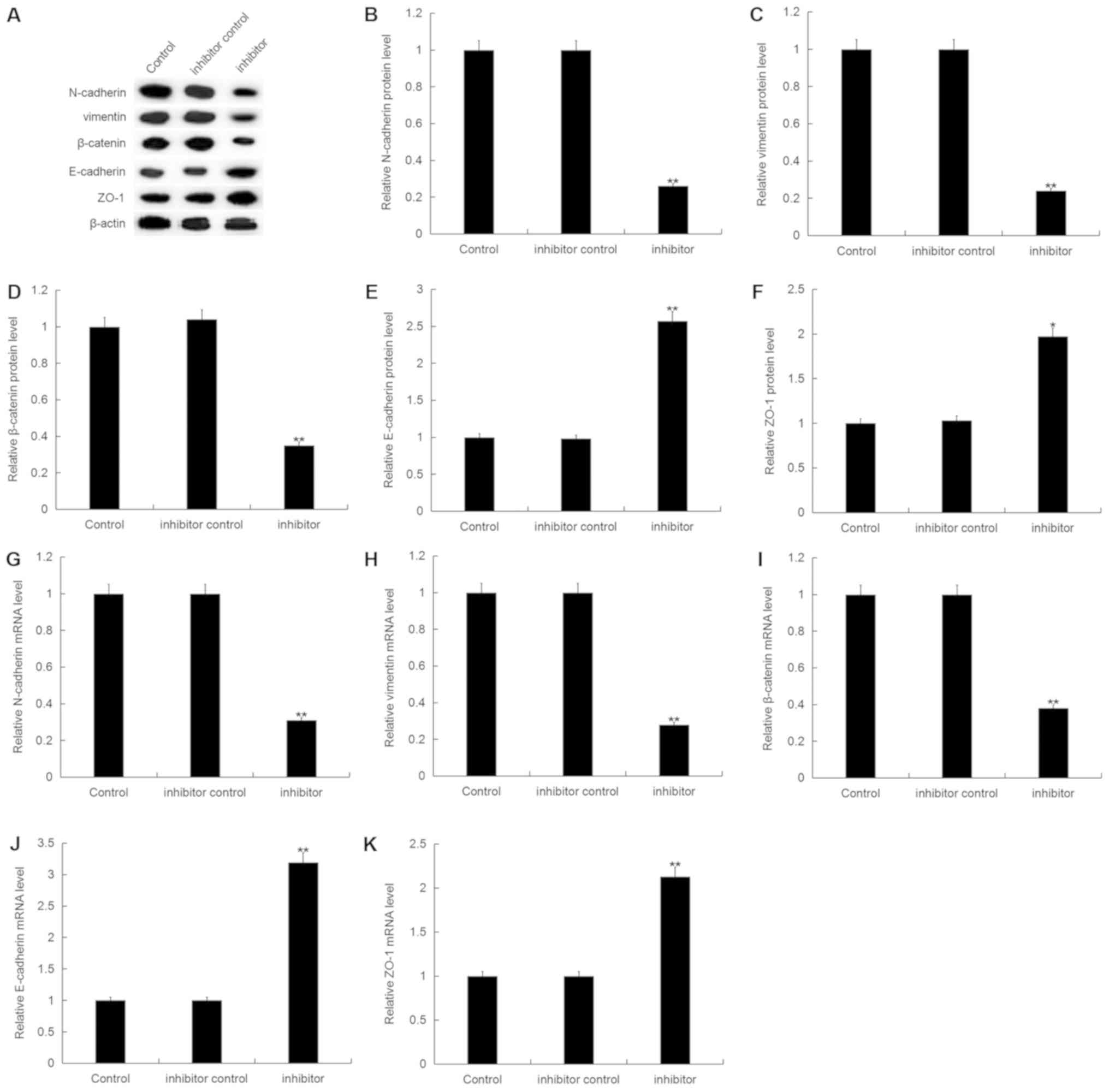 | Figure 6.Effect of miR-150-5p inhibition on
epithelial-mesenchymal transition in breast cancer cells. (A) The
relative protein expression level of N-cadherin, vimentin,
β-catenin, E-cadherin and ZO-1 were determined by western blot
analysis in MDA-MB-468 cells following 48-h transfection with
miR-150-5p inhibitor and inhibitor control. Quantification of (B)
N-cadherin, (C) vimentin, (D) β-catenin, (E) E-cadherin and (F)
ZO-1 protein expression. The relative (G) N-cadherin, (H) vimentin,
(I) β-catenin, (J) E-cadherin and (K) ZO-1 mRNA expression level
was determined by RT-qPCR in MDA-MB-468 cells following 48-h
transfection with miR-150-5p inhibitor and inhibitor control. Data
are presented as the mean ± standard deviation. *P<0.05 and
**P<0.01 vs. Control. miR, microRNA; ZO-1, tight junction
protein ZO-1; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction. |
Discussion
Breast cancer is currently a major health threat to
women worldwide. In China, there were ~268,600 new breast cancer
cases and 69,500 breast cancer-related mortalities in women in 2015
(31). Although there are numerous
drug combinations and treatment regimens, patients with advanced
breast cancer develop resistance to treatment. Resistance to
chemotherapy is a major obstacle to the effective treatment of
breast cancer (32). Therefore, it
is necessary to develop a novel treatment strategy for breast
cancer with an enhanced therapeutic effect and decreased drug
resistance.
