Introduction
Phthalates are a group of industrial chemicals that
are used as plasticizers, lubricants and solvents, and impart
favorable characteristics to various consumer products, including
food packaging, pharmaceuticals, medical devices, construction
materials and household furnishings (1). In 2006, the total production of
plasticizer phthalates was ~4.06 million metric tons (2). As a result of ubiquitous exposure,
phthalates and their metabolites are detected in biological samples
from humans, including urine and serum (3). Individuals are continuously exposed to
phthalates via ingestion, inhalation, intravenous injection and
skin absorption, beginning in utero, and dietary intake from
contaminated foods is thought to be the most important means of
exposure (4–6). Di-(2-ethylhexyl) phthalate (DEHP) is
the most abundant phthalate in the environment (7). Phthalates may provoke various adverse
effects, including endocrine disruption, as well as developmental
and reproductive dysfunction (8).
The mechanisms of their toxic effects on the reproductive system
have remained to be fully elucidated; however, certain effects are
potentially associated with anti-androgenic activity (9).
The molecular mechanisms of phthalate toxicity have
been heavily investigated; however, previous studies have only
focused on one or several cellular and molecular biological
processes, or the expression of a small number of genes/proteins.
It is difficult to collate this information to systematically
explain the molecular mechanisms of phthalate toxicity. In recent
years, next-generation sequencing (NGS) technology has emerged to
provide an effective method for high-throughput sequence
determination, and has markedly improved the speed and efficiency
of the identification of novel key genes or biomarkers (10,11).
Previous studies have indicated that phthalates
decrease the number of Sertoli cells per tubule in testis
cross-sections in a dose dependent manner, promote disruption of
cellular tight junction proteins (12), and decrease androgen receptor
(13) and follicle-stimulating
hormone (FSH) receptor expression. Sertoli cells appear to be the
primary target of DEHP and mono-(2-ethylhexyl) phthalate (MEHP), an
active metabolite of DEHP, in rodents (14–16). TM4
is an immature Sertoli cell line derived from the testis of an
immature BALB/c mouse, and it is a particularly good model for
investigating endocrine disruption, including the effects of
phthalates, as the cells maintain the ability to respond to FSH
stimulation and express androgen and estrogen receptors (17). Considering that NGS technology is
recognized as a reliable method to determine mechanisms of
drug-induced activity and to identify biomarkers (18), the present study performed
whole-transcriptome sequencing of control- and MEHP-treated TM4
cells to identify differentially expressed genes (DEGs) and to
provide novel insight into the toxic mechanisms of phthalates.
Materials and methods
Materials and chemicals
The TM4 normal mouse Sertoli cell line was obtained
from the American Type Culture Collection (Manassas, VA, USA).
Fetal bovine serum (FBS) was purchased from Thermo Fisher
Scientific, Inc. (Waltham, MA, USA). MEHP standard reagent was
obtained from Shanghai Aladdin Biochemical Technology Co., Ltd.
(Shanghai, China). The Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) apoptosis kit (cat. no. AD10) and Cell
Counting Kit-8 (CCK-8; cat. no. CK04) were obtained from Dojindo
Molecular Technologies, Inc. (Kumamoto, Japan). The
TruSeq® RNA LT Sample Prep Kit v2 (cat. no. RS-122),
ruSeq PE Cluster Kit v3-cBot-HS (cat. no. GD-401-3001) and TruSeq
SBS Kit v3-HS (cat. no. FC-401) used for RNA library construction
were obtained from Illumina, Inc. (San Diego, CA, USA). The SYBR
Premix Ex Taq reagent kit (cat. no. RR420) and PrimeScript RT
reagent kit (cat. no. RR037) were purchased from Takara
Biotechnology Co., Ltd. (Dalian, China).
Cell culture
TM4 cells were cultured at 37°C with 5%
CO2 in Dulbecco's modified Eagle's medium supplemented
with 10% FBS and 0.5% penicillin-streptomycin.
Apoptosis analysis
The cells (>1×106) were seeded on a
6-well cell culture plate. Following cell attachment at 60–70%,
MEHP dissolved in dimethylsufloxide (DMSO) was added. The final
concentrations used were 1×10−4, 1×10−5 and
1×10−6 mol/l (1% v/v DMSO). An equal volume of DMSO was
added to the control group. Each treatment group was set up in
three wells. After 48 h, apoptosis was detected using a flow
cytometer (FACS Calibur; Becton, Dickinson and Company, Franklin,
Lakes, NJ, USA) according to the instructions of the Annexin V
FITC/PI apoptosis kit.
Cell proliferation assay
TM4 cells were seeded in each well of a 96-well
plate (3,000 cells per well) and incubated for 24 h at 37°C to
allow for cell attachment. After 24 h, cells were exposed to
serially diluted concentrations of MEHP (1×10−3,
5×10−4, 2.5×10−4, 1.25×10−4,
6.25×10−5, 3.13×10−5, 1.56×10−5,
7.81×10−6, 3.91×10−6 and 1.95×10−6
mmol/l; con1-con10, respectively). Each treatment group was set up
in three wells. The cells were cultured in medium containing 10%
FBS and 1% v/v DMSO. After 24 or 48 h, the relative cell number was
estimated using a CCK-8 proliferation kit. The absorbance at 450 nm
was read using an Envision plate reader (PerkinElmer, Inc.,
Waltham, MA, USA) for cell counting.
RNA library construction and
sequencing
Based on the results of the proliferation assay, TM4
cells were exposed to 1×10−3 (con1) and
2.5×10−4 (con3) mmol/l MEHP for 48 h. The cells were
then subjected to transcriptome sequencing. Total RNA from TM4
cells in the treated and control group was isolated using TRIzol
reagent (Thermo Fisher Scientific, Inc.). The RNA quantity and
quality were examined using a Nanodrop 2000 instrument (Nanodrop
Technologies; Thermo Fisher Scientific, Inc.) and electrophoresis,
respectively. A total of three RNA libraries were constructed using
Illumina TruSeq Stranded mRNA library prep kits according to the
manufacturer's protocols and all sequencing was performed using the
Illumina Hiseq2500 (Illumina, Inc.). The experimental results were
processed using FastQC0.11.2 software (Babraham Bioinformatics,
Cambridge, UK).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
DEGs were selected from the results of the
transcriptome sequencing (P<0.005) and verified by RT-qPCR.
Primers (details in Table II) were
designed to span intron boundaries to avoid amplification of
genomic DNA and to amplify all known isoforms of each gene based on
the National Center for Biotechnology Information (NCBI) Reference
Sequence Database (RefSeq; http://www.ncbi.nlm.nih.gov/refseq/). Total RNA from
TM4 cells in the treated and control group was isolated using
TRIzol reagent (Thermo Fisher Scientific, Inc.; cat. no. 15596026).
RNA was transcribed into DNA using Transcriptor First Stand cDNA
Synthesis kit (Roche Diagnostics, Basel, Switzerland; cat. no.
