Introduction
Rheumatoid arthritis (RA) is a common systemic,
non-infectious autoimmune disease that is mainly characterized by
synovitis. The pathological characteristics of RA include
autoimmune response and chronic inflammation-induced synovial
hyperplasia, angiogenesis and cartilage destruction (1,2). In
Europe, a preliminary understanding of this disease entity
prevailed as early as the 17th century, but it was not until 1859,
when it was named RA by Garrod, that it was more comprehensively
understood (3,4). The incidence rate of RA in China is
~30% (~4 million patients) and the disability rate (60%) is
relatively high (5). One of the most
important pathophysiological processes is the formation of the
synovial pannus, which erodes and destroys adjacent articular
cartilage and bone, resulting in joint remodeling, distortion and
ankylosis (6). There is currently no
effective treatment available for this common disease, and the
underlying mechanisms of the exited targeting drug remain
elusive.
Triptolide is a non-steroidal anti-inflammatory
agent and immunosuppressant. Previous clinical studies have
reported that triptolide possesses anti-rheumatic, anti-oxidative
and anti-cancer properties, and is the most commonly used Chinese
medicine in the treatment of RA. Triptolide, the major active
component of Tripterygium wilfordii, is a diterpene lactone
epoxide compound whose specific molecular targets in RA have
remained to be fully determined (7–9). Only
few studies have assessed the mechanisms and pathways associated
with the action of triptolide. In order to further elucidate the
therapeutic mechanisms and targets of triptolide in RA, the present
study used a rat model of RA to establish a basis for clinical
treatment and to provide fundamental evidence supporting the use of
this Traditional Chinese Medicine (TCM) component in the treatment
of RA (10,11).
Many human infectious diseases are typically
associated with the deregulation of gene expression, and even
normal tissues are accompanied by changes in gene expression
patterns (12). Gene chip analysis
is a milestone technological innovation in the field of molecular
biology and one of the most advanced and effective methods. It
allows researchers to determine how thousands of genes act
simultaneously, quantify certain genes in patients to identify a
disease and provides novel targets for drug design and disease
treatment (13,14). However, the application of gene chip
technology to assess the treatment effect of triptolide in RA has
not been previously reported. By analyzing the differences in the
gene expression profile in synovial tissues of model animals with
and without drug administration, the differential gene expression
spectrum of synovial tissue in RA may be established and the
mechanism of the triptolide treatment effect on RA may be
qualitatively and quantitatively analyzed at the gene level.
The aim of the present study was to determine the
differentially expressed genes (DEGs) in synovial tissues of RA
rats by using the gene chip technique and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR), and
to explore the targets of the DEGs following treatment of RA by
triptolide, including the direct molecular targets of triptolide
and the effects of RA declining as a result of triptolide
treatment.
Materials and methods
Animals
A total of 20 pathogen-free male adult Wistar rats
(age, 8 weeks; weight, 150±10 g) were provided by the Shanghai
Experimental Animal Center of the Chinese Academy of
Sciences/Shanghai Shrek Experimental Animal Co., Ltd. [animal
certificate no. SCXK (Shanghai 2007-0005)]. All of the rats were
housed at the Animal Center of Zhejiang Chinese Medical University
(Hangzhou, China) at a constant temperature of 23±1°C, humidity of
40–70% and under a 12-h light/dark cycle, with free access to food
and water. All procedures conformed to the guidelines of the
National Institutes of Health Guide for the Care and Use of
Laboratory Animals and were approved by the Ethics Committee for
the Use of Experimental Animals of Zhejiang Chinese Medical
University (Hangzhou, China; animal permit no. SYXK (Zhejiang
Province 2008-0115).
Experimental groups and adjuvant
arthritis (AA) model
The 20 rats used in the present study were randomly
divided into two groups, namely the AA model group and the
triptolide treatment (TP) group (n=10 per group). The AA model was
induced by daily injecting Freund's complete adjuvant (0.1 ml) into
the muscle of the hind paw with a microsyringe for ~18 days. The
activity of the animals and ankle swelling were observed (10,15). At
day 18, the successful establishment of this model was evaluated by
determining the arthritis index (AI) (16,17). The
standard grading of AI was as follows: 0, no redness; 1, mild
swelling of the toe joint, inflammation in a single area of the paw
or foot pad; 2, mild redness of the joint, inflammation in >2
areas of the paw and foot pad or ankle; 3, moderate redness and
mild dysfunction of the joint; 4, severe redness of the joint,
rigidity or even malformation, severe dysfunction. Each paw scored
4 points, adding up to a total of 8 points. The model was
considered to be successfully established if the AI score was >4
points (16,17).
Drug administration and sample
collection
After model establishment, the rats in the TP group
were administered the medicine from the 18th day onwards as
follows: First, the foot pad of the hind leg was routinely
sterilized, and each rat received a muscle injection with
triptolide at a dose of 0.4 mg/kg daily for 14 days (18,19). The
rats in the model group were injected with an equal volume of 0.9%
physiological saline. All of the rats were sacrificed on day 14,
and the hind leg and ankle joint were resected and processed.
Following anesthesia by intraperitoneal injection of 10% chloral
hydrate (400 mg/kg) and after disinfection with alcohol, the skin
was incised along the middle of the ankle joint until an area ~2×2
cm was exposed. The joint cavity was opened, and the synovial
tissue around the ankle was carefully resected with a surgical
blade. The synovial tissue specimen was rinsed with diethyl
pyrocarbonate, added into a cryopreservation tube and placed in
liquid nitrogen to be used for gene chip detection and qPCR
analysis (20,21). The rats were sacrificed by
dislocation of the neck at 30 min after anesthesia.
