Introduction
Lung cancer was reported to be the most common
cancer type in males in 2016, and is associated with a poor
prognosis (1). Non-small cell lung
cancer (NSCLC) accounts for 80–85% of all cases of lung cancer,
with squamous cell carcinoma (SCC) and adenocarcinoma (ADE) being
the most common histological subtypes (2,3). Despite
advances in the early detection and therapeutic techniques, the
overall 5-year survival rate is low. Consequently, further
investigations into the mechanisms of initiation and progression,
and the development of prognostic biomarkers and drug targets are
required in order to improve the prognosis for NSCLC and allow for
personalized therapy.
Heat shock protein 90 (HSP90) is one of the most
widespread heat-associated proteins. HSP90 forms flexible
homodimers and its basic structure comprises three parts: The
N-terminal domain, the middle domain and the C-terminal domain.
HSP90 is overexpressed in various cancer types, including
pancreatic, ovarian, breast, lung and endometrial cancer, as well
as oropharyngeal SCC and multiple myeloma (4–7). High
expression of HSP90 was indicated to be a marker of poor prognosis
in lung cancer, esophageal cancer, bladder cancer, melanoma and
leukemia (8–11). Combination therapy with HSP90
inhibitors and conventional photon radiation delays tumor growth
more effectively than radiotherapy alone (12). The HSP90 inhibitor NVP-AUY922 induces
cell death in the lung and is currently undergoing clinical trials
in lung cancer patients (13).
However, the prognostic value of HSP90 in NSCLC has remained to be
determined.
In the present study, the Kaplan-Meier (KM) plotter
database (http://kmplot.com/analysis/index.php?p=service&cancer=lung)
was employed to assess the correlation between HSP90 mRNA
expression and overall survival (OS). At present, the KM plotter
database comprises gene expression information and clinical outcome
parameters of various types of cancer (14–18). The
KM plotter database may be used to analyze individual genes that
may correlate with the OS of NSCLC patients. To date, the KM
plotter database has been used to identify and validate numerous
genes involved in NSCLC (15–18). In
the present study, this database was used to evaluate the
prognostic value of individual HSP90 protein members in patients
with NSCLC.
Materials and methods
The information on the NSCLC patients contained in
the KM plotter database was extracted from the Gene Expression
Omnibus (GEO), the Cancer Biomedical Informatics Grid and The
Cancer Genome Atlas database. The following NSCLC datasets were
obtained from the GEO database: GSE14814, GSE19188, GSE29013,
GSE30219, GSE31210, GSE3141, GSE31908, GSE37745, GSE43580, GSE4573,
GSE50081 and GSE8894 (http://kmplot.com/analysis/index.php?p=service&cancer=lung)
(19). The KM plotter database was