04897030001). The RT-Primers were Anchored-oligo(dT)18 Primer and
Random Hexamer Primer. Primers were synthesized by Sigma-Aldrich
(Merck KGaA, Darmstadt, Germany). qPCR was performed in triplicate
using 1 µl diluted complementary (c)DNA template (400 ng) in a µl
total volume of 10 µl. Reactions were performed in a 384-well plate
format using the ABI Viia7 Real-time PCR system (Thermo Fisher
Scientific, Inc.) and Sensifast SYBR Lo-ROX Master Mix (Bioline;
Meridian Life Science, Inc., Memphis, TN, USA). A third-step
thermocycling protocol was used, with an initial hold at 95°C for 2
min to activate the polymerase, followed by 95°C for 2 min, 40
cycles of 95°C for 5 sec, 60°C for 20 sec, 72°C for 20 sec. The
2−ΔΔCq method was used for data normalization
(19). GAPDH was used as the
housekeeping gene.
 | Table II.Primers used for polymerase chain
reaction analysis. |
Table II.
Primers used for polymerase chain
reaction analysis.
| Primer name | Primer sequence
(5′-3′) | Product length
(bp) |
|---|
| COL1A1-S |
TCAGCTTTGTGGACCTCCG | 197 |
| COL1A1-A |
ATTGCATTGCACGTCATCG |
|
| ACTN1-S |
AGGACACCTTCATCGTACATACCAT | 198 |
| ACTN1-A |
CCTGAGGCGTGATGGTTGTATAG |
|
| VCL-S |
GTTGGAAAAGAGACTGTTCAGACCA | 171 |
| VCL-A |
CCTAGAGCCGTCAATGAGGTAATC |
|
| CAV1-S |
GCAGACGAGGTGACTGAGAAGC | 224 |
| CAV1-A |
AAGATCGTAGACAACAAGCGGTAA |
|
| MYL9-S |
ACTTTTCTTCTCTGCAGCAGGG | 253 |
| MYL9-A |
ATACTCGTCTGTGGGGTTCTTCC |
|
| THBS1-S |
TTCCTGATGGTGAATGCTGCC | 158 |
| THBS1-A |
GCCCTCGCATCTGTTGTTGA |
|
| COL3A1-S |
GTTTCTTCTCACCCTTCTTCATCC | 247 |
| COL3A1-A |
AGGCTGTGGGCATATTGCA |
|
| SDC4-S |
CCCTCAGAGCCCAAGGAACT | 179 |
| SDC4-A |
AAGAGGATGCCTACCACGCC |
|
| CD44-S |
ATCCCTCCGTTTCATCCAGC | 226 |
| CD44-A |
CATGGTGGGTAAGGTACTGTTGAA |
|
| FDPS-S |
AGGAGGTCCTAGAGTACAATGCC | 195 |
| FDPS-A |
TGAGGGAAGAGTCCATGATGTC |
|
| IDI1-S |
CCATTAAGTAACCCAGGCGAG | 162 |
| IDI1-A |
ACCATCAGATTGGGCCTTGT |
|
| CYP51-S |
ATTTGGAGCTGGGCGTCAT | 262 |
| CYP51-A |
TTCTCAGAGGCTTCATTCTTTGC |
|
| SQLE-S |
ACCCGGAAGTGATCATCGTG | 203 |
| SQLE-A |
ATGGGCATTGAGACCTTCTACTGT |
|
| SPP1-S |
TGTCCTCTGAAGAAAAGGATGACTT | 238 |
| SPP1-A |
TCGACTGTAGGGACGATTGGAG |
|
| GAPDH-S |
GCCTTCCGTGTTCCTACC | 183 |
| GAPDH-A |
AGAGTGGGAGTTGCTGTTG |
Analysis of transcriptome sequencing
results
Sequencing data were checked for sequencing quality
using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Adaptors and poor-quality sequences were then removed using Trim
Galore v0.3.7 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/).
Pairs of trimmed reads were aligned against the human genome (hg19
build) using Tophat (v 2.1; http://ccb.jhu.edu/software/tophat/index.shtml).
Mapped fragments per kilobase of transcript per
million (FPKM) of each gene or transcript variant were calculated
using Cufflink software (http://cole-trapnell-lab.github.io/cufflinks/).
Cuffdiff software produces the P-value and q-value. The number of
DEGs screened by the Q-value may be too small for enrichment
analysis. More DEGs may be obtained by using the P-value, which is
conducive to subsequent enrichment analysis. Furthermore, certain
randomly emerging DEGs can be filtered out by enrichment analysis
to avoid their influence on the experimental result. Therefore, the
P-value was selected as a screening index, with a cutoff criterion
of P < 0.05, which was used to screen for DEGs. Functional
enrichment analysis of DEGs was performed using Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway analysis (www.genome.jp/kegg) and gene ontology (GO) analysis
(http://www.geneontology.org/).
Statistical analysis
All data were analyzed using SPSS 18.0 software
(SPSS, Inc., Chicago, IL, USA). Fisher's exact test was used to
identify the DEGs. The Benjamini-Hochberg correction was used to
control for false-positives. Normally distributed data are
presented as the mean ± standard deviation, while a Kruskal-Wallis
test was used to analyze data with an abnormal distribution, which
are presented as the median and interquartile range (Q).
Shapiro-Wilk test was used to examine whether data were normally
distributed. First, one-way analysis of variance was used to
determine significant differences among all groups, including one
control group and two treated groups. If the difference was
statistically significant, a Fisher's least-significant differences
test was used for further mutual and random comparison among the
three groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Analysis of apoptosis
TM4 cells treated with different concentrations of
MEHP for 48 h were analyzed using flow cytometry to detect
apoptosis. The results indicated that MEHP at concentrations of
≤1×10−4 mol/l did not significantly affect the viability
of the cells (χ2=7.421; P=0.06; Table I). Therefore, MEHP was used at this
non-toxic concentration range for the subsequent cell proliferation
assays.
 | Table I.Results of the flow cytometric early
apoptosis (%) assay. |
Table I.