Total RNA extraction and
purification
Total RNA was extracted with TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA);
50–100 mg tissue/ml TRIzol was used in the present study and all
procedures were performed according to the manufacturer's protocol.
Quality control of the RNA samples was performed with a Nanodrop
ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.) by
measuring the absorbance of each sample at 260 nm (21). The RNA samples were then subjected to
1% agarose gel electrophoresis. In order to obtain high-quality
purified RNA samples, and reach the labeling efficiency of the
probe and optimal results of hybridization in the subsequent
experiment, the Qiagen RNeasy kit (Qiagen GmBH, Hilden, Germany)
was used to purify total RNA, according to the manufacturer's
protocol (22,23).
RNA labeling and microarray
hybridization
The mRNA microarray profile (ID: 014879) was
provided by Agilent Technologies, Inc. (Santa Clara, CA, USA).
Total RNA was amplified and labeled using the Low-Input Quick Amp
Labeling kit, One-Color (cat. no. 5190–2305; Agilent Technologies,
Inc.), according to the manufacturer's protocol. Labeled
complementary RNA (cRNA) was purified with the RNeasy mini kit
(cat. no. 74106; Qiagen GmBH). Each slide was hybridized with 1.65
µg cyanine 3-labeled cRNA using the Gene Expression Hybridization
kit (cat. no. 5188–5242; Agilent Technologies, Inc.) in a
hybridization oven (cat. no. G2545A; Agilent Technologies, Inc.)
according to the manufacturer's protocol. After 17 h of
hybridization, the slides were washed in staining dishes (cat. no.
121; Shandon™; Thermo Fisher Scientific, Inc.) with a Gene
Expression Wash Buffer kit (cat. no. 5188–5327; Agilent
Technologies, Inc.) according to the manufacturer's protocol
(24,25). The microarray data were deposited in
the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124639).
Screening of DEGs
The raw microarray data were processed using
GeneSpring software version 12.6.1 (Agilent Technologies, Inc.).
The data were normalized and the gene expression was compared
between the two groups. The gene chip data were screened for DEGs
according to different selection criteria (fold-change >2 for
upregulated and <0.5 for downregulated genes; P<0.01).
Gene ontology (GO) and pathway
analysis
The GO (http://www.geneontology.org) database was used to
identify functional terms enriched by the DEGs. GO analysis
determines the accumulation of DEGs in terms of three independent
categories, namely biological process, cellular component and
molecular function. The DEGs are used as input in the GO analysis.
In the analysis of DEGs from gene expression profiles, GO may be
used to provide information on the functions, structural
characterization and gene function classification labeling.
Generation of the interaction
network
The pathway analysis was performed using the and
Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg), which provides a
systematic analysis of gene function, associated genome information
and functional information. Cell biochemical processes, including
membrane transport, metabolism, cell cycle and signal transmission,
may be fully elucidated by searching the more advanced KEGG pathway
database. Each node in the database was mapped to calculate the
number of genes per node.
RT-qPCR
Total RNA samples from each group were
reverse-transcribed into complementary (c)DNA. RT was performed
with the Maxima First Strand cDNA Synthesis kit for RT-qPCR (cat.
no. K1641; Thermo Fisher Scientific, Inc.). qPCR analysis was
performed using the DyNAmo Flash SYBR Green qPCR kit (cat. no.
F415XL; Thermo Fisher Scientific, Inc.) to evaluate vascular
endothelial growth factor (VEGF) A and C1q and tumor necrosis
factor related protein 3 (C1QTNF3) levels using β-actin as the
internal control. Each sample was analyzed in triplicate. The PCR
consisted of an initial denaturation step at 94°C for 5 min,
followed by an amplification with 35 reaction cycles, including
denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec, and
final extension at 72°C for 1 min (26). The relative mRNA expression levels
were calculated using the quantification cycle (Cq) method. The
primers used were as follows: VEGFA, forward
5′-GAGGAAAGGGAAAGGGTCAAA-3′ and reverse
5′-CACAGTGAACGCTCCAGGATT-3′; C1QTNF3, forward
5′-GTGCTCAGAAATAATTGGCTCCT-3′ and reverse
5′-AAGGTGTGGCAAGCCAAATG-3′; and β-actin, forward
5′-TCTGTGTGGATTGGTGGCTCTA-3′ and reverse
5′-CTGCTTGCTGATCCACATCTG-3′.