Several studies have demonstrated that miRNAs serve
a role in the development and progression of several types of
cancer (8–14). Previous studies demonstrated that
miR-150 expression was decreased in several types of human cancer,
including lymphoma, liver, colon and bladder cancer (33–38).
miR-150, as a tumor suppressor gene, could inhibit the
proliferation, migration and invasion of cancer cells via several
mechanisms; for example, through regulating the expression of
oncogenes or tumor suppressor genes and cell cycle checkpoints
(33–38). A previous study indicated that
miRNA-150 inhibits the proliferation and tumorigenicity of
nasopharyngeal carcinoma cells (5).
By contrast, several studies have demonstrated that miR-150 was
up-regulated in several types of cancer, including chronic
lymphocytic leukemia (39),
non-small cell lung cancer (40) and
gastric cancer (41). As an
oncogene, miR-150 could promote cancer cell proliferation and
metastasis (40,41,26). Cao
et al (26) revealed that
miR-150 was highly expressed in lung cancer cells, and miR-150
overexpression could promote lung cancer cell proliferation.
Whether miR-150 is an oncogene or a tumor suppressor gene mostly
depends on the type of cancer, as miR-150 serves a different role
in different types of cancer (42).
However, the role of miR-150 and its underlying mechanism in breast
cancer remains unknown.
The current study demonstrated that miR-150-5p
expression is up-regulated in triple-negative breast cancer tissue
compared with adjacent healthy tissue samples. In addition, the
level of miR-150-5p expression was significantly increased in all
breast cancer cell lines (MCF7, MDA-MB-468, MDA-MB-231 and
MDA-MB-157) compared with the normal human breast epithelial cell
line MCF10A, and the greatest increase was observed in MDA-MB-468
cells. Therefore, the triple negative breast cancer cell line
MDA-MB-468 for chosen for all subsequent experimentation. Further
analysis revealed that the miR-150-5p mimic significantly increased
MDA-MB-468 cell viability, invasion and migration ability. By
contrast, the miR-150-5p inhibitor attenuated MDA-MB-468 cell
viability, invasion and migration ability. These results suggest
that miR-150-5p may be acting as an oncogene in breast cancer.
To further investigate the underlying mechanism of
miR-150-5p in breast cancer, bioinformatics analysis was used to
predict SRCIN1 as a direct target of miR-150-5p. Src is a
non-receptor tyrosine kinase, which is mainly expressed in brain,
testis and certain epithelial-rich organs including the mammary
glands, lungs, colon and kidneys (43). Yang et al (44) recently reported that SRCIN1
expression was significantly decreased in breast cancer tissues and
cells, and that SRCIN1 expression was inversely correlated with
miR-346 expression. Di Stefano et al indicated that SRCIN1
overexpression inhibited cell proliferation, migration and invasion
in breast cancer cells (23).
Furthermore, a previous study demonstrated that there was a
negative correlation between SRCIN1 and breast cancer (22). Consistent with previous studies, the
current study demonstrated that SRCIN1 was significantly
down-regulated in both breast cancer tissues and cell lines. In
addition, SRCIN1 was identified as a direct target gene of
miR-150-5p, and SRCIN1 was negatively regulated by miR-150-5p.
To further confirm the role of miR-150-5p in
promoting the invasion and migration of breast cancer cells, the
expression of EMT-related proteins was analyzed. The results
demonstrated that miR-150-5p significantly increased the expression
of mesenchymal cell markers and reduced the expression of
epithelial cell markers at both the protein and mRNA level.