Results of the flow cytometric early
apoptosis (%) assay.
| MEHP, mol/l | M, %
(Xmin, Xmax) | Q (%) | χ2 | P-value |
|---|
| 0 (control) | 0.39 (0.34,
0.45) | 0.06 | 7.42 | 0.06 |
|
1×10−4 | 0.49 (0.47,
0.98) | 0.14 |
|
|
|
1×10−5 | 0.48 (0.20,
0.55) | 0.16 |
|
|
|
1×10−6 | 0.46 (0.38,
0.56) | 0.14 |
|
|
Cell proliferation
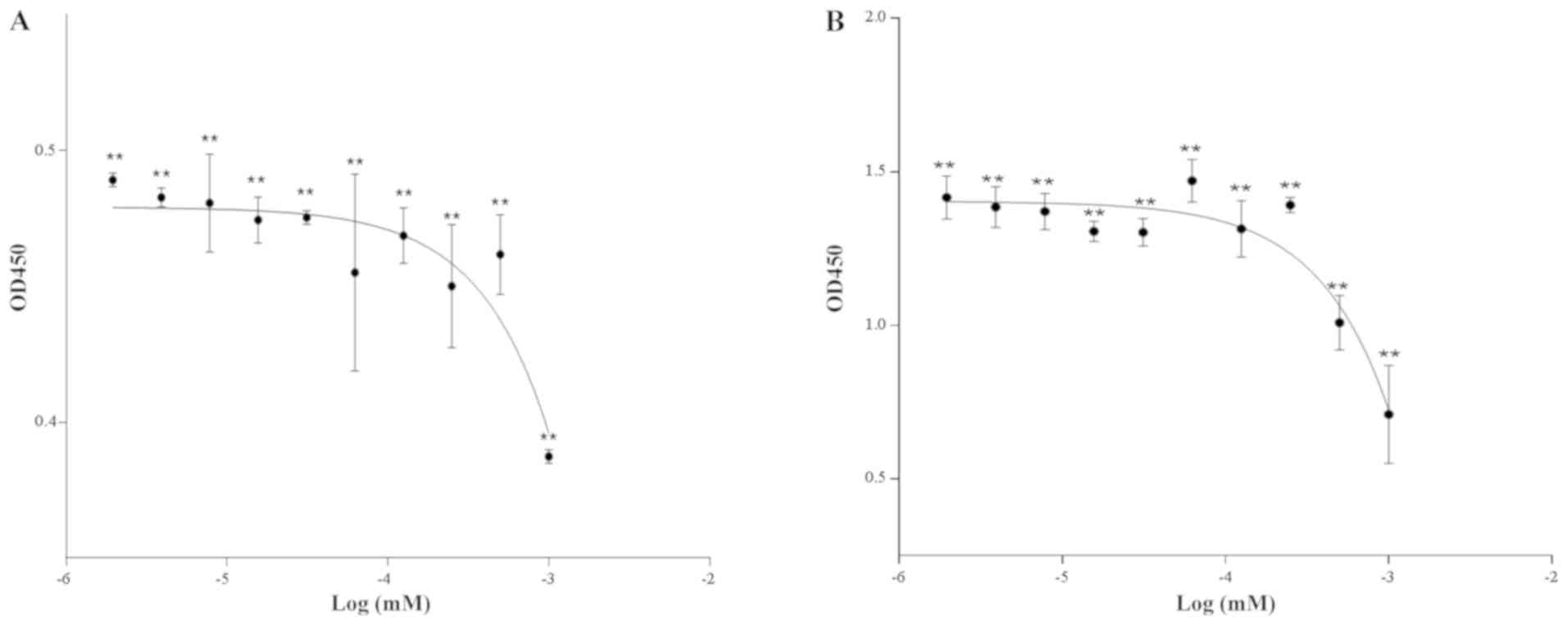
The cell growth curves indicated that the
differences in cell proliferation among the groups were more marked
after 48 h of exposure compared with those at 24 h. The cell
proliferation rates of the con1 and con3 groups were significantly
lower than those in the other groups (Fig. 1). In order to better reflect the
differences in phenotypic changes of the cells, con1 and con3 were
used as the high and low doses, respectively, in the transcriptome
sequencing experiments and the exposure time was set at 48 h.
Transcriptome sequencing and DEG
analysis
To determine the effects of MEHP on Sertoli cells,
transcriptome sequencing was performed using a cDNA library
generated from an equal amount of RNA isolated from the control or
MEHP-treated TM4 cells. An average of 10,947 genes (with a required
FPKM value of >0.7) were detected among the different
groups.
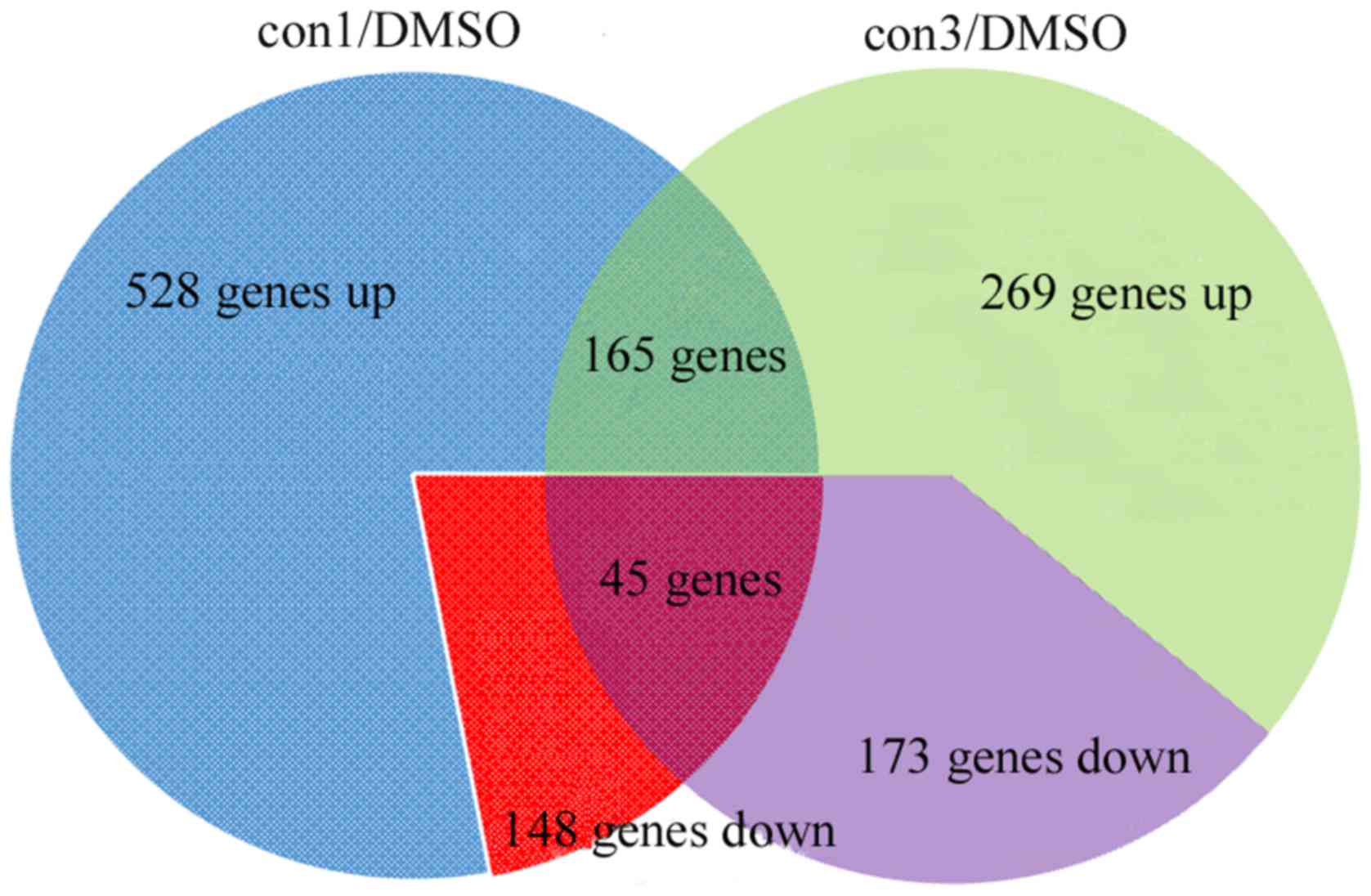
A total of 528 and 269 genes were identified to be
upregulated in the con1 and con3 group cells, respectively,
compared with the control group (DMSO); in addition, 148 and 173
genes were downregulated, respectively (Fig. 2). Among these genes, 165 were
consistently upregulated and 45 were consistently downregulated in
the two MEHP-treated groups (con1 and con3). These 210 DEGs were
regarded as candidates for further investigation.