Statistical analysis
SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was
used for statistical analysis. One-way analysis of variance and the
Student-Newman-Keuls test were used to analyze the differences
between the experimental groups. Values are expressed as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
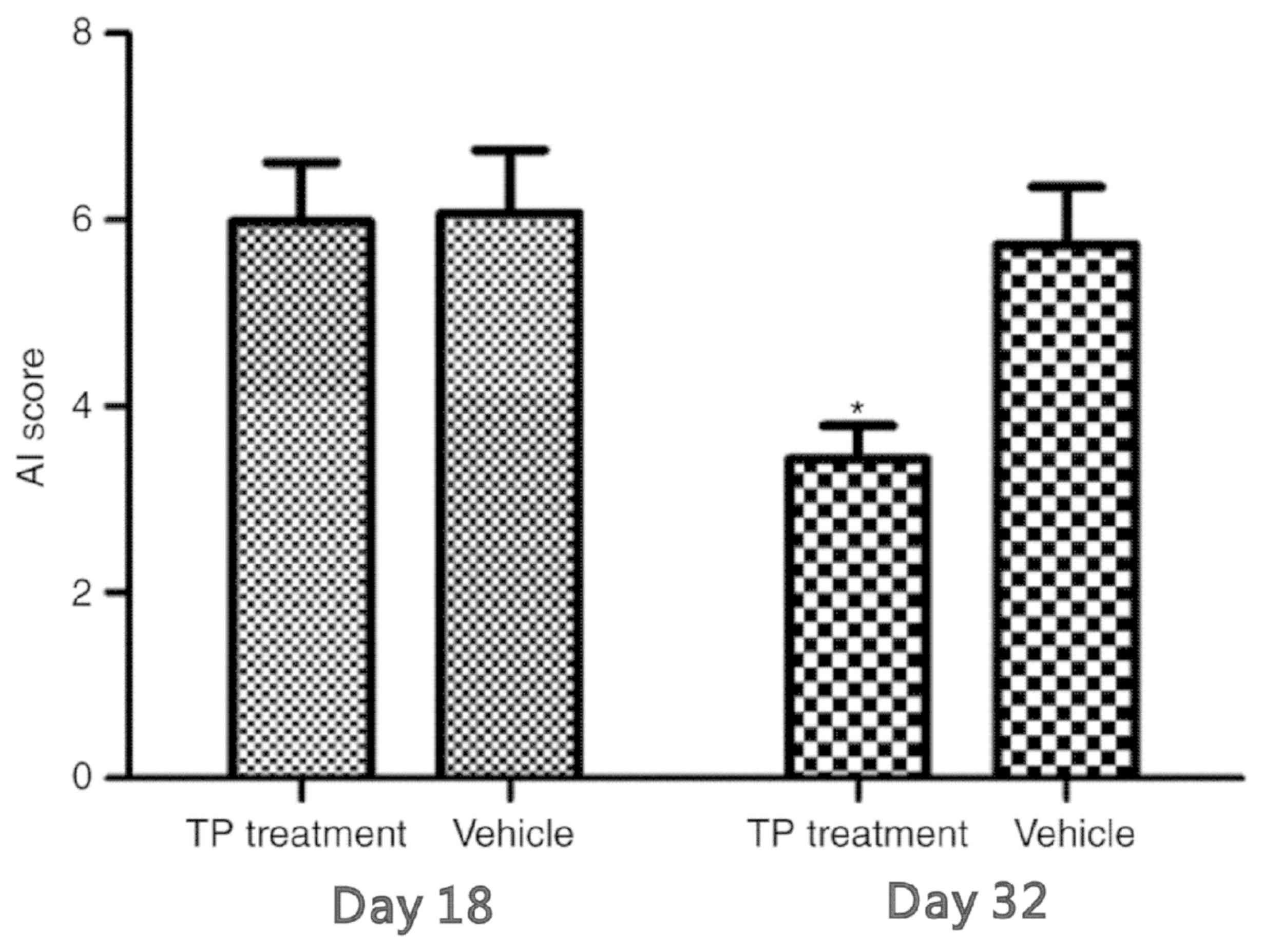
AI score
The AI scores in the rats of the present study was
determined to confirm successful model establishment. AI scores of
>4 on the 18th day after modeling were considered to indicate
successful establishment of the model. A comparison of the AI
scores in the model and TP groups is presented in Fig. 1. In the TP group, the AI score was
significantly decreased following TP treatment, while that in the
model/vehicle-treated group was unchanged.
Screening of DEGs between the
treatment and model groups
The gene chip data were screened according to the
abovementioned fold change/significance criteria for DEGs. A number
of DEGs were screened out presented in the list, the data of which
are presented in Fig. 2. There were
6 upregulated DEGs and 5 downregulated DEGs listed, including VEGFA
and C1QTNF3.