In conclusion, the results of the current study
demonstrated that miR-150-5p may promote cell viability, invasion
and migration by directly targeting SRCIN1 and thereby identifying
a potentially novel therapeutic target in the treatment of breast
cancer.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
All data sets used and/or generated during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
QL designed the study. QL and ZG analyzed the data
and prepared the manuscript. HQ analyzed the data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Soochow University,
and written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Althuis MD, Dozier JM, Anderson WF, Devesa
SS and Brinton LA: Global trends in breast cancer incidence and
mortality 1973–1997. Int J Epidemiol. 34:405–412. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martin AM, Cagney DN, Catalano PJ, Warren
LE, Bellon JR, Punglia RS, Claus EB, Lee EQ, Wen PY, Haas-Kogan DA,
et al: Brain metastases in newly diagnosed breast cancer: A
population-based study. JAMA Oncol. 3:1069–1077. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
de la Mare JA, Contu L, Hunter MC, Moyo B,
Sterrenberg JN, Dhanani KC, Mutsvunguma LZ and Edkins AL: Breast
cancer: Current developments in molecular approaches to diagnosis
and treatment. Recent Pat Anticancer Drug Discov. 9:153–175. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li X, Liu F, Lin B, Luo H, Liu M, Wu J, Li
C, Li R, Zhang X, Zhou K and Ren D: miR-150 inhibits proliferation
and tumorigenicity via retarding G1/S phase transition in
nasopharyngeal carcinoma. Int J Oncol. 10:Mar;2017.(Epub ahead of
print).
|
|
6
|
Khew-Goodall Y and Goodall GJ:
Myc-modulated miR-9 makes more metastases. Nat Cell Biol.
12:209–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ren D, Wang M, Guo W, Huang S, Wang Z,
Zhao X, Du H, Song L and Peng X: Double-negative feedback loop
between ZEB2 and miR-145 regulates epithelial-mesenchymal
transition and stem cell properties in prostate cancer cells. Cell
Tissue Res. 358:763–778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Baranwal S and Alahari SK: miRNA control
of tumor cell invasion and metastasis. Int J Cancer. 126:1283–1290.
2010.PubMed/NCBI
|
|
10
|
Ren D, Wang M, Guo W, Zhao X, Tu X, Huang
S, Zou X and Peng X: Wild-type p53 suppresses the
epithelial-mesenchymal transition and stemness in PC-3 prostate
cancer cells by modulating miR-145. Int J Oncol. 42:1473–1481.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in Cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Liu J, Zang D, Wu S, Liu A, Zhu
J, Wu G, Li J and Jiang L: Upregulation of miR-572
transcriptionally suppresses SOCS1 and p21 and contributes to human
ovarian cancer progression. Oncotarget. 6:15180–15193.
2015.PubMed/NCBI
|
|
13
|
Wang M, Ren D, Guo W, Wang Z, Huang S, Du
H, Song L and Peng X: Loss of miR-100 enhances migration, invasion,
epithelial-mesenchymal transition and stemness properties in
prostate cancer cells through targeting Argonaute 2. Int J Oncol.
45:362–372. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang S, Guo W, Tang Y, Ren D, Zou X and
Peng X: miR-143 and miR-145 inhibit stem cell characteristics of
PC-3 prostate cancer cells. Oncol Rep. 28:1831–1837. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang D, Qiu C, Zhang H, Wang J, Cui Q and
Yin Y: Human microRNA oncogenes and tumor suppressors show
significantly different biological patterns: From functions to
targets. PLoS One. 5(pii): e130672010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et
al: A MicroRNA signature associated with prognosis and progression
in chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang L, Ye Y, Tu H, Hildebrandt MA, Zhao
L, Heymach JV, Roth JA and Wu X: MicroRNA-related genetic variants
in iron regulatory genes, dietary iron intake, microRNAs and lung
cancer risk. Ann Oncol. 28:1124–1129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Di Stefano P, Leal MP, Tornillo G, Bisaro
B, Repetto D, Pincini A, Santopietro E, Sharma N, Turco E, Cabodi S
and Defilippi P: The adaptor proteins p140CAP and p130CAS as
molecular hubs in cell migration and invasion of cancer cells. Am J
Cancer Res. 1:663–673. 2011.PubMed/NCBI
|
|
22
|
Damiano L, Di Stefano P, Camacho Leal MP,
Barba M, Mainiero F, Cabodi S, Tordella L, Sapino A, Castellano I,
Canel M, et al: p140Cap dual regulation of E-cadherin/EGFR
cross-talk and Ras signalling in tumour cell scatter and
proliferation. Oncogene. 29:3677–3690. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Di Stefano P, Damiano L, Cabodi S, Aramu
S, Tordella L, Praduroux A, Piva R, Cavallo F, Forni G, Silengo L,
et al: p140Cap protein suppresses tumour cell properties,
regulating Csk and Src kinase activity. EMBO J. 26:2843–2855. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun W, Wang X, Li J, You C, Lu P, Feng H,
Kong Y, Zhang H, Liu Y, Jiao R, et al: MicroRNA-181a promotes
angiogenesis in colorectal cancer by targeting SRCIN1 to promote
the SRC/VEGF signaling pathway. Cell Death Dis. 9:4382018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen R, Liao JY, Huang J, Chen WL, Ma XJ
and Luo XD: Downregulation of SRC kinase signaling inhibitor 1
(SRCIN1) expression by MicroRNA-32 promotes proliferation and
epithelial-mesenchymal transition in human liver cancer cells.