Enrichment analysis of DEGs
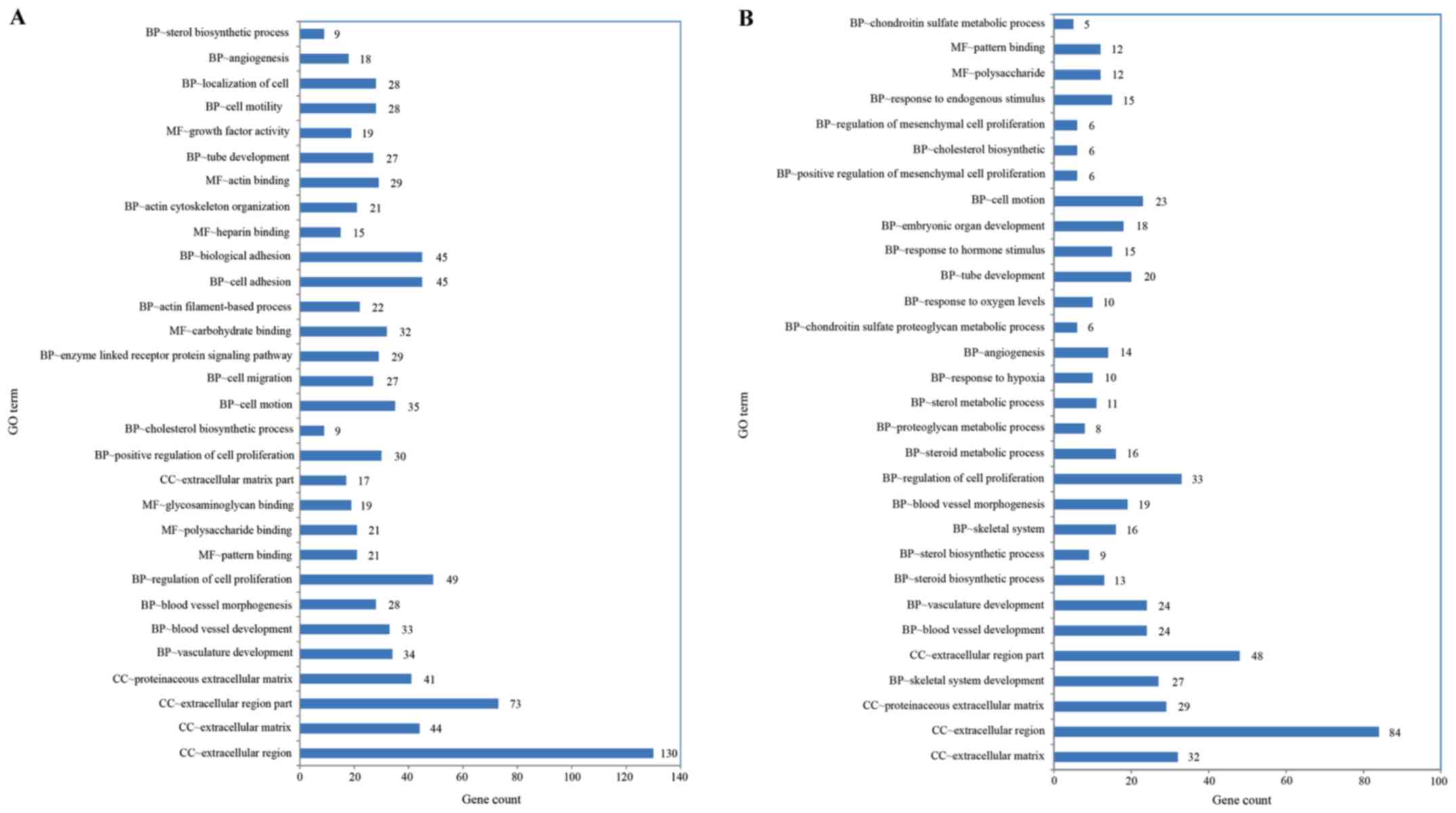
Functional enrichment analysis of DEGs was performed
using the GO and KEGG methods. GO analysis of the con1 group
provided significant enrichment of DEGs in 119 terms in the
biological process (BP), 20 in the cellular component (CC) and 16
in the molecular function (MF) category (P<0.01). For the con3
group, the DEGs were significantly enriched in 116 BP, 6 CC and 12
MF terms (P<0.01). The top 30 GO categories with significant
enrichment of DEGs (con1 and con3 vs. DMSO group) are presented in
Fig. 3A and B, respectively. Based
on the level of significance, the top term was ‘extracellular
region’ in the CC domain in the con1 and con3 groups.
In the KEGG analysis, 22 signaling pathways were
significantly enriched by the DEGs from the con1 group (P<0.05;
Fig. 4A). For the con3 group, 10
signaling pathways were significantly enriched by the DEGs
(P<0.05; Fig. 4B). The pathways
were mainly associated with tumors, steroid synthesis and cell
adhesion. According to the statistical significance, the top three
pathways were ‘focal adhesion’, ‘extracellular matrix
(ECM)-receptor interaction’ and ‘terpenoid backbone
biosynthesis’.