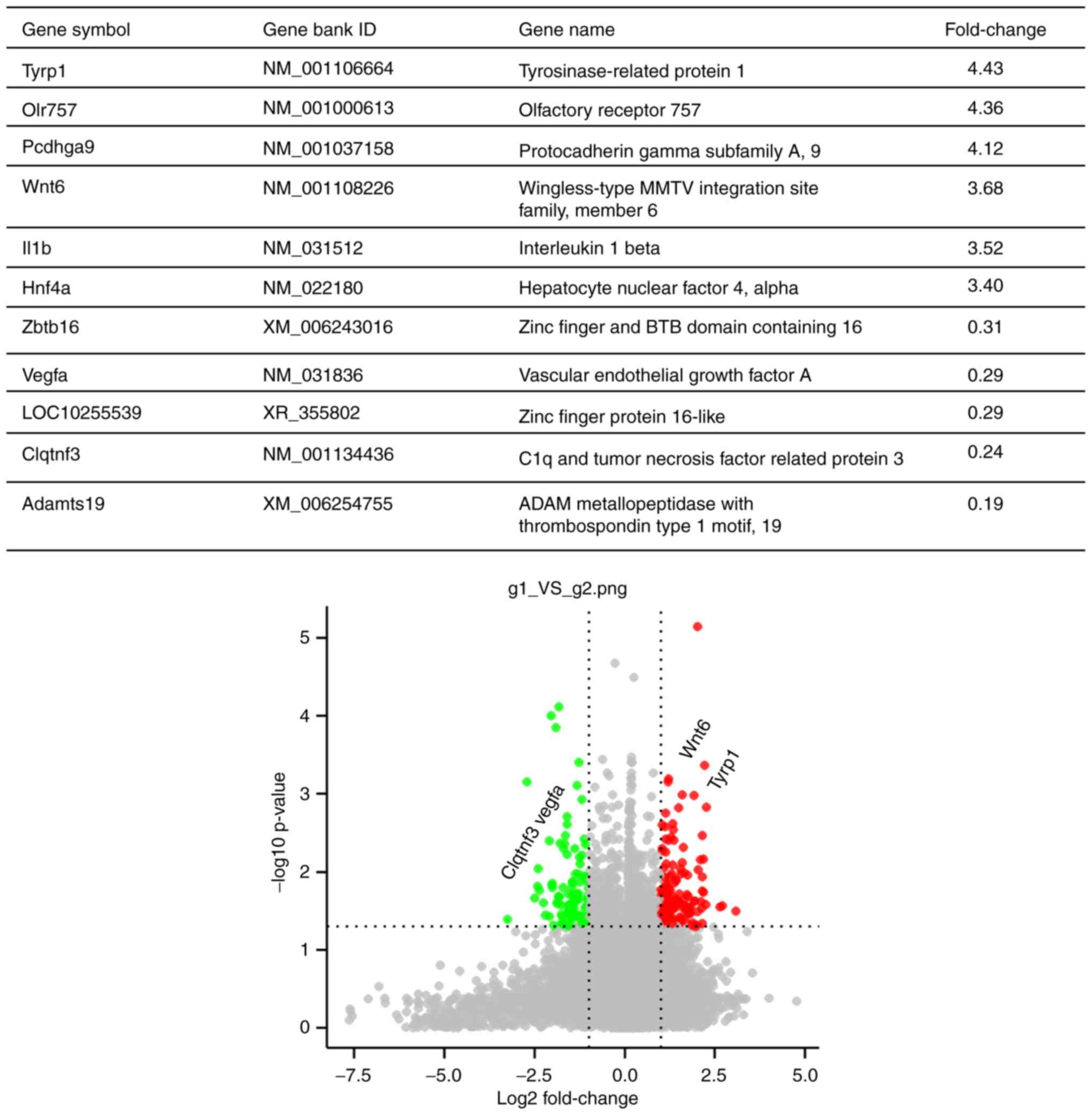 | Figure 2.DEGs in the joints of triptolide- vs.
vehicle-treated rats with rheumatoid arthritis. The gene chip data
were screened according to the screening criteria of DEGs
(fold-change >2 for upregulated and <0.5 for downregulated
genes). Among the significantly upregulated DEGs are TYRP1, OLR757,
PCDHGA9, WNT6, IL1Band HNF4A, while ZBTB16, VEGFA, LOC10255539,
C1QTNF3 and Adamts19 are among the downregulated DEGs. DEG,
differentially expressed gene; VEGFA, vascular endothelial growth
factor A; C1QTNF3, C1q and tumor necrosis factor related protein
3. |
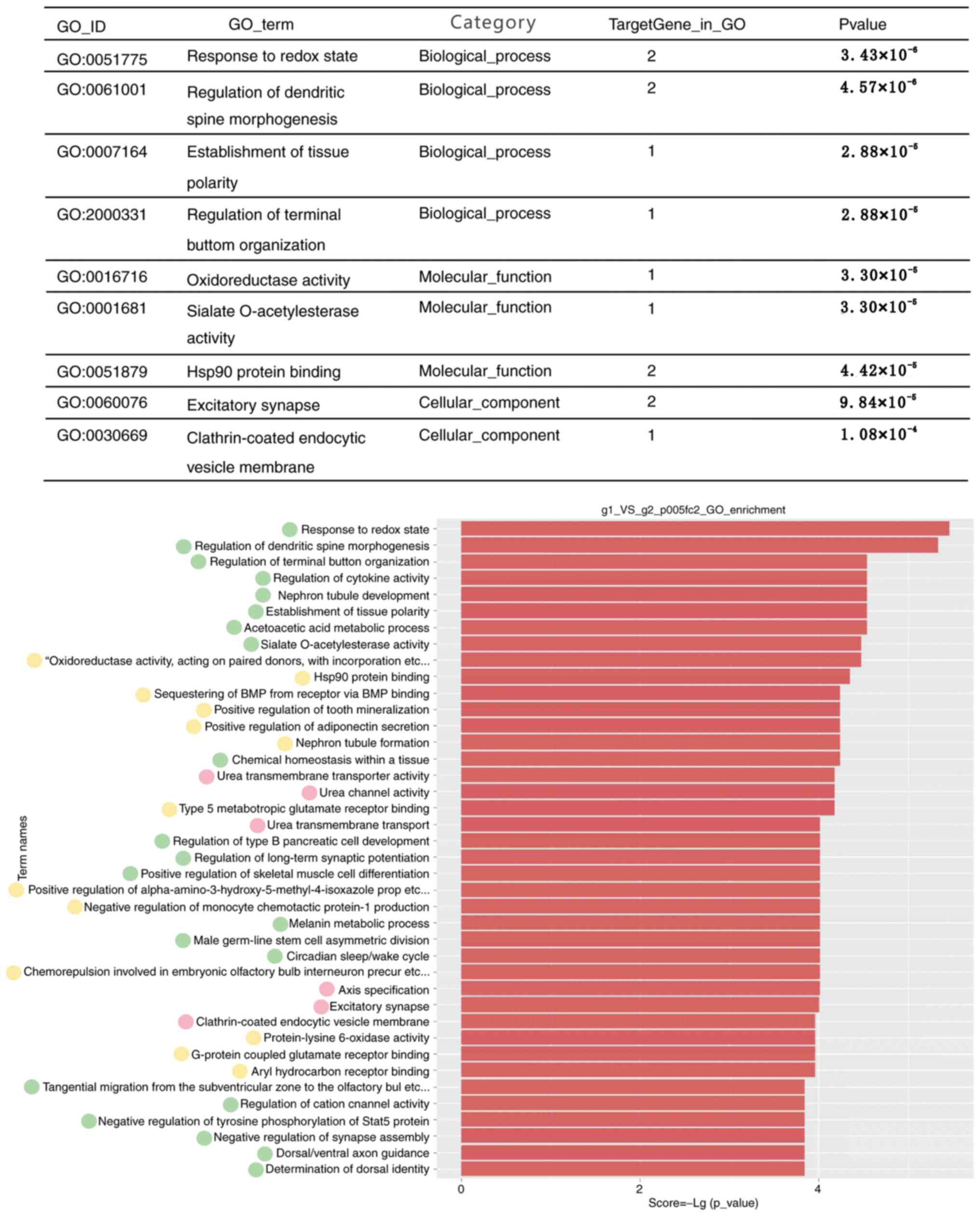
GO analysis
The DEGs were subjected to a GO functional
enrichment analysis. It was revealed that the DEGs were implicated
in a wide range of molecular functions, including tumor-associated
pathways, cell cycle regulation and interaction between cell
receptors and extracellular matrix. Details of the association
between RA and the GO functional terms associated with the genes
affected by triptolide treatment in RA are presented in Fig. 3. The top GO terms include response to
redox state, regulation of dendritic spine morphogenesis and
establishment of tissue polarity.