used to analyze the association of the mRNA expression of
individual HSP90 members with relapse-free survival. WinStat 2013
software was used as an analysis tool. Data regarding age, sex,
smoking history, histology, stage, success of surgery, radiotherapy
and applied chemotherapy were recorded for all patients. In
general, five HSP90 subfamily members [HSP90AA1, HSP90AA2,
HSP90AB1, HSP90B1 and tumor necrosis factor receptor-associated
protein 1 (TRAP1)] were included in the KM plotter analysis to
acquire KM survival plots, in which the number of patients at risk
for certain time-points is compared between subgroups with
different gene expression status. The hazard ratio (HR) and 95%
confidence intervals (CI) and log-rank P-values were determined. A
total of 1,926 lung cancer patients were included in the study.
When the P-value was <0.05, the difference was regarded as
statistically significant.
The Database for Annotation, Visualization and
Integrated Discovery (DAVID) version 6.8 (https://david.ncifcrf.gov/) was used to perform gene
ontology (GO) functional annotation analysis and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway analysis of various HSP90
members. Gene-gene interaction networks were generated with
GeneMANIA (http://genemania.org/). Protein-protein
interaction networks were constructed with the Search Tool for the
Retrieval if Interacting Genes and Proteins (STRING; http://string-db.org/).
Results
Prognostic value of HSP90 members in
NSCLC
The HSP90 family comprises five sub-members
(HSP90AA1, HSP90AA2, HSP90AB1, HSP90B1 and TRAP1). Of these, only
HSP90AA2 was not included in the KM plot database, possibly due to
its low levels of expression.
First, the prognostic value of the mRNA expression
of HSP90AA1 was evaluated in the dataset. The valid gene Affymetrix
ID was 211969_at (HSP90AA1). Survival curves were generated to draw
preliminary conclusions regarding the influence of HSP90AA1 on the
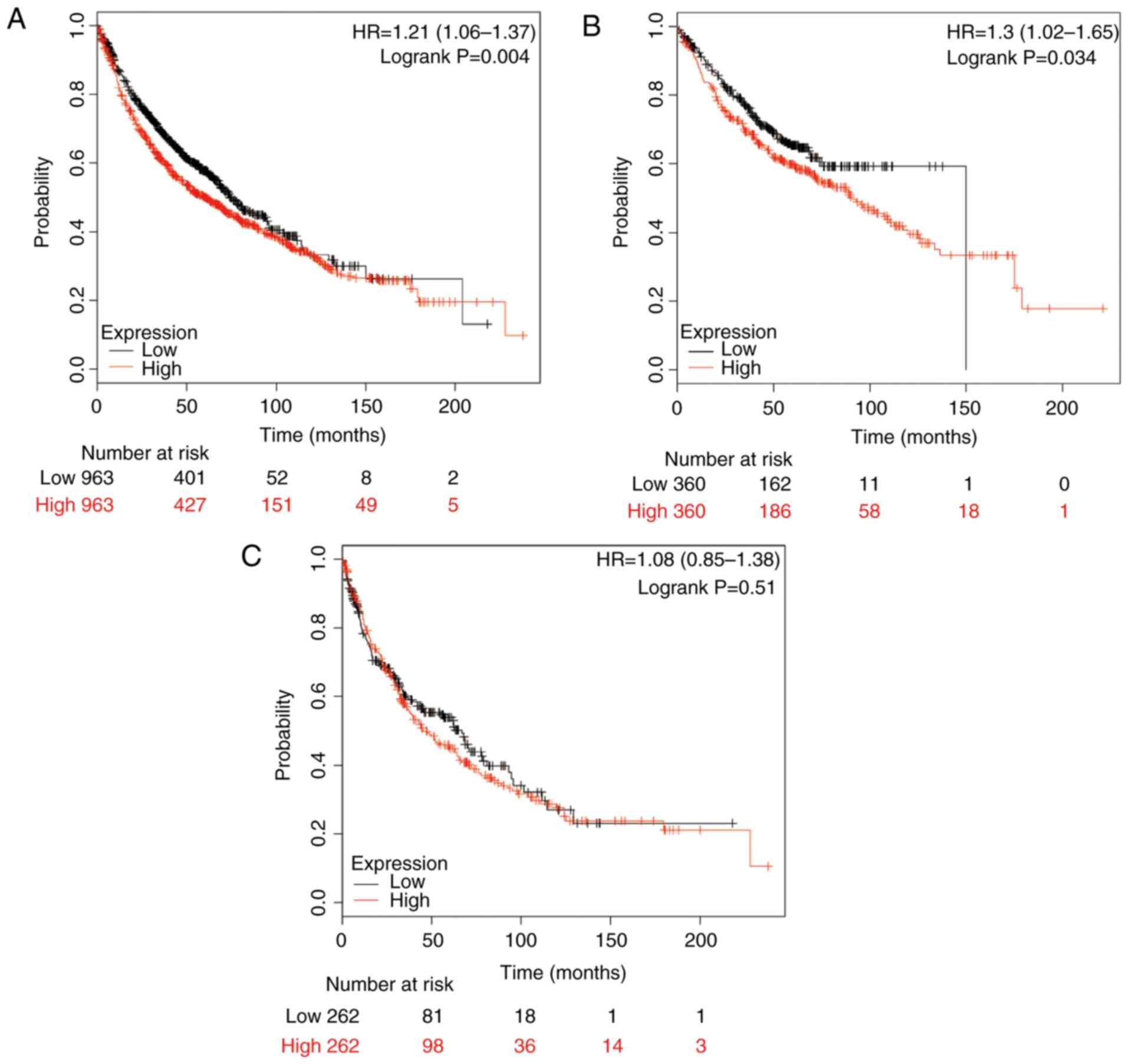
prognosis of patients with NSCLC (n=1,926; Fig. 1A), ADE (n=720; Fig. 1B) and SCC (n=524; Fig. 1C). A high expression of HSP90AA1 mRNA
was significantly associated with a worse OS for all NSCLC cases
(HR, 1.21, 95% CI: 1.06–1.37; P=0.004; Fig. 1A). High expression of HSP90AA1 mRNA
was also significantly associated with poor rates of OS in ADE
patients (HR, 1.30; 95% CI: 1.02–1.65; P=0.034; Fig. 1B), while the association in SCC
patients was not significant (HR, 1.08; 95% CI, 0.85–1.38; P=0.51;
Fig. 1C).
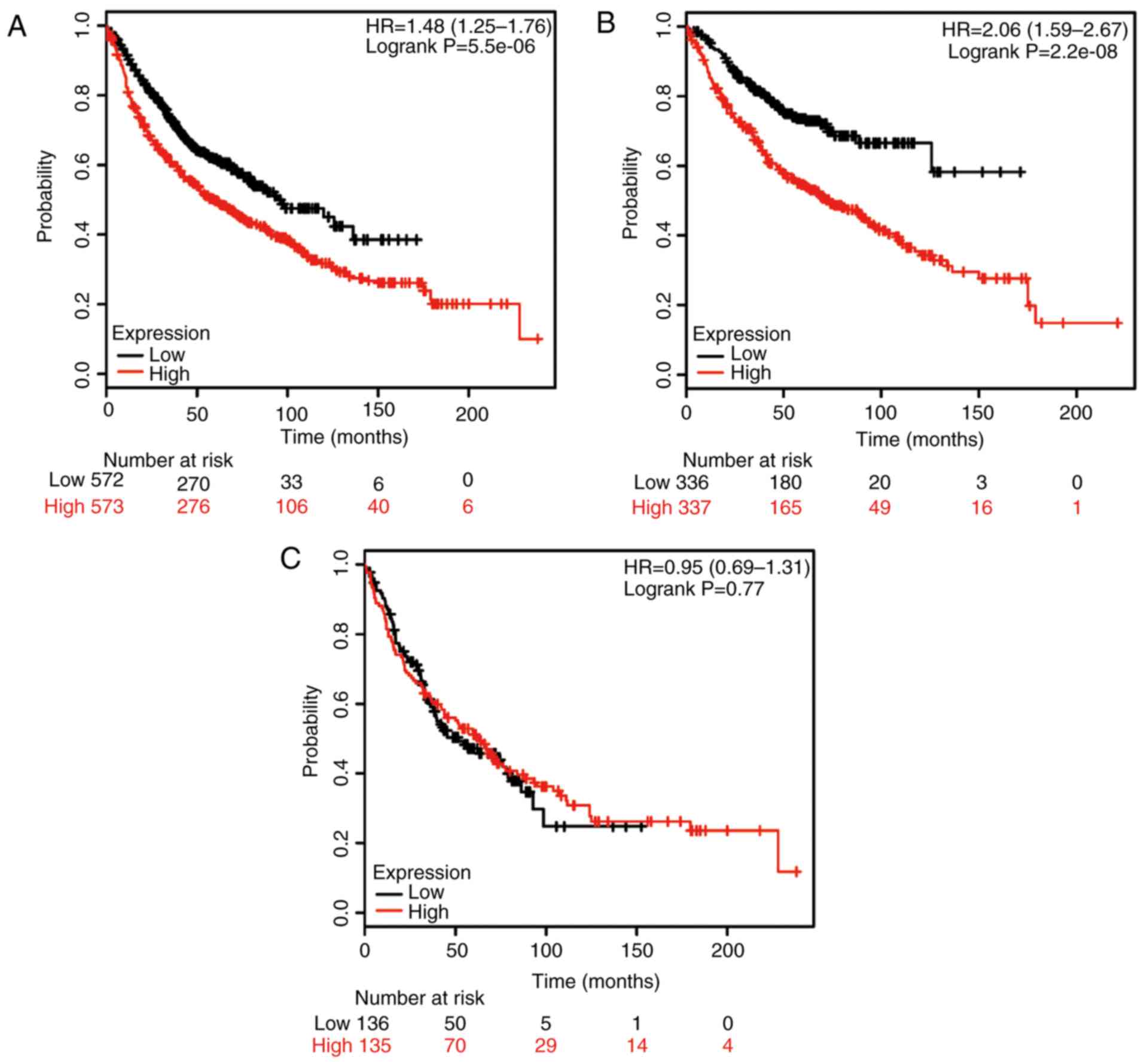
Next, the potential prognostic role of HSP90AB1 mRNA
expression was evaluated using the database. The valid gene
Affymetrix ID was 1557910_at (HSP90AB1). High expression of
HSP90AB1 mRNA was significantly associated with poor rates of OS
for all NSCLC patients (HR, 1.48; 95% CI: 1.25–1.76;
P=5.5×10−6; Fig. 2A).