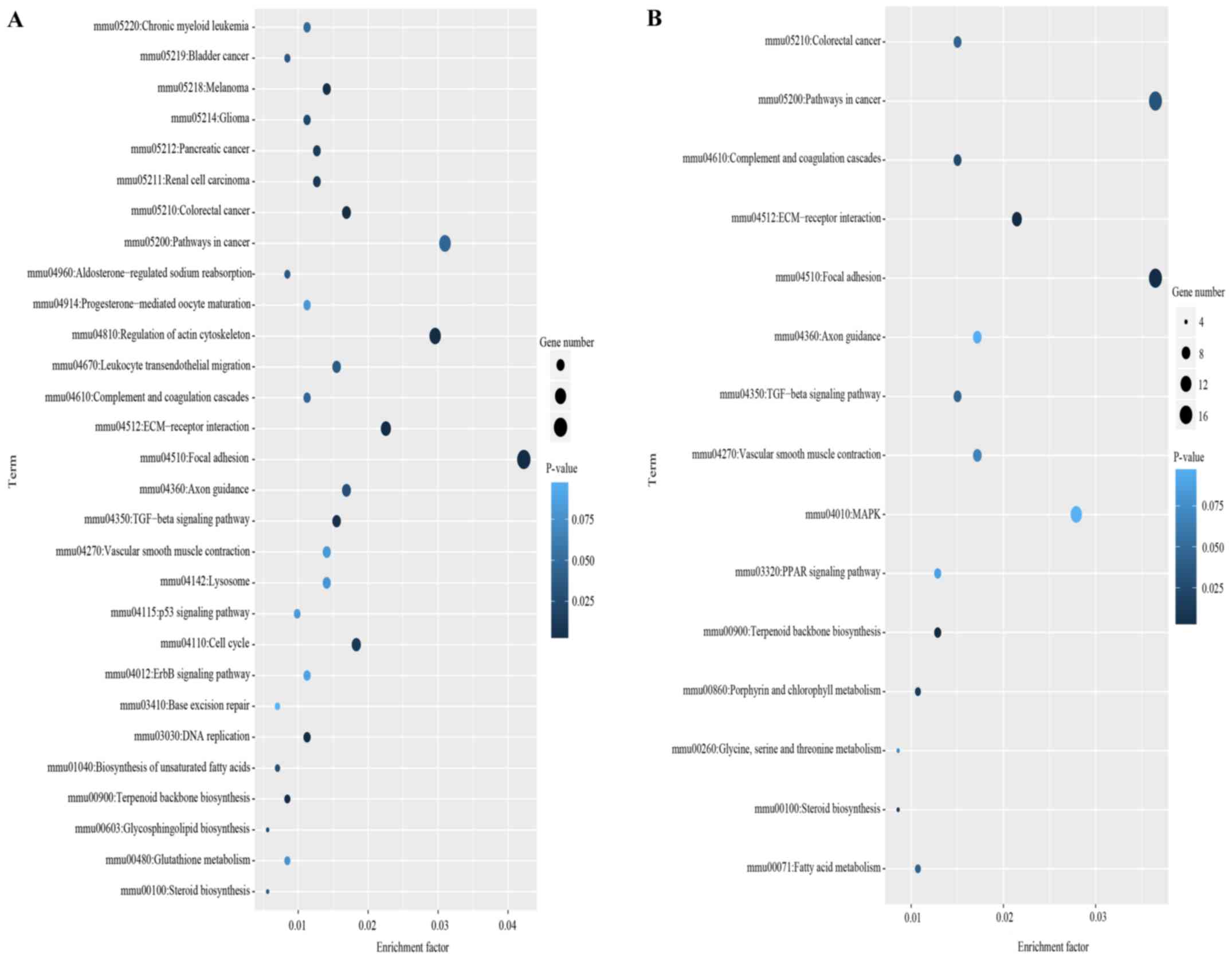 | Figure 4.Scatterplot for most enriched KEGG
pathways of DEGs in the MEHP-treated group compared with control
cells. (A) Differentially enriched KEGG pathways in the con1 group
compared with the control group. (B) Differentially enriched KEGG
pathways in the con3 group compared with the control group.
Enrichment factor was the ratio of the number of DEGs to the total
gene number. Smaller P-values indicate a higher degree of
enrichment. KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs,
differentially expressed genes; MEHP, mono-(2-ethylhexyl)
phthalate; ECM, extracellular matrix; TGF, transforming growth
factor; MAPK, mitogen-activated protein kinase; mmu, Mus
musculus; con 1, con1, MEHP at 1×10−3 mmol/l; con3,
MEHP at 2.5×10−4 mmol/l. |
Compared with the control group, there were eight
common enriched pathways in the high- and low-dose groups,
including ‘terpenoid backbone biosynthesis’, ‘focal adhesion’,
‘ECM-receptor interaction’, ‘pathways in cancer’, ‘colorectal
cancer’, ‘transforming growth factor-β signaling pathway’,
‘vascular smooth muscle contraction’ and ‘axon guidance’.
RT-qPCR validation of transcriptome
sequencing
To validate the results of the transcriptome
sequencing, transcripts of 14 key DEGs associated with the enriched
KEGG pathways were detected using RT-qPCR (Fig. 5). The analysis confirmed the aberrant
expression of collagen type I α 1 chain (COL1A1), actinin α 1
(ACTN1), vinculin (VCL), caveolin 1 (CAV1) and myosin light chain 9
(MYL9) for the ‘focal adhesion’ pathway, thrombospondin 1 (THBS1),
COL3A1, secreted phosphoprotein 1 (SPP1), syndecan 4 (SDC4) and
CD44 for the ‘ECM-receptor interaction’, and farnesyl diphosphate
synthase (FDPS), isopentenyl-diphosphate δ isomerase 1 (IDI1),
cytochrome P450 family 51 (CYP51) and squalene epoxidase (SQLE) for
‘terpenoid backbone biosynthesis and steroid biosynthesis’. Please
clarify whether this only applies to SQLE.
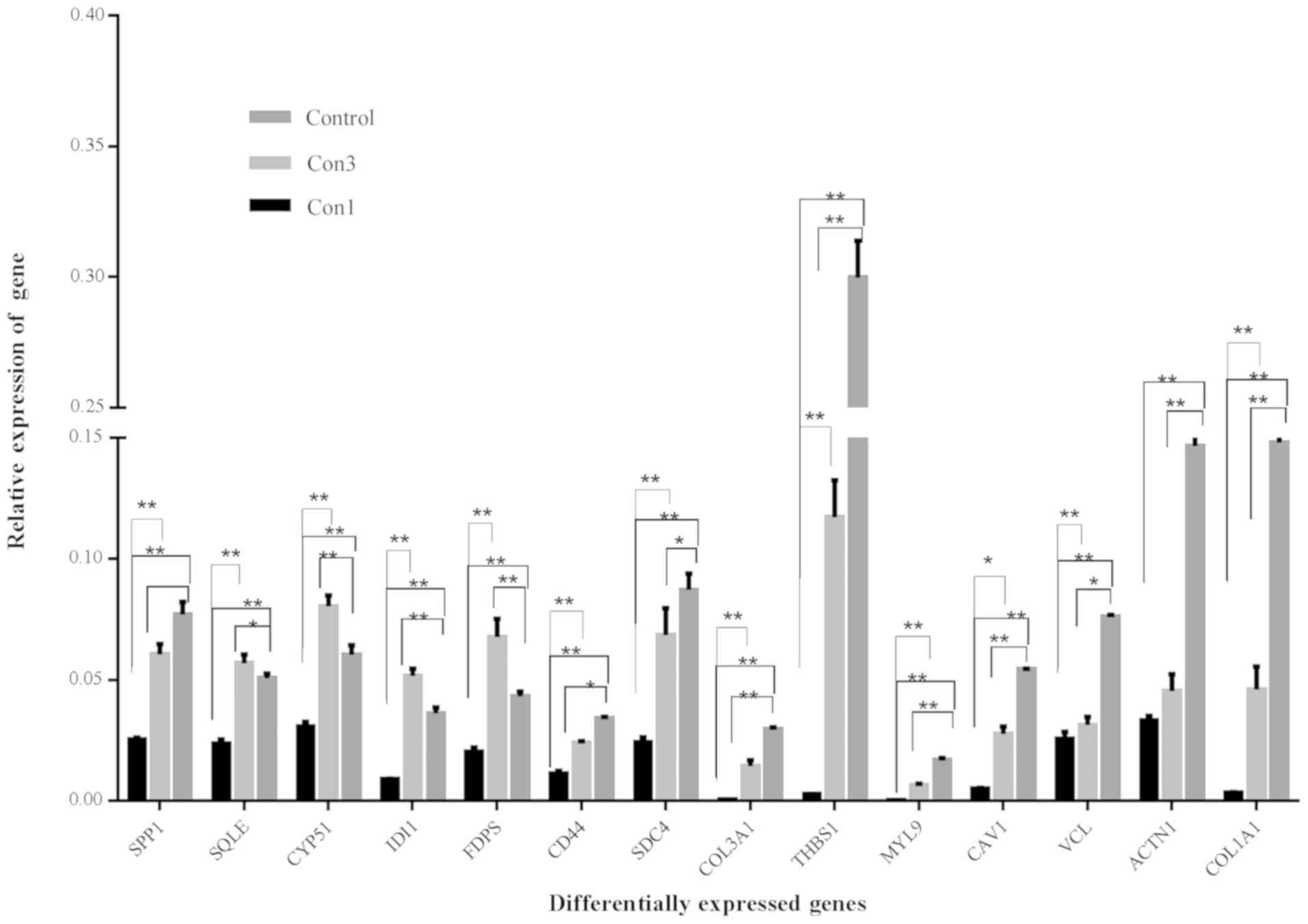 | Figure 5.mRNA expression of 14 genes in the
MEMP-treated TM4 cells. *P<0.05, **P<0.01. MEHP,
mono-(2-ethylhexyl) phthalate; con1, MEHP at 1×10−3
mmol/l; con3, MEHP at 2.5×10−4 mmol/l; COL1A1, collagen
type 1 α1 chain; ACTN1, actin α1; VCL, vinculin; CAV1, caveolin 1;
MYL9, myosin light chain 9; THBS1, thrombospondin 1; SDC4, syndecan
4; FDPS, farnesyl diphosphate synthase; IDI1, isopentyl-diphosphate
δ isomerase 1; CYP51, cytochrome P 450 family 51; SQLE, squalene
epoxidase; SPP, secreted phosphoprotein 1. |
The mRNA levels of COL1A, VCL, ACTN1, CAV1, MYL9,
THBS1, COL3A, SDC4, CD44 and SPP1 were significantly decreased in
the low-does (con3) and high-dose (con1) groups. However, four
genes, FDPS, IDI1, CYP51 and SQLE, were increased in the low-dose
group and decreased in the high-dose group.
Discussion
In the last 70 years, the use of phthalates has
changed the materials science substantially, which has made
exposure to phthalates unavoidable. In view of the harm that
phthalates may cause to the body, particularly in sensitive
populations, including infants and young children, the damaging
effects of phthalates have received increasing attention (20–24).
However, the internal exposure of the human body to phthalates was
not as high as expected. Studies have reported that the mean
concentrations of MEHP in the cord blood of neonates, the group
considered to be most sensitive to the developmental and
reproductive toxicity of phthalates, were 2×10−6 and
1.1×10−8 M (4,25). Although exposure to small amounts
does not directly cause cell death, it may have sub-lethal
cytological effects, including genetic damage and malfunction of
cellular metabolism (26). MEHP is
the major toxic metabolite of DEHP and is known to have
reproductive and developmental toxicity (27–29);
thus, in the present study, a low exposure dose of MEHP that was
not lethal to TM4 cells was used in order to realistically mimic