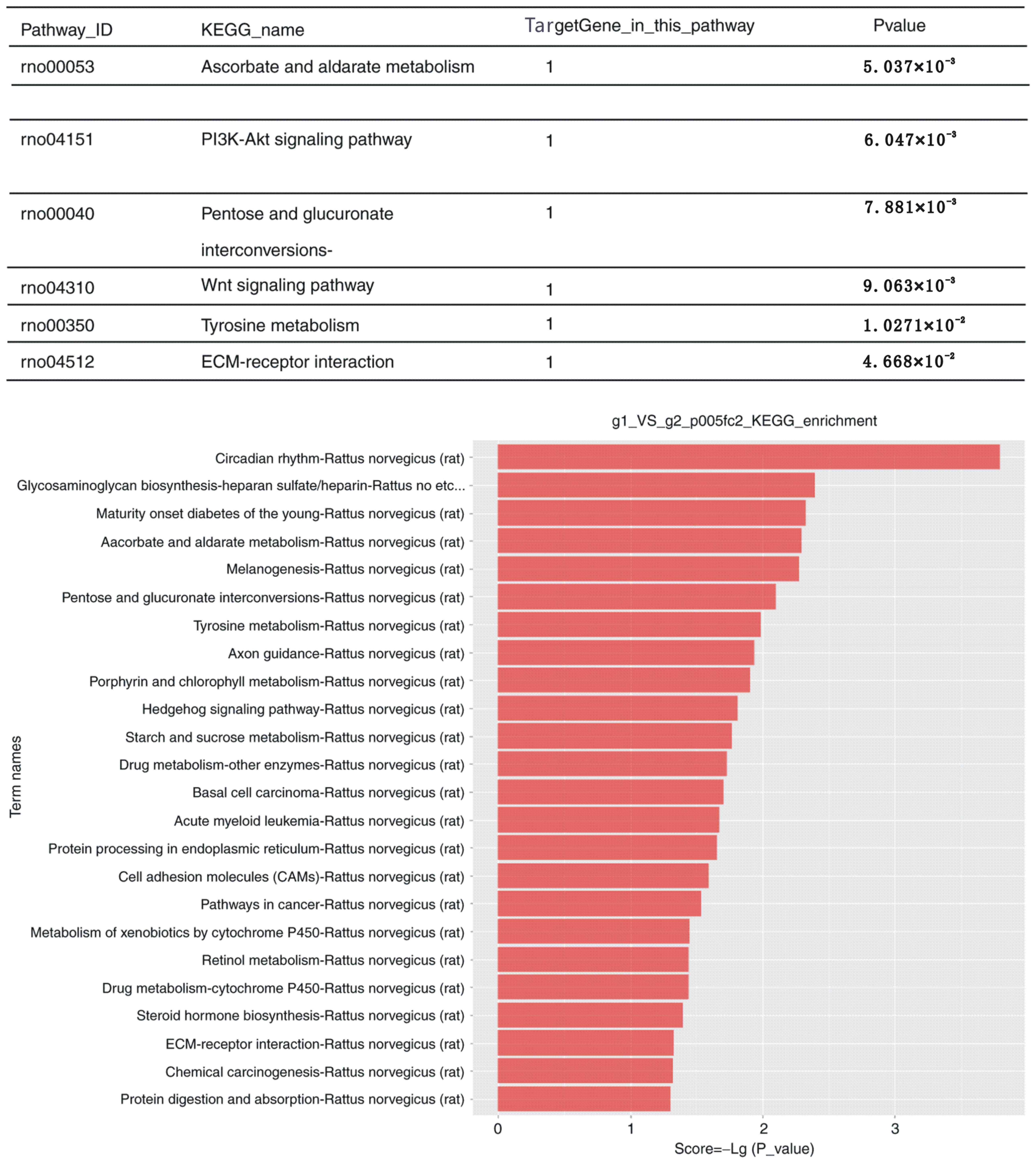
Pathway analysis
Using KEGG pathway enrichment analysis, the DEGs
were identified to participate in various signaling pathways. The
detailed information is presented in Fig. 4. The most significant pathways are
listed in the top panel of the figure. The pathway analysis
suggested that the phosphoinositide-3 kinase (PI3K)/AKT signaling
pathway has a key role in the proliferation and apoptosis of
synovial cells of RA joints. Furthermore, other pathways were
listed that could be potentially involved in the whole
regulation.