High expression of HSP90AB1 mRNA was also associated with poor
rates of OS in ADE patients (HR, 2.06; 95% CI: 1.59–2,67;
P=2.2×10−8; Fig. 2B), but
the correlation with OS rates in SCC patients was not significant
(HR, 0.95; 95% CI: 0.69–1.31; P=0.77; Fig. 2C).
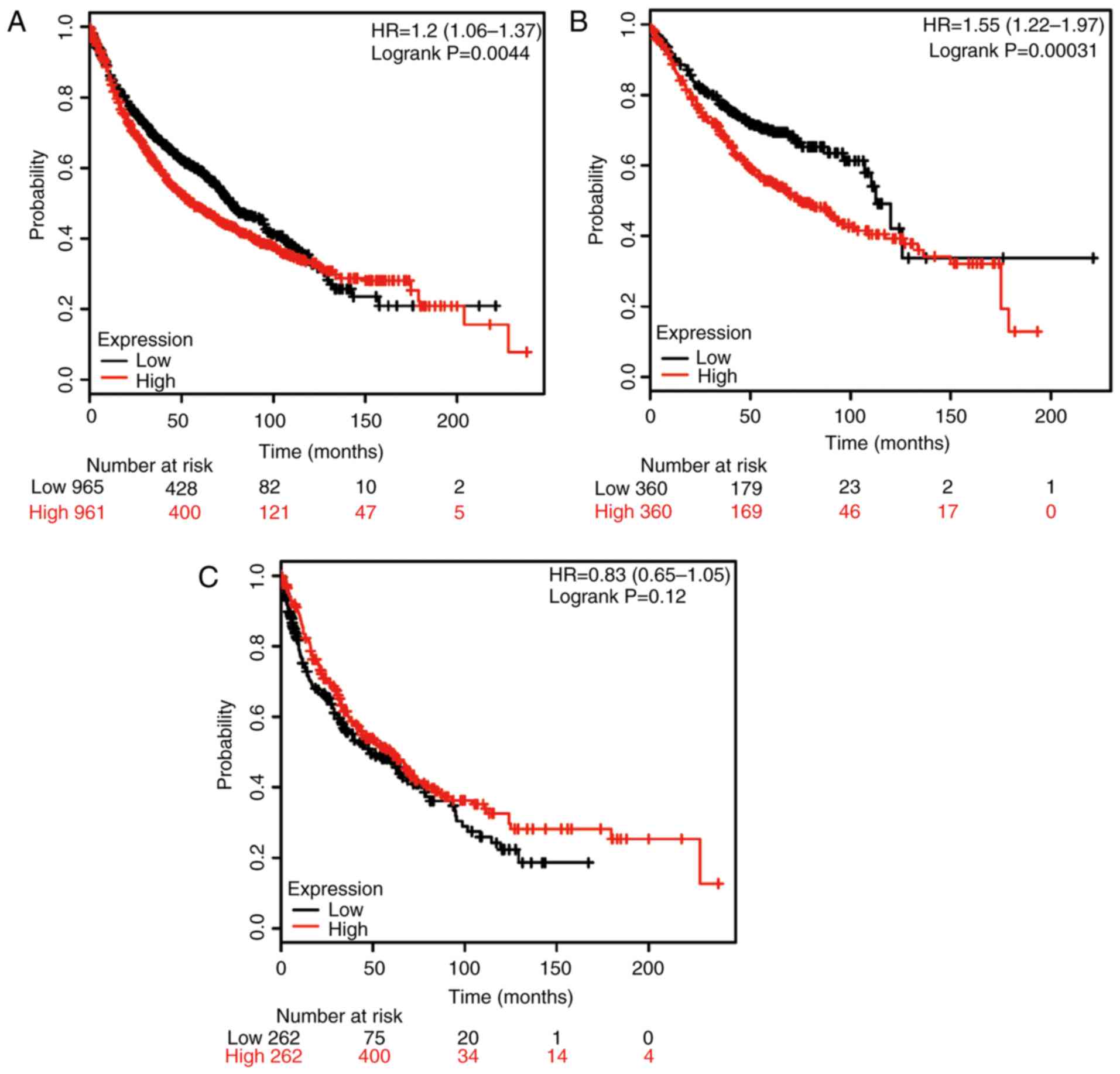
Furthermore, the potential prognostic value of
HSP90B1 mRNA expression was evaluated using the database (Fig. 3). The valid gene Affymetrix ID was
216449_x_at (HSP90B1). High expression of HSP90B1 mRNA was
significantly associated with poor rates of OS for all NSCLC
patients (HR, 1.20; 95% CI: 1.06–1.37; P=0.0044; Fig. 3A), as well as for ADE patients (HR,
1.55; 95% CI: 1.22–1.97; P=0.00031; Fig.
3B); however, no significant association was obtained for SCC
patients (HR, 0.83; 95% CI: 0.65–1.05; P=0.12; Fig. 3C).