human internal exposure to DEHP, which is the most widely used
phthalate. The appropriate dose of MEHP, which was not lethal to
TM4 cells, was determined in a cell apoptosis assay. The
concentration and exposure time used in the transcriptome
sequencing experiment were determined from the results of a cell
proliferation assay. Transcriptome sequencing of TM4 cells was
performed following treatment with MEHP or control treatment. The
combination of next-generation transcriptome sequencing technology
and bioinformatics analysis provides a useful method to analyze the
mechanisms of action of toxic chemical agents, and to identify
potential hazardous compounds. To the best of our knowledge, the
present study was the first to use transcription sequencing to
investigate gene expression changes in the TM4 Sertoli cell line
following treatment with MEHP.
Transcriptomics approaches have previously been used
to investigate the toxic effects of DEHP (30). Studies have revealed that phthalates
have effects on peroxisome proliferator-activated receptors
(PPARs), tumor necrosis factor signaling pathways, calcium binding
and numerous myosin proteins. In the present study, 165 genes were
upregulated and 45 were downregulated in the two MEHP-treated
groups of TM4 cells. The genes with the greatest change in
expression out of the 210 DEGs may potentially be used as
biomarkers of DEHP-induced damage and as a useful tool to further
improve the identification of developmental toxicants (31). The DEGs of the high- and low-dose
groups had 8 pathways in common. In the GO analysis, the terms with
the highest significance, in the con1 and con3 groups, was
‘extracellular region’ in the CC domain. In the KEGG analysis, the
top three pathways (P<0.01) were ‘focal adhesion’, ‘regulation
of actin cytoskeleton’ and ‘ECM receptor interaction’. The results
were consistent for the responses of TM4 cells to 1×10−3
and 2.5×10−4 mmol/l MEHP. In the study by Nardelli et
al (32), 13 DEGs associated
with PPAR and cholesterol biosynthesis signaling pathways were
identified using a gene expression microarray. The present study
provides evidence supporting the roles of genes associated with
‘focal adhesion’, ‘regulation of actin cytoskeleton’ and
‘ECM-receptor interaction’ as the key functional genes that mediate
the effects of low-dose MEHP in Sertoli cells. Furthermore, the
RT-qPCR results confirmed that the expression of genes with roles
in ‘focal adhesion’ and ‘ECM-receptor interaction’ was decreased
with the levels of MEHP exposure increasing, indicating a
dose-response effect.
KEGG analysis revealed that the ‘focal adhesion’
pathway was enriched in the largest number of DEGs in the high- and
low-dose groups, and RT-qPCR analysis confirmed that the gene
expression of COL1A1, ACTN1, VCL, CAV1 and MYL9 was decreased
following MEHP treatment in a dose-dependent manner. Ectoplasmic
specialization (ES) is a unique cellular structure that maintains
cell shape and connections between Sertoli cells. Actinin, fimbrin,
espin and vinculin are all involved in ES (33,34). It
was previously reported that MEHP is able to destroy the ES between
rat Sertoli cells and spermatogenic cells, causing a release of the
immature germ cells into the seminiferous tubule, which weakens
reproductive function (35,36). The present study indicated that the
expression of the genes that encode actinin and vinculin was
decreased by MEHP, which was similar to the results of a previous
study, in which Syrian hamster embryo cells were exposed to 50 µM
DEHP for 24 h, revealing that the downregulated genes were
associated with focal adhesion or cell junctions (37). This indicates that MEHP may affect
the ECM-receptor interaction and focal adhesions to disrupt
intercellular junctions and the formation of fissures between
Sertoli cells and spermatogenic cells, resulting in the loss of
spermatogenic cells from the seminiferous epithelium.