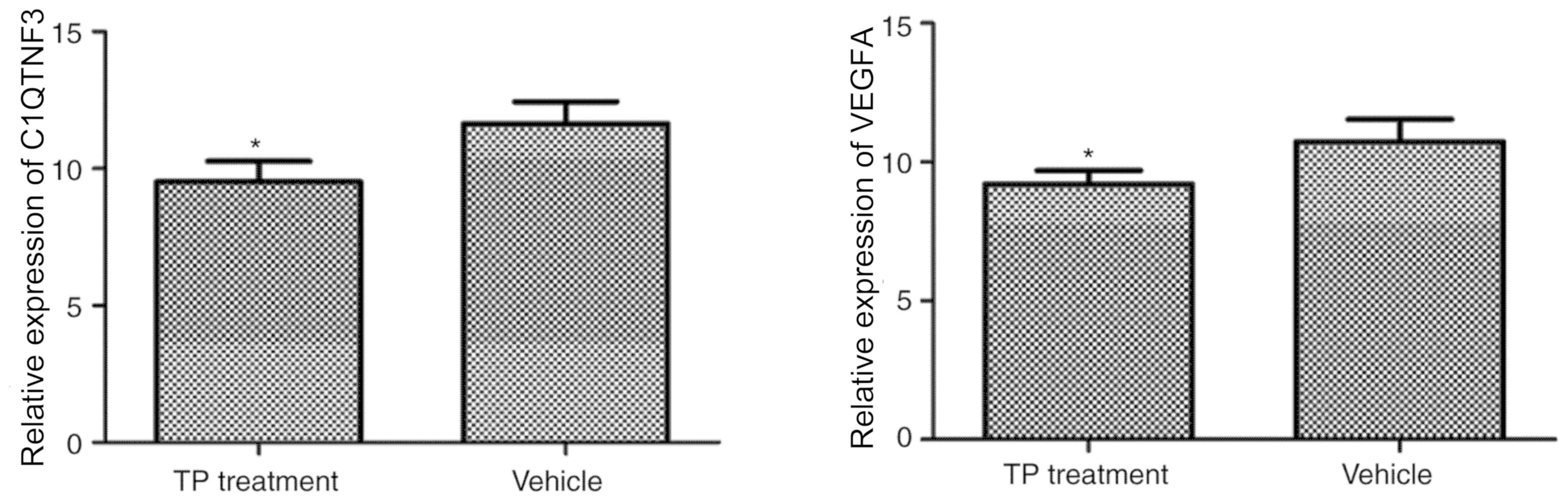
RT-qPCR analysis
The distinct DEGs VEGFA and C1QTNF3 were selected
from the microarray and RT-qPCR was used for detection using the
same RNA samples. Thereby, the results obtained with the gene chip
method were verified. The mRNA expression of VEGFA (27) and C1QTNF3 (28) was significantly downregulated in the
TP treatment group compared with that in the vehicle-treated group
(P<0.05; Fig. 5). The expression
of VEGFA and C1QTNF3 in the vehicle (model) and TP groups was
therefore consistent with that determined by gene chip
detection.
Discussion
The present study investigated the effect of
triptolide treatment on the gene expression profiles in the joint
tissue of RA model rats. The major manifestations of RA are joint
pain, edema, limited range of flexion and extension, morning
stiffness, joint deformities and even joint swelling and burning
(1). Since RA is a lingering disease
that is associated with a high disability rate, the primary purpose
of treatment is to relieve these symptoms and prevent the
progression of the disease. TCM generally treats the body as a
whole, applying the principles of syndrome differentiation
(29,30). It helps relieve the pain and enables
functional recovery of patients with joint swelling and pain, and
is associated with only few adverse reactions, effectively reducing
the risk of disability (31).
It was previously demonstrated that triptolide
significantly reduces paw edema induced by carrageenan or egg white
in AA rats. Its anti-inflammatory effects are mainly mediated by
inhibiting the release of prostaglandins during the inflammatory
response, and by decreasing the permeability of blood vessels and
reducing the concentration of platelets, as well as the
proliferation of fibers in the later stages of inflammation
(32,33). The AA model is an ideal animal model
for investigating RA and screening of drugs, as its pathological
changes and cellular immune abnormalities exhibit several
similarities with RA (34).
Therefore, AA rats were used as a model to study the effect of
triptolide on the gene expression profile in joint tissues, which
may provide an experimental basis for the treatment of RA in the
future. However, one limitation was that the mechanisms of action
of triptolide were not assessed via a comparison between RA model
rats, triptolide-treated RA model rats and normal rats.
As the in vivo pathway and specific molecular
mechanisms of action of triptolide in the prevention and treatment
of RA have remained elusive, the present study investigated the
pharmacological mechanism and effect of triptolide at the molecular
level by employing the gene chip method. The present results
identified distinct DEGs (VEGFA and C1QTNF3) that were selected
from the microarray. It has been reported that activation of the
PI3K/AKT pathway has a negative role in modulating genes that
promote inflammation, thrombogenicity and vascular permeability,
and thereby protect vascular function. Furthermore, the
neuroprotective role of the PI3K/AKT pathway in other disease
models, including cerebral ischemia, has been widely studied
(35). In the present study, GO and
KEGG pathway analysis suggested that the PI3K/AKT signaling pathway
has a key role in the proliferation and apoptosis of synovial cells
in RA joints with triptolide treatment. A previous study has
revealed that an imbalance of the phosphatase and tensin
homolog/PI3K/AKT pathway in rats with AA is one of the mechanisms
of synovial neovasculization (36).