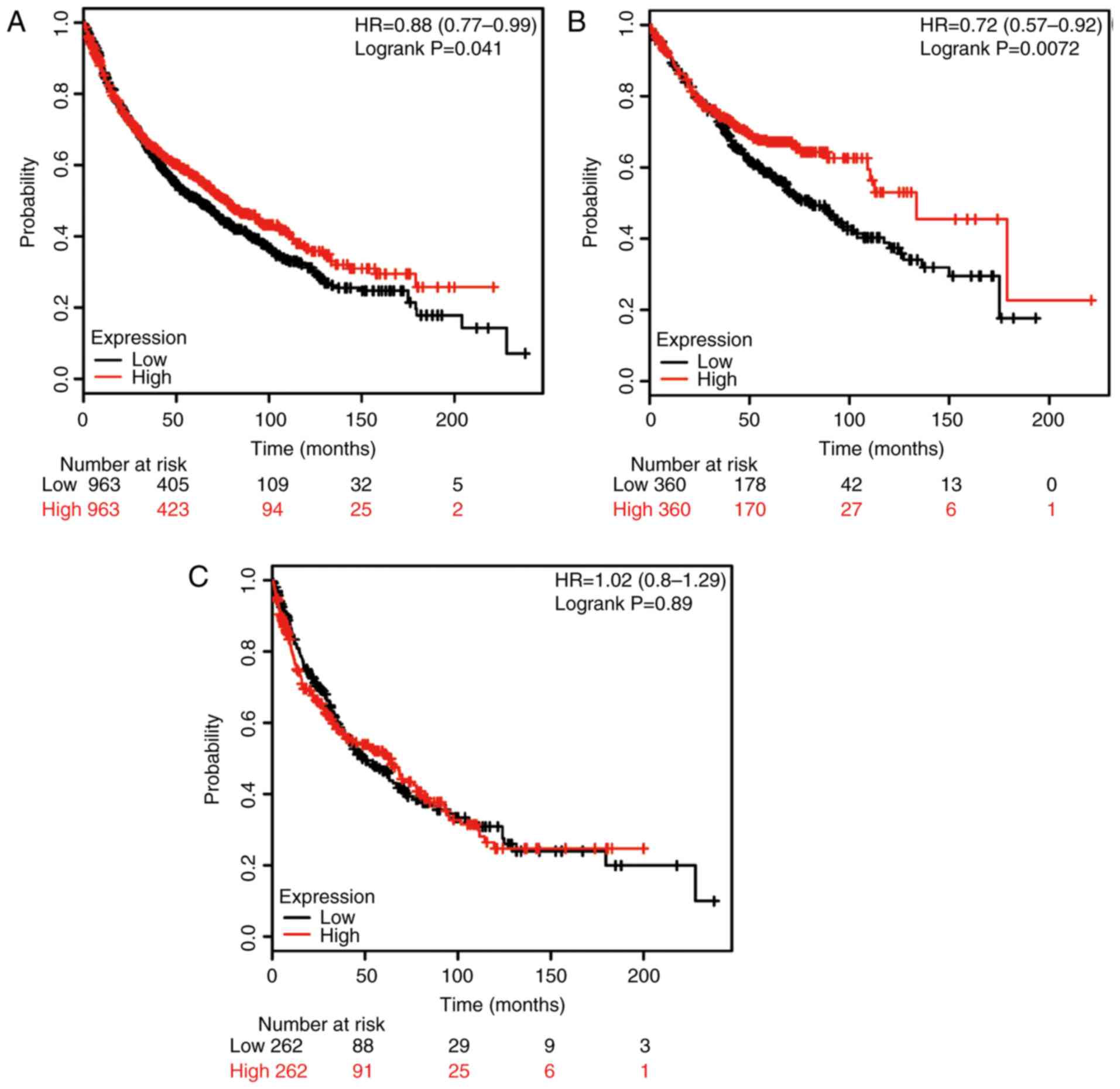
Finally, the potential prognostic role of TRAP1 mRNA
expression was evaluated using the database. The valid gene
Affymetrix ID was 201391_at (TRAP1). High expression of TRAP1 mRNA
was significantly associated with higher OS rates for all patients
(HR, 0.88; 95% CI: 0.77–0.99; P=0.041; Fig. 4A), as well as for ADE patients (HR,
0.72; 95% CI: 0.57–0.92; P=0.0072; Fig.
4B); however, no significant association was identified for SCC
patients (HR, 1.02; 95% CI: 0.80–1.29; P=0.89; Fig. 4C).
Next, to assess whether the prognostic value of the
mRNA expression status of individual HSP90 members depends on other
clinicopathological factors, the influence of high vs. low
expression on OS was determined for NSCLC patients stratified based
on the following parameters: The patients' smoking history,
clinical stage of the disease and chemotherapy status. As presented
in Table I, high expression of
HSP90AA1, HSP90B1 and TRAP1 mRNA had no significant prognostic
value in patients with or without a history of smoking. However, a
high expression of HSP90AB1 was identified in patients with a
smoking history and HSP90B1 in those that did not smoke. Table II reveals that there was only a
significant prognostic value for HSP90AA1 and HSP90AB1 in stage I
patients, and for HSP90B1 in stage I and II patients, but not in
patients at stage III. No significant prognostic values were
observed in all stages of TRAP1 in NSCLC. No significant
association with OS was identified for any of the HSP90 members in
patients that did or did not undergo chemotherapy (Table III).
 | Table I.Influence of HSP90 mRNA expression on
the prognosis of non-small cell lung cancer patients with a history
of smoking (n=820) and those who never smoked (n=205). |
Table I.
Influence of HSP90 mRNA expression on
the prognosis of non-small cell lung cancer patients with a history
of smoking (n=820) and those who never smoked (n=205).
| Gene/smoking
history | HR | 95% CI | P-value |
|---|
| HSP90AA1 |
|
|
|
| Never
smoked | 0.81 | 0.46–1,42 | 0.46 |
|
Smoked | 1.14 | 0.93–1.40 | 0.22 |
| HSP90AB1 |
|
|
|
| Never
smoked | 1.89 | 0.84–4.33 | 0.12 |
|
Smoked | 1.59 | 1.05–2.42 |
0.029 |
| HSP90B1 |
|
|
|
| Never
smoked | 3.51 | 1.86–6.62 |
3.40×10−5 |
|
Smoked | 1.11 | 0.90–1.37 | 0.31 |
| TRAP1 |
|
|
|
| Never
smoked | 0.73 | 0.42–1.28 | 0.27 |
|
Smoked | 0.92 | 0.75–1.13 | 0.43 |
 | Table II.Influence of HSP90 mRNA expression on
the prognosis of non-small cell lung cancer patients with the
clinical stage of I (n=577), II (n=244) and III (n=70). |
Table II.
Influence of HSP90 mRNA expression on
the prognosis of non-small cell lung cancer patients with the
clinical stage of I (n=577), II (n=244) and III (n=70).
| Gene/clinical
stage | HR | 95% CI | P-value |
|---|
| HSP90AA1 |
|
|
|
| I | 1.56 | 1.29–1.88 |
2.70×10−6 |
| II | 1.18 | 0.82–1.71 | 0.38 |
|
III | 1.02 | 0.58–1.79 | 0.94 |
| HSP90AB1 |
|
|
|
| I | 3.28 | 2.29–4.7 |
7.40×10−12 |
| II | 1.66 | 1.04–2.66 |
0.033 |
|
III | 1.34 | 0.64–2.79 | 0.44 |
| HSP90B1 |
|
|
|
| I | 1.86 | 1.40–2.47 |
1.5×10−5 |
| II | 1.1 | 0.76–1.59 | 0.61 |
|
III | 0.92 | 0.54–1.59 | 0.78 |
| TRAP1 |
|
|
|
| I | 0.83 | 0.63–1.09 | 0.17 |
| II | 1.14 | 0.79–1.65 | 0.48 |
|
III | 0.68 | 0.39–1.17 | 0.16 |
 | Table III.Influence of HSP90 mRNA expression on
the prognosis of non-small cell lung cancer patients with
chemotherapy (n=176) and without chemotherapy (n=310). |
Table III.