Previous studies have reported that target genes
altered by phthalates are involved in several important signaling
pathways, including steroid synthesis, as well as lipid and
cholesterol homeostasis (38,39).
Testosterone is the major androgen and anabolic hormone, with an
important role in the growth and development of male animals. It
has been previously demonstrated that DEHP is able to mimic or
antagonize the actions of steroid hormones (40), and the effects of DEHP on
testosterone and estrogen-like activity have also been reported
(41). Considering the fundamental
role of steroid hormones in reproductive and developmental
functions, an imbalance in the synthesis and/or signaling of these
hormones may adversely affect different aspects of sexual
development (42). Of note, in the
present study, the enrichment of two pathways, ‘terpenoid backbone
biosynthesis’ and ‘steroid biosynthesis’, was higher in the
low-dose MEHP group than the high-dose group and the control group,
indicating that different doses of MEHP may exhibit opposing
effects on the synthesis of steroidal compounds in Sertoli cells,
which is potentially associated with the excitatory effect of MEHP.
Biphasic effects of di (n-butyl) phthalate were observed on
cholesterol side-chain cleavage enzyme and 3β-hydroxysteroid
dehydrogenase mRNA, which were generally increased at a low dose of
10 mg/kg, and at higher doses (50–400 mg/kg), an apparent
dose-dependent decrease was obtained (43). Hormesis is a concept describing a
dose-response association where there is a stimulation at a low
dose and inhibition at a high dose (44). Hormetic effects are considered as an
adaptive response to a moderate stress induced by the stimulus
(45). A study has proved that the
developmental toxicity of DEHP may be hermetic by using a score
test (46). In this light, the
present results suggest a hormetic-like biphasic dose-response
association of MEHP in Sertoli cells. Cell signaling-mediated
bidirectional control of gene expression has been considered as the
major hormetic mechanism triggered by exposure to xenobiotics
(47). This indicates that
‘terpenoid backbone biosynthesis’ and ‘steroid biosynthesis’
signaling pathways were activated in response to this low level of
exposure. This indicates that MEHP may affect testosterone
synthesis via multiple mechanisms to cause reproductive
toxicity.
The results of the present study demonstrated that
the mechanisms of the toxicity of MEHP are complex, including
effects on reproduction, development and metabolism involving a
numerous different genes, cellular processes and signaling
pathways. The interaction of these factors ultimately leads to the
complex toxic effects of MEHP. Although the present study is
limited to gene expression changes, the results of the present
study provide a foundation for further examination of the toxic
effects of low-level phthalate exposure. More gene and protein
expression validation and functional analyses should be performed
to fully elucidate the molecular mechanisms of phthalate
toxicity.
Acknowledgements
Not applicable.
Funding
This work was supported by funding from the National
Natural Science Foundation of China (grant no. 81202208).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are not publicly available due to ongoing research;
however, data are available from the corresponding author on
reasonable request.
Authors' contributions
LW was responsible for experimental design,
implementation and writing of article. TD was responsible for
experimental data processing. SL was responsible for experimental
design and revision of the article. YL was responsible for
experimental design and revision of the article. All authors have
read and approved the manuscript.
Ethics approval and informed consent
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schettler T: Human exposure to phthalates
via consumer products. Int J Androl. 29:134–139; discussion
181–185. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Markarian J: PVC additives-What lies
ahead? Plast Addit Compound. 9:22–25. 2007. View Article : Google Scholar
|
|
3
|
Högberg J, Hanberg A, Berglund M,
Skerfving S, Remberger M, Calafat AM, Filipsson AF, Jansson B,
Johansson N, Appelgren M and Håkansson H: Phthalate diesters and
their metabolites in human breast milk, blood or serum, and urine
as biomarkers of exposure in vulnerable populations. Environ Health
Perspect. 116:334–339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Latini G, De Felice C, Presta G, Del
Vecchio A, Paris I, Ruggieri F and Mazzeo P: In utero exposure to
di-(2-ethylhexyl)phthalate and duration of human pregnancy. Environ
Health Perspect. 111:1783–1785. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Latini G, De Felice C, Presta G, Del
Vecchio A, Paris I, Ruggieri F and Mazzeo P: Exposure to
Di(2-ethylhexyl)phthalate in humans during pregnancy. A preliminary
report. Biol Neonate. 83:22–24. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Swan SH: Prenatal phthalate exposure and
anogenital distance in male infants. Environ Health Perspect.
114:A88–A89. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fay M, Donohue JM and De Rosa C: ATSDR
evaluation of health effects of chemicals. VI.
Di(2-ethylhexyl)phthalate. Agency for toxic substances and disease
registry. Toxicol Ind Health. 15:651–746. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lyche JL, Gutleb AC, Bergman A, Eriksen
GS, Murk AJ, Ropstad E, Saunders M and Skaare JU: Reproductive and
developmental toxicity of phthalates. J Toxicol Environ Health B
Crit Rev. 12:225–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fratantoni JC: Review: The platelet
storage lesion: Possible role of plasticizers? Blood Cells.
18:435–440; discussion 441–443. 1992.PubMed/NCBI
|
|
10
|
Hayes DN and Kim WY: The next steps in
next-gen sequencing of cancer genomes. J Clin Invest. 125:462–468.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Q, Lu Q and Zhao H: A review of study
designs and statistical methods for genomic epidemiology studies
using next generation sequencing. Front Genet. 6:1492015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fiorini C, Tilloy-Ellul A, Chevalier S,
Charuel C and Pointis G: Sertoli cell junctional proteins as early
targets for different classes of reproductive toxicants. Reprod
Toxicol. 18:413–421. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rodriguez-Sosa JR, Bondareva A, Tang L,
Avelar GF, Coyle KM, Modelski M, Alpaugh W, Conley A, Wynne-Edwards
K, França LR, et al: Phthalate esters affect maturation and
function of primate testis tissue ectopically grafted in mice. Mol
Cell Endocrinol. 398:89–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sjöberg P, Bondesson U, Kjellen L,
Lindquist NG, Montin G and Plöen L: Kinetics of di-(2-ethylhexyl)
phthalate in immature and mature rats and effect on testis. Acta
Pharmacol Toxicol (Copenh). 56:30–37. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sjöberg P, Lindqvist NG and Plöen L:
Age-dependent response of the rat testes to di(2-ethylhexyl)
phthalate. Environ Health Perspect. 65:237–242. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chapin RE, Gray TJ, Phelps JL and Dutton
SL: The effects of mono-(2-ethylhexyl)-phthalate on rat Sertoli
cell-enriched primary cultures. Toxicol Appl Pharmacol. 92:467–479.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Galdieri M, Ziparo E, Palombi F, Russo MA
and Stefanini M: Pure sertoli cell cultures: A new model for the
study of somatic-Germ cell interactions. J Androl. 2:249–254. 1981.