In the present study, the gene microarray profiles
of the treatment and the model group were determined, from which
the DEGs between the two groups were determined. The analysis
demonstrated that triptolide affected the gene expression profile
of AA model rats. A total of 48 DEGs were identified, among which
32 genes were upregulated and 16 were downregulated. The GO terms
of these DEGs were determined, and included transcription factor
regulation, kinase, cytoskeleton and protein synthesis, transport
function, modification of cytokines, inflammatory response, immune
response, cell proliferation and invasion and cell cycle
regulation. The KEGG signaling pathways involved are mainly
associated with angiogenesis, apoptosis, proliferation, oxidative
stress and inflammation. The DEGs were determined to be involved in
tumor-associated pathways, cell cycle regulation and cell receptor
interaction with the extracellular matrix. According to the
Bioinformatics analysis, certain significant DEGs, the key genes,
identified from the microarray results were associated with
tumor-like properties, and the genes located at the key signaling
pathway nodes were determined. Two genes, VEGFA and C1QTNF3, were
selected for verification of their expression levels by RT-qPCR.
The results demonstrated that the expression of the two genes in
the treatment group was lower compared with that in the model
group, which was consistent with the results of the microarray,
indicating that the reproducibility and reliability of the
experiment was satisfactory.
Acknowledgements
Not applicable.
Funding
The present study was supported by Zhejiang
Provincial Natural Science Foundation of China (grant no.
LY14H060005).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124639)
and from the corresponding author on reasonable request.
Authors' contributions
YZ and WH established the animal model, calculated
the arthritis index score and performed drug administration and
tissue collection. WH performed reverse transcription-quantitative
polymerase chain reaction and gene chip array. YZ performed data
analysis and drafted the manuscript. Both authors reviewed and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The animal experiment of the present study was
approved by the Ethics Committee for Use of Experimental Animals in
Zhejiang Chinese Medical University (Hangzhou, China); animal
permit code, SYXK (Zhejiang Province 2008–0115).
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Aletaha D, Neogi T, Silman AJ, Funovits J,
Felson DT, Bingham CO III, Birnbaum NS, Burmester GR, Bykerk VP,
Cohen MD, et al: 2010 Rheumatoid arthritis classification criteria:
An American college of rheumatology/European league against
rheumatism collaborative initiative. Arthritis Rheum. 62:2569–2581.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marrelli A, Cipriani P, Liakouli V,
Carubbi F, Perricone C, Perricone R and Giacomelli R: Angiogenesis
in rheumatoid arthritis: A disease specific process or a common
response to chronic inflammation? Autoimmun Rev. 10:595–598. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kong X, Zhang Y, Liu C, Guo W, Li X, Su X,
Wan H, Sun Y and Lin N: Anti-angiogenic effect of triptolide in
rheumatoid arthritis by targeting angiogenic cascade. PLoS One.
8:e775132013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Klareskog L, Catrina AI and Paget S:
Rheumatoid arthritis. Lancet. 373:659–672. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
The Chinese herbal remedy Tripterygium
wilfordii Hook F in the treatment of rheumatoid arthritis. Ann
Intern Med. 151:I–36. 2009.
|
|
6
|
Goldbach-Mansky R, Wilson M, Fleischmann
R, Olsen N, Silverfield J, Kempf P, Kivitz A, Sherrer Y, Pucino F,
Csako G, et al: Comparison of Tripterygium wilfordii Hook F versus
sulfasalazine in the treatment of rheumatoid arthritis: A
randomized trial. Ann Intern Med. 151:229–240, W49-W51. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu T, Zong M, Fan S, Lu Y, Yu S and Fan L:
Thioredoxin 1 is associated with the proliferation and apoptosis of
rheumatoid arthritis fibroblast-like synoviocytes. Clin Rheumatol.
37:117–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan D, He X, Bian Y, Guo Q, Zheng K, Zhao
Y, Lu C, Liu B, Xu X, Zhang G and Lu A: Triptolide modulates TREM-1
signal pathway to inhibit the inflammatory response in rheumatoid
arthritis. Int J Mol Sci. 17:4982016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu C, Zhang Y, Kong X, Zhu L, Pang J, Xu
Y, Chen W, Zhan H, Lu A and Lin N: Triptolide prevents bone
destruction in the collagen-induced arthritis model of rheumatoid
arthritis by targeting RANKL/RANK/OPG signal pathway. Evid Based
Complement Alternat Med. 2013:6260382013.PubMed/NCBI
|
|
10
|
Lin N, Liu C, Xiao C, Jia H, Imada K, Wu H
and Ito A: Triptolide, a diterpenoid triepoxide, suppresses
inflammation and cartilage destruction in collagen-induced
arthritis mice. Biochem Pharmacol. 73:136–146. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen BJ: Triptolide, a novel
immunosuppressive and anti-inflammatory agent purified from a
Chinese herb Tripterygium wilfordii Hook F. Leuk Lymphoma.
42:253–265. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Glehr M, Fritsch-Breisach M, Lohberger B,
Walzer SM, Moazedi-Fuerst F, Rinner B, Gruber G, Graninger W,
Leithner A and Windhager R: Influence of resveratrol on rheumatoid
fibroblast-like synoviocytes analysed with gene chip transcription.
Phytomedicine. 20:310–318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li ZJ, Wang DQ, Hu SP, Wang XF, He Y and
Zhang ZJ: A gene chip study of ‘Jun Du Yan Bingzhi’ on liver in a
sepsis model of rat. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue.
21:44–47. 2009.(In Chinese). PubMed/NCBI
|
|
14
|
Collins JF: Gene chip analyses reveal
differential genetic responses to iron deficiency in rat duodenum
and jejunum. Biol Res. 39:25–37. 2006.PubMed/NCBI
|
|
15
|
Saeki Y, Matsui T, Saisho K and Tohma S:
Current treatments of rheumatoid arthritis: From the ‘NinJa’
registry. Expert Rev Clin Immunol. 8:455–465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Wei D, Lai Z and Le Y: Triptolide
inhibits CC chemokines expressed in rat adjuvant-induced arthritis.