Influence of HSP90 mRNA expression on
the prognosis of non-small cell lung cancer patients with
chemotherapy (n=176) and without chemotherapy (n=310).
|
Genes/chemotherapy | HR | 95% CI | P-value |
|---|
| HSP90AA1 |
|
|
|
| No | 1.17 | 0.84–1.64 | 0.36 |
|
Yes | 0.91 | 0.60–1.39 | 0.67 |
| HSP90AB1 |
|
|
|
| No | 0.79 | 0.16–3.98 | 0.78 |
|
Yes | 0.73 | 0.23–2.33 | 0.59 |
| HSP90B1 |
|
|
|
| No | 0.86 | 0.62–1.20 | 0.38 |
|
Yes | 0.87 | 0.58–1.32 | 0.52 |
| TRAP1 |
|
|
|
| No | 1.16 | 0.83–1.62 | 0.39 |
|
Yes | 1.27 | 0.84–1.89 | 0.25 |
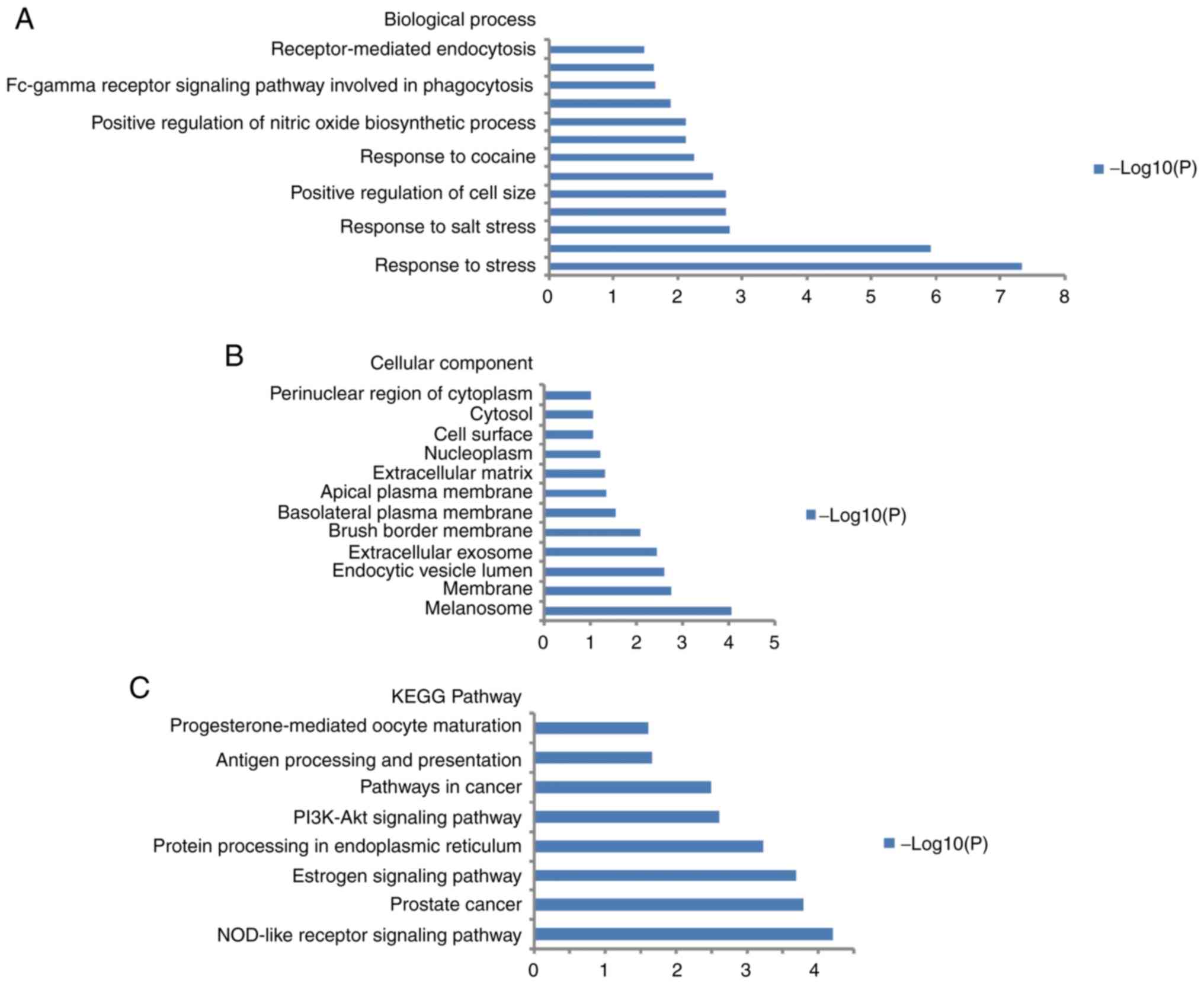
GO functional annotation analysis and
KEGG pathway analysis of HSP90 members
DAVID was used to investigate the biological
functions of the HSP90 members (HSP90AA1, HSP90B1, HSP90AB1 and
TRAP1). GO enrichment analysis in the categories cellular component
and biological process was performed (Fig. 5A and B). The results revealed that
cell size was positively regulated and the response to stress was
enriched. Enrichment of certain genes interacting with HSP90 family
members in certain KEGG pathways was also identified (Fig. 5C) and included the
phosphatidylinositol 3′kinase (PI3K)-Akt signaling pathway in
cancer.
Gene-gene interaction networks were constructed with
GeneMANIA (Fig. 6A) and
protein-protein interaction networks were constructed using STRING
(Fig. 6B). It was indicated that the
HSP90 family has complex interactions, and that sub-members of the
HSP90 family are co-expressed with each other.