View Article : Google Scholar
|
|
18
|
van Dijk EL, Auger H, Jaszczyszyn Y and
Thermes C: Ten years of next-generation sequencing technology.
Trends Genet. 30:418–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Casas L, Fernández MF, Llop S, Guxens M,
Ballester F, Olea N, Irurzun MB, Rodríguez LS, Riaño I, Tardón A,
et al: Urinary concentrations of phthalates and phenols in a
population of Spanish pregnant women and children. Environ Int.
37:858–866. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Frederiksen H, Aksglaede L, Sorensen K,
Skakkebaek NE, Juul A and Andersson AM: Urinary excretion of
phthalate metabolites in 129 healthy Danish children and
adolescents: Estimation of daily phthalate intake. Environ Res.
111:656–663. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo Y and Kannan K: Comparative assessment
of human exposure to phthalate esters from house dust in China and
the United States. Environ Sci Technol. 45:3788–3794. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Romero-Franco M, Hernández-Ramírez RU,
Calafat AM, Cebrián ME, Needham LL, Teitelbaum S, Wolff MS and
López-Carrillo L: Personal care product use and urinary levels of
phthalate metabolites in Mexican women. Environ Int. 37:867–871.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Woodruff TJ, Zota AR and Schwartz JM:
Environmental chemicals in pregnant women in the United States:
NHANES 2003–2004. Environ Health Perspect. 119:878–885. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin S, Ku HY, Su PH, Chen JW, Huang PC,
Angerer J and Wang SL: Phthalate exposure in pregnant women and
their children in central Taiwan. Chemosphere. 82:947–955. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Midic U, Vincent KA, VandeVoort CA and
Latham KE: Effects of long-term endocrine disrupting compound
exposure on Macaca mulatta embryonic stem cells. Reprod Toxicol.
65:382–393. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Moss EJ, Cook MW, Thomas LV and Gray TJ:
The effect of mono-(2-ethylhexyl) phthalate and other phthalate
esters on lactate production by Sertoli cells in vitro. Toxicol
Lett. 40:77–84. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lamb JC IV and Chapin RE: Testicular and
germ cell toxicity: In vitro approaches. Reprod Toxicol. 7 (Suppl
1):S17–S22. 1993. View Article : Google Scholar
|
|
29
|
Davis BJ, Weaver R, Gaines LJ and Heindel
JJ: Mono-(2-ethylhexyl) phthalate suppresses estradiol production
independent of FSH-cAMP stimulation in rat granulosa cells. Toxicol
Appl Pharmacol. 128:224–228. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stenz L, Escoffier J, Rahban R, Nef S and
Paoloni-Giacobino A: Testicular dysgenesis syndrome and
long-lasting epigenetic silencing of mouse sperm genes involved in
the reproductive system after prenatal exposure to DEHP. PLoS One.
12:e01704412017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van Dartel DA, Pennings JL, Robinson JF,
Kleinjans JC and Piersma AH: Discriminating classes of
developmental toxicants using gene expression profiling in the
embryonic stem cell test. Toxicol Lett. 201:143–151. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nardelli TC, Erythropel HC and Robaire B:
Toxicogenomic screening of replacements for di(2-Ethylhexyl)
phthalate (DEHP) using the immortalized TM4 sertoli cell line. PLoS
One. 10:e01384212015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vogl AW, Pfeiffer DC and Redenbach DM:
Ectoplasmic (‘junctional’) specializations in mammalian Sertoli
cells: Influence on spermatogenic cells. Ann NY Acad Sci.
637:175–202. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vogl AW, Pfeiffer DC, Mulholland D, Kimel
G and Guttman J: Unique and multifunctional adhesion junctions in
the testis: Ectoplasmic specializations. Arch Histol Cytol.
63:1–15. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li LH, Jester WF Jr and Orth JM: Effects
of relatively low levels of mono-(2-ethylhexyl) phthalate on
cocultured Sertoli cells and gonocytes from neonatal rats. Toxicol
Appl Pharmacol. 153:258–265. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yao PL, Lin YC and Richburg JH:
Mono-(2-ethylhexyl) phthalate-induced disruption of junctional
complexes in the seminiferous epithelium of the rodent testis is
mediated by MMP2. Biol Reprod. 82:516–527. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Landkocz Y, Poupin P, Atienzar F and
Vasseur P: Transcriptomic effects of di-(2-ethylhexyl)-phthalate in
Syrian hamster embryo cells: An important role of early
cytoskeleton disturbances in carcinogenesis? BMC Genomics.
12:5242011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Thompson CJ, Ross SM and Gaido KW:
Di(n-butyl) phthalate impairs cholesterol transport and
steroidogenesis in the fetal rat testis through a rapid and
reversible mechanism. Endocrinology. 145:1227–1237. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu K, Lehmann KP, Sar M, Young SS and
Gaido KW: Gene expression profiling following in utero exposure to
phthalate esters reveals new gene targets in the etiology of
testicular dysgenesis. Biol Reprod. 73:180–192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sharpe RM: Hormones and testis development
and the possible adverse effects of environmental chemicals.
Toxicol Lett. 120:221–232. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Akingbemi BT, Ge R, Klinefelter GR, Zirkin
BR and Hardy MP: Phthalate-induced Leydig cell hyperplasia is
associated with multiple endocrine disturbances. Proc Natl Acad Sci
USA. 101:775–780. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gunnarsson D, Leffler P, Ekwurtzel E,
Martinsson G, Liu K and Selstam G: Mono-(2-ethylhexyl) phthalate
stimulates basal steroidogenesis by a cAMP-independent mechanism in
mouse gonadal cells of both sexes. Reproduction. 135:693–703. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bello UM, Madekurozwa MC, Groenewald HB,
Aire TA and Arukwe A: The effects on steroidogenesis and
histopathology of adult male Japanese quails (Coturnix coturnix
japonica) testis following pre-pubertal exposure to di(n-butyl)
phthalate (DBP). Comp Biochem Physiol C Toxicol Pharmacol.
166:24–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Calabrese EJ and Baldwin LA: Hormesis:
U-shaped dose responses and their centrality in toxicology. Trends
Pharmacol Sci. 22:285–291. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Calabrese EJ: Overcompensation
stimulation: A mechanism for hormetic effects. Crit Rev Toxicol.
31:425–470. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hunt D and Rai SN: Testing threshold and
hormesis in a random effects dose-response model applied to
developmental toxicity data. Biom J. 47:319–328. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen F, Liu SS, Yu M, Qu R and Wang MC:
Blocking the entrance of AMP pocket results in hormetic stimulation
of imidazolium-based ionic liquids to firefly luciferase.
Chemosphere. 132:108–113. 2015. View Article : Google Scholar : PubMed/NCBI
|