Oncol Res. 26:573–579. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao M, Hou D, Liang H, Gong F, Wang Y, Yan
X, Jiang X, Wang C, Zhang J, Zen K, et al: miR-150 promotes the
proliferation and migration of lung cancer cells by targeting SRC
kinase signalling inhibitor 1. Eur J Cancer. 50:1013–1024. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. 232:3261–3272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shenoy AK, Jin Y, Luo H, Tang M, Pampo C,
Shao R, Siemann DW, Wu L, Heldermon CD, Law BK, et al:
Epithelial-to-mesenchymal transition confers pericyte properties on
cancer cells. J Clin Invest. 126:4174–4186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
DeMichele A, Yee D and Esserman L:
Mechanisms of resistance to neoadjuvant chemotherapy in breast
cancer. N Engl J Med. 377:2287–2289. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Watanabe A, Tagawa H, Yamashita J, Teshima
K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T,
et al: The role of microRNA-150 as a tumor suppressor in malignant
lymphoma. Leukemia. 25:1324–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun W, Zhang Z, Wang J, Shang R, Zhou L,
Wang X, Duan J, Ruan B, Gao Y, Dai B, et al: microRNA-150
suppresses cell proliferation and metastasis in hepatocellular
carcinoma by inhibiting the GAB1-ERK axis. Oncotarget.
7:11595–11608. 2016.PubMed/NCBI
|
|
35
|
Ito M, Teshima K, Ikeda S, Kitadate A,
Watanabe A, Nara M, Yamashita J, Ohshima K, Sawada K and Tagawa H:
MicroRNA-150 inhibits tumor invasion and metastasis by targeting
the chemokine receptor CCR6, in advanced cutaneous T-cell lymphoma.
Blood. 123:1499–1511. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ma Y, Zhang P, Wang F, Zhang H, Yang J,
Peng J, Liu W and Qin H: miR-150 as a potential biomarker
associated with prognosis and therapeutic outcome in colorectal
cancer. Gut. 61:1447–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Srivastava SK, Bhardwaj A, Singh S, Arora
S, Wang B, Grizzle WE and Singh AP: MicroRNA-150 directly targets
MUC4 and suppresses growth and malignant behavior of pancreatic
cancer cells. Carcinogenesis. 32:1832–1839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wuerkenbieke D, Wang J, Li Y and Ma C:
miRNA-150 downregulation promotes pertuzumab resistance in ovarian
cancer cells via AKT activation. Arch Gynecol Obstet.
292:1109–1116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stamatopoulos B, Van Damme M, Crompot E,
Dessars B, Housni HE, Mineur P, Meuleman N, Bron D and Lagneaux L:
Opposite prognostic significance of cellular and serum circulating
MicroRNA-150 in patients with chronic lymphocytic leukemia. Mol
Med. 21:123–133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yin QW, Sun XF, Yang GT, Li XB, Wu MS and
Zhao J: Increased expression of microRNA-150 is associated with
poor prognosis in non-small cell lung cancer. Int J Clin Exp
Pathol. 8:842–846. 2015.PubMed/NCBI
|
|
41
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: MiR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Summy JM and Gallick GE: Src family
kinases in tumor progression and metastasis. Cancer Metastasis Rev.
22:337–358. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang F, Luo LJ, Zhang L, Wang DD, Yang SJ,
Ding L, Li J, Chen D, Ma R, Wu JZ and Tang JH: MiR-346 promotes the
biological function of breast cancer cells by targeting SRCIN1 and
reduces chemosensitivity to docetaxel. Gene. 600:21–28. 2017.
View Article : Google Scholar : PubMed/NCBI
|