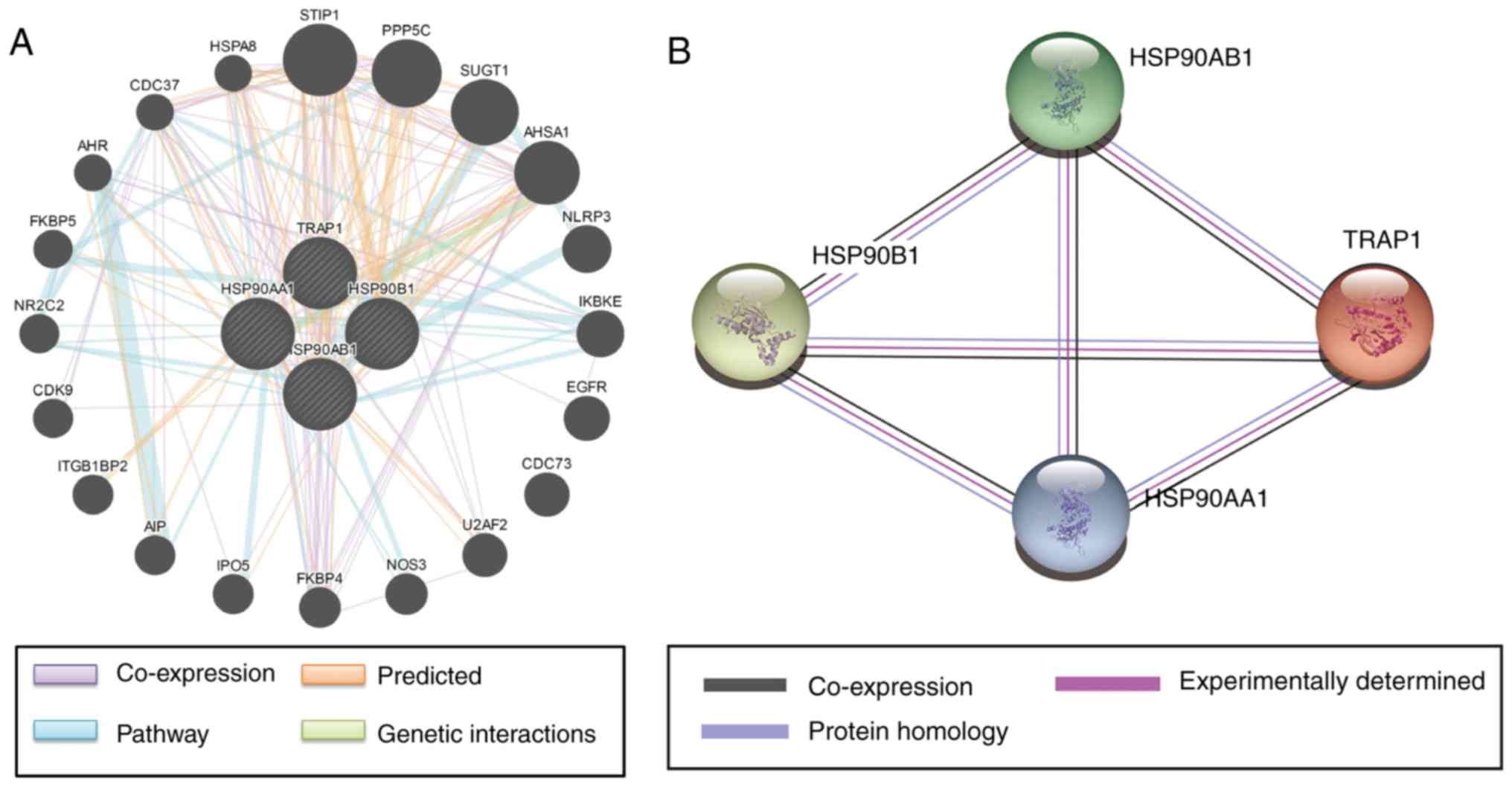 | Figure 6.(A) Gene-gene interaction networks
among selected genes constructed by GeneMANIA. (B) Protein-protein
interaction networks among selected proteins constructed with the
Search Tool for the Retrieval if Interacting Genes and Proteins.
The size of the circles indicates the degree of interaction. HSP,
heat shock protein; TRAP1, tumor necrosis factor
receptor-associated protein 1; STIP1, stress induced phosphoprotein
1; PPP5C, protein phosphatase 5 catalytic; SUGT1, suppressor of G2
allele of sKP1; AHSA1, activator of HSP90 ATPase activity 1; NLRP3,
NLR family pyrin domain containing 3; IKBKE, inhibitor of nuclear
factor kappa B kinase subunit epsilon; EGFR, epidermal growth
factor receptor; CDC73, cell division cycle 73; U2AF2, U2 small
nuclear RNA auxiliary factor 2; NOS3, nitric oxide synthase 3;
FKBP4, FK506 binding protein 4; IPO5, importin 5; AIP, aryl
hydrocarbon receptor interacting protein; ITGB1BP2, integrin
subunit beta 1 binding protein 2; CDK9, cyclin dependent kinase 9;
NR2C2, nuclear receptor subfamily 2 group C member 2; FKBP5, FK506
binding protein 5; AHR, aryl hydrocarbon receptor; CDC37, cell
division cycle 37; HSPA8, heat shock protein family A member 8. |
Discussion
NSCLC is a highly malignant type of cancer with a
poor 5-year survival rate (3,20). HSP90
has been reported to have a role in numerous types of cancer, and
overexpression of this protein has been proposed to enhance
carcinogenesis and affect the prognosis of patients (21–25).
HSP90α, the protein encoded by HSP90AA1, is considered to have an
important regulatory role in tumor invasion and migration (26,27). The
present study indicated an influence of the HSP90 expression status
on the OS rate of NSCLC patients. An association between HSP90 and
the clinical features of NSCLC was also observed.
HSP90α is secreted extracellularly and has an
important role in wound healing and inflammation. These two
processes are frequently hijacked by cancer, leading to malignant
cell motility, metastasis and extravasion (28). HSP90α modulates growth and lung
metastasis by affecting the proliferation, migration and invasion
of isolated primary carcinoma cells (29). One study indicated that the level of
HSP90α in the plasma of patients with the advanced stages of
malignant tumors was relatively high (30). The expression of HSP90α has been
reported to correlate with the degree of malignancy of esophageal
SCC and the prognosis of affected patients (8). Furthermore, the present study suggested
that a high expression of HSP90AA1 mRNA was associated with a poor
prognosis in ADE patients, but not in SCC patients. Overall, it may
be concluded that high expression of HSP90α mRNA correlates with
poorer rates of OS in all NSCLC patients combined.
In previous study, the expression rate of HSP90AB1
in lung ADE tissues was determined to be obviously higher than that
in lung SCC tissues, and its overexpression was associated with
poor prognosis for lung ADE patients (31). In the present study, high expression
of HSP90AB1 mRNA was significantly associated with a poorer OS rate
in ADE patients but not in SCC patients.
HSP90B1 is essential for mediating the contribution
of tumor-associated macrophages to inflammatory colon tumorigenesis
in mice (32). Overexpression of
HSP90β leads to significantly increased proliferation, migration,
invasion and tube formation of human endothelial cells. HSP90β was
reported to promote endothelial cell-dependent tumor angiogenesis
by increasing vascular endothelial growth factor receptor
expression (33). Serum HSP90B1
levels were reported to be significantly elevated in dogs with
mammary tumors compared with those in healthy controls (34). Another study demonstrated that
HSP90B1 is an oncogenic chaperone in hepatocyte carcinogenesis
(35). Overexpression of HSP90B1 may
be associated with the poor survival rate of hepatocellular
carcinoma patients (36). In
patients with early-stage liver cancer, a considerable increase in
HSP90B1 is associated with tumor metastasis and recurrence
(37). Analysis of survival rates
demonstrated that HSP expression was an independent unfavorable
prognostic factor for gall bladder cancer (38). Furthermore, HSP90B1 overexpression
was indicated to be an accurate predictive factor for shorter OS
and disease-free survival in NSCLC patients (39).
High TRAP1 expression has previously been reported
to be an adverse prognostic factor for patients with NSCLC
(40,41). Conversely, the present results
indicated that TRAP1 mRNA expression is correlated with an improved
prognosis in all NSCLC patients combined, as well as ADE patients
alone. The significance of TRAP1 in NSCLC therefore requires
further investigation.
Nicotine, a major addictive component of tobacco
smoke, has been reported to induce cell cycle progression,
angiogenesis and metastasis during cancer (42,43).
However, the potential role of nicotine in HSP90 activation in
NSCLC patients has not been previously addressed, to the best of
our knowledge. Nicotine is known to induce apoptosis in human
cells, possibly through increased HSP90α expression (44). In the present study, a high
expression of HSP90AB1 mRNA was identified to be a significant
prognostic factor in NSCLC patients with a smoking history, but not
in those who never smoked. However, high expression of HSP90B1 mRNA
was identified to be a significant prognostic factor in NSCLC
patients who never smoked, but not in those with a smoking history.
Further research is required to confirm these results and identify
the possible underlying mechanisms.
Previous studies have proposed that HSP90 may be a
crucial drug target for the treatment of NSCLC. For instance,
inhibition of the mitochondrial HSP90 network may be an effective
treatment for highly refractory tumors (45), and topical application of the HSP90
inhibitor 17AAG is effective in preventing epidermal hyperplasia
and SCC (46).
The current study has some limitations that need
clarification. A stratified analysis for all NSCLC patients
combined was performed, but included patients with SCC, which
demonstrated no significance. Therefore, a stratified survival
analysis for each type of NSCLC should be performed separately with
a large sample size in the future.
In conclusion, the results of the present study
suggest that HSP90AA1, HSP90AB1 and HSP90B1 may be potential
biomarkers for the prognosis of NSCLC. However, the present results
were obtained using bioinformatics methods and further research is
required to explore the underlying molecular mechanisms in the
future.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National
Natural Science Foundation of China (grant nos. 81460460, 81360405
and 81760542), the Foundation of Sharing Project Based on Tumor
Precise Radiotherapy (grant no. ZY18076006), the China Postdoctoral
Science Foundation (grant no. 2016M602918XB) and the Research
Foundation of the Science and Technology Department of Guangxi
Province, China (grant nos. 2016GXNSFAA380252 and
2014GXNSFBA118114).
Authors' contributions
RW designed the study and performed bioinformatics
analysis. KL analyzed the data and wrote the manuscript. MK, WQ and
JL analyzed the data. All authors read and approved the final
manuscript.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Ethical approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Minna JD, Roth JA and Gazdar AF: Focus on
lung cancer. Cancer Cell. 1:49–52. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burrows F, Zhang H and Kamal A: Hsp90
activation and cell cycle regulation. Cell Cycle. 3:1530–1536.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kolosenko I, Grander D and Tamm KP: IL-6
activated JAK/STAT3 pathway and sensitivity to Hsp90 inhibitors in
multiple myeloma. Curr Med Chem. 21:3042–3047. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Patel K, Wen J, Magliocca K, Muller S, Liu
Y, Chen ZG, Saba N and Diaz R: Heat shock protein 90 (HSP90) is
overexpressed in p16-negative oropharyngeal squamous cell
carcinoma, and its inhibition in vitro potentiates the effects of
chemoradiation. Cancer Chemother Pharmacol. 74:1015–1022. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shi Y, Liu X, Lou J, Han X, Zhang L, Wang
Q, Li B, Dong M and Zhang Y: Plasma levels of heat shock protein 90
alpha associated with lung cancer development and treatment
responses. Clin Cancer Res. 20:6016–6022. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang T, Chen S, Han H, Li H, Huang Z,
Zhang J, Yin Q, Wang X, Ma X, Dai P, et al: Expression of
Hsp90alpha and cyclin B1 were related to prognosis of esophageal
squamous cell carcinoma and keratin pearl formation. Int J Clin Exp
Pathol. 7:1544–1552. 2014.PubMed/NCBI
|
|
9
|
Tian WL, He F, Fu X, Lin JT, Tang P, Huang
YM, Guo R and Sun L: High expression of heat shock protein 90 alpha
and its significance in human acute leukemia cells. Gene.
542:122–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McCarthy MM, Pick E, Kluger Y,
Gould-Rothberg B, Lazova R, Camp RL, Rimm DL and Kluger HM: HSP90
as a marker of progression in melanoma. Ann Oncol. 19:590–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Záčková M, Moučková D, Lopotová T,
Ondračková Z, Klamová H and Moravcová J: Hsp90-a potential
prognostic marker in CML. Blood Cells Mol Dis. 50:184–189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hirakawa H, Fujisawa H, Masaoka A, Noguchi
M, Hirayama R, Takahashi M, Fujimori A and Okayasu R: The
combination of Hsp90 inhibitor 17AAG and heavy-ion irradiation
provides effective tumor control in human lung cancer cells. Cancer
Med. 4:426–436. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johnson ML, Yu HA, Hart EM, Weitner BB,
Rademaker AW, Patel JD, Kris MG and Riely GJ: Phase I/II study of
HSP90 inhibitor AUY922 and erlotinib for EGFR-mutant lung cancer
with acquired resistance to epidermal growth factor receptor
tyrosine kinase inhibitors. J Clin Oncol. 33:1666–1673. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Györffy B, Lanczky A, Eklund AC, Denkert
C, Budczies J, Li Q and Szallasi Z: An online survival analysis
tool to rapidly assess the effect of 22,277 genes on breast cancer
prognosis using microarray data of 1,809 patients. Breast Cancer
Res Treat. 123:725–731. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang ZC, Li H, Sun ZQ, Zheng J, Zhao RK,
Chen J, Sun SG and Wu CJ: Distinct prognostic roles of HSPB1
expression in non-small cell lung cancer. Neoplasma. 65:161–166.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schmidt LH, Brand C, Stucke-Ring J,
Schliemann C, Kessler T, Harrach S, Mohr M, Gorlich D, Marra A,
Hillejan L, et al: Potential therapeutic impact of CD13 expression
in non-small cell lung cancer. PLoS One. 12:e01771462017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie S, Shen C, Tan M, Li M, Song X and
Wang C: Systematic analysis of gene expression alterations and
clinical outcomes of adenylate cyclase-associated protein in
cancer. Oncotarget. 8:27216–27239. 2017.PubMed/NCBI
|
|
18
|
Hou GX, Liu P, Yang J and Wen S: Mining
expression and prognosis of topoisomerase isoforms in
non-small-cell lung cancer by using Oncomine and Kaplan-Meier
plotter. PLoS One. 12:e01745152017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Győrffy B, Surowiak P, Budczies J and
Lanczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lukas RV, Gondi V, Kamson DO, Kumthekar P
and Salgia R: State-of-the-art considerations in small cell lung
cancer brain metastases. Oncotarget. 8:71223–71233. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Calderwood SK and Neckers L: Hsp90 in
cancer: Transcriptional roles in the nucleus. Adv Cancer Res.
129:89–106. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang B, Wang J, Huang Z, Wei P, Liu Y,
Hao J, Zhao L, Zhang F, Tu Y and Wei T: Aberrantly upregulated
TRAP1 is required for tumorigenesis of breast cancer. Oncotarget.
6:44495–44508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Palmieri C, Mancini M, Benazzi C and Della
Salda L: Heat shock protein 90 is associated with hyperplasia and
neoplastic transformation of canine prostatic epithelial cells. J
Comp Pathol. 150:393–398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Badowska-Kozakiewicz AM and Malicka E:
Immunohistochemical evaluation of expression of heat shock proteins
HSP70 and HSP90 in mammary gland neoplasms in bitches. Pol J Vet
Sci. 15:209–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee JH, Kang KW, Kim JE, Hwang SW, Park
JH, Kim SH, Ji JH, Kim TG, Nam HY, Roh MS, et al: Differential
expression of heat shock protein 90 isoforms in small cell lung
cancer. Int J Clin Exp Pathol. 8:9487–9493. 2015.PubMed/NCBI
|
|
26
|
Wong DS and Jay DG: Emerging roles of
extracellular Hsp90 in cancer. Adv Cancer Res. 129:141–163. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sims JD, McCready J and Jay DG:
Extracellular heat shock protein (Hsp)70 and Hsp90α assist in
matrix metalloproteinase-2 activation and breast cancer cell
migration and invasion. PLoS One. 6:e188482011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eustace BK, Sakurai T, Stewart JK,
Yimlamai D, Unger C, Zehetmeier C, Lain B, Torella C, Henning SW,
Beste G, et al: Functional proteomic screens reveal an essential
extracellular role for hsp90 alpha in cancer cell invasiveness. Nat
Cell Biol. 6:507–514. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vartholomaiou E, Madon-Simon M, Hagmann S,
Muhlebach G, Wurst W, Floss T and Picard D: Cytosolic Hsp90α and
its mitochondrial isoform Trap1 are differentially required in a
breast cancer model. Oncotarget. 8:17428–17442. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Song X, Zhuo W, Fu Y, Shi H, Liang
Y, Tong M, Chang G and Luo Y: The regulatory mechanism of
Hsp90alpha secretion and its function in tumor malignancy. Proc
Natl Acad Sci USA. 106:21288–21293. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang M, Feng L, Li P, Han N, Gao Y and
Xiao T: Hsp90AB1 protein is overexpressed in non-small cell lung
cancer tissues and associated with poor prognosis in lung
adenocarcinoma patients. Zhongguo Fei Ai Za Zhi. 19:64–69. 2016.(In
Chinese). PubMed/NCBI
|
|
32
|
Morales C, Rachidi S, Hong F, Sun S,
Ouyang X, Wallace C, Zhang Y, Garret-Mayer E, Wu J, Liu B and Li Z:
Immune chaperone gp96 drives the contributions of macrophages to
inflammatory colon tumorigenesis. Cancer Res. 74:446–459. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng J, Liu Y, Han J, Tan Q, Chen S, Qiao
K, Zhou H, Sun T and Yang C: Hsp90β promoted endothelial
cell-dependent tumor angiogenesis in hepatocellular carcinoma. Mol
Cancer. 16:722017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sunil Kumar BV, Bhardwaj R, Mahajan K,
Kashyap N, Kumar A and Verma R: The overexpression of Hsp90B1 is
associated with tumorigenesis of canine mammary glands. Mol Cell
Biochem. 440:23–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rachidi S, Sun S, Wu BX, Jones E, Drake
RR, Ogretmen B, Cowart LA, Clarke CJ, Hannun YA, Chiosis G, et al:
Endoplasmic reticulum heat shock protein gp96 maintains liver
homeostasis and promotes hepatocellular carcinogenesis. J Hepatol.
62:879–888. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang Z, Zhuang L, Szatmary P, Wen L, Sun
H, Lu Y, Xu Q and Chen X: Upregulation of heat shock proteins
(HSPA12A, HSP90B1, HSPA4, HSPA5 and HSPA6) in tumour tissues is
associated with poor outcomes from HBV-related early-stage
hepatocellular carcinoma. Int J Med Sci. 12:256–263. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hou J, Li X, Li C, Sun L, Zhao Y, Zhao J
and Meng S: Plasma membrane gp96 enhances invasion and metastatic
potential of liver cancer via regulation of uPAR. Mol Oncol.
9:1312–1323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Y, Chen C, Ma C, Sun S, Zhang J and
Sun Y: Expression of heat-shock protein gp96 in gallbladder cancer
and its prognostic clinical significance. Int J Clin Exp Pathol.
8:1946–1953. 2015.PubMed/NCBI
|
|
39
|
Xu Y, Chen Z, Zhang G, Xi Y, Sun R, Wang
X, Wang W, Chai F and Li X: HSP90B1 overexpression predicts poor
prognosis in NSCLC patients. Tumour Biol. 37:14321–14328. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Agorreta J, Hu J, Liu D, Delia D, Turley
H, Ferguson DJ, Iborra F, Pajares MJ, Larrayoz M, Zudaire I, et al:
TRAP1 regulates proliferation, mitochondrial function, and has
prognostic significance in NSCLC. Mol Cancer Res. 12:660–669. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sisinni L, Maddalena F, Condelli V,
Pannone G, Simeon V, Li Bergolis V, Lopes E, Piscazzi A, Matassa
DS, Mazzoccoli C, et al: TRAP1 controls cell cycle G2-M transition
through the regulation of CDK1 and MAD2 expression/ubiquitination.
J Pathol. 243:123–134. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schaal C and Chellappan SP:
Nicotine-mediated cell proliferation and tumor progression in
smoking-related cancers. Mol Cancer Res. 12:14–23. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cardinale A, Nastrucci C, Cesario A and
Russo P: Nicotine: Specific role in angiogenesis, proliferation and
apoptosis. Crit Rev Toxicol. 42:68–89. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu YP, Kita K and Suzuki N: Involvement of
human heat shock protein 90 alpha in nicotine-induced apoptosis.
Int J Cancer. 100:37–42. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Siegelin MD: Inhibition of the
mitochondrial Hsp90 chaperone network: A novel, efficient treatment
strategy for cancer? Cancer Lett. 333:133–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Singh A, Singh A, Sand JM, Bauer SJ,
Hafeez BB, Meske L and Verma AK: Topically applied Hsp90 inhibitor
17AAG inhibits UVR-induced cutaneous squamous cell carcinomas. J
Invest Dermatol. 135:1098–1107. 2015. View Article : Google Scholar : PubMed/NCBI
|