Int Immunopharmacol. 6:1825–1832. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yifan W, Dengming W, Zheng L, Yanping L
and Junkan S: Triptolide inhibits CCR5 expressed in synovial tissue
of rat adjuvant-induced arthritis. Pharmacol Rep. 59:795–799.
2007.PubMed/NCBI
|
|
18
|
Li S, Chen JW, Xie X, Tian J, Deng C, Wang
J, Gan HN and Li F: Autophagy inhibitor regulates apoptosis and
proliferation of synovial fibroblasts through the inhibition of
PI3K/AKT pathway in collagen-induced arthritis rat model. Am J
Transl Res. 9:2065–2076. 2017.PubMed/NCBI
|
|
19
|
Xiao C, Zhou J, He Y, Jia H, Zhao L, Zhao
N and Lu A: Effects of triptolide from Radix Tripterygium wilfordii
(Leigongteng) on cartilage cytokines and transcription factor
NF-kappaB: A study on induced arthritis in rats. Chin Med.
4:132009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiao Y, Ding H, Huang S, Liu Y, Sun X, Wei
W, Ma J and Zheng F: Bcl-XL and Mcl-1 upregulation by calreticulin
promotes apoptosis resistance of fibroblast-like synoviocytes via
activation of PI3K/Akt and STAT3 pathways in rheumatoid arthritis.
Clin Exp Rheumatol. 36:841–849. 2018.PubMed/NCBI
|
|
21
|
Zhou J, Xiao C, Zhao L, Jia H, Zhao N, Lu
C, Yang D, Tang JC, Chan AS and Lu AP: The effect of triptolide on
CD4+ and CD8+ cells in Peyer's patch of SD rats with collagen
induced arthritis. Int Immunopharmacol. 6:198–203. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Evans SJ, Datson NA, Kabbaj M, Thompson
RC, Vreugdenhil E, De Kloet ER, Watson SJ and Akil H: Evaluation of
affymetrix gene chip sensitivity in rat hippocampal tissue using
SAGE analysis. Serial analysis of gene expression. Eur J Neurosci.
16:409–413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Jiang Z, Ji J, Wang X, Wang T,
Zhang Y, Tai T, Chen M, Sun L, Li X and Zhang L: Gene expression
profiling and pathway analysis of hepatotoxicity induced by
triptolide in Wistar rats. Food Chem Toxicol. 58:495–505. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang J, Wang A, Zeng H, Liu L, Jiang W,
Zhu Y and Xu Y: Effect of triptolide on T-cell receptor beta
variable gene mRNA expression in rats with collagen-induced
arthritis. Anat Rec (Hoboken). 295:922–927. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qu Y, Wu J, Deng JX, Zhang YP, Liang WY,
Jiang ZL, Yu QH and Li J: MicroRNA-126 affects rheumatoid arthritis
synovial fibroblast proliferation and apoptosis by targeting PIK3R2
and regulating PI3K-AKT signal pathway. Oncotarget. 7:74217–74226.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang M, Chen DQ, Chen L, Liu D, Zhao H,
Zhang ZH, Vaziri ND, Guo Y, Zhao YY and Cao G: Novel RAS inhibitors
poricoic acid ZG and poricoic acid ZH attenuate renal fibrosis via
a Wnt/β-catenin pathway and targeted phosphorylation of smad3
signaling. J Agric Food Chem. 66:1828–1842. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ramirez-Bello J, Cadena-Sandoval D,
Fragoso JM, Barbosa-Cobos RE, Moreno-Eutímio MA, Saavedra-Salinas
MÁ, Valencia-Pacheco G, López-Villanueva RF and Jiménez-Morales S:
The VEGFA-1154G/A polymorphism is associated with reduced risk of
rheumatoid arthritis but not with systemic lupus erythematosus in
Mexican women. J Gene Med. 20:e30242018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Murayama MA, Kakuta S, Maruhashi T,
Shimizu K, Seno A, Kubo S, Sato N, Saijo S, Hattori M and Iwakura
Y: CTRP3 plays an important role in the development of
collagen-induced arthritis in mice. Biochem Biophys Res Commun.
443:42–48. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Salliot C and van der Heijde D: Long-term
safety of methotrexate monotherapy in patients with rheumatoid
arthritis: A systematic literature research. Ann Rheum Dis.
68:1100–1104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Malemud CJ: Negative regulators of
JAK/STAT signaling in rheumatoid arthritis and osteoarthritis. Int
J Mol Sci. 18:4842017. View Article : Google Scholar
|
|
31
|
Wang J, Li Y, Yang Y, Du J, Zhao M, Lin F,
Zhang S and Wang B: Systems pharmacology dissection of multiscale
mechanisms of action for herbal medicines in treating rheumatoid
arthritis. Mol Pharm. 14:3201–3217. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tu S, Hu Y, Zeng K, Zhang M, Lai X and
Weichen Z: Effects of triptolide on the expression and activity of
NF-kappaB in synovium of collagen-induced arthritis rats. J
Huazhong Univ Sci Technolog Med Sci. 25:543–545. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Matta R, Wang X, Ge H, Ray W, Nelin LD and
Liu Y: Triptolide induces anti-inflammatory cellular responses. Am
J Transl Res. 1:267–282. 2009.PubMed/NCBI
|
|
34
|
Yang M, Huang J, Pan HZ and Jin J:
Triptolide overcomes dexamethasone resistance and enhanced
PS-341-induced apoptosis via PI3k/Akt/NF-kappaB pathways in human
multiple myeloma cells. Int J Mol Med. 22:489–496. 2008.PubMed/NCBI
|
|
35
|
Liu YY, Jiao ZY, Li W and Tian Q: PI3K/AKT
signaling pathway activation in a rat model of migraine. Mol Med
Rep. 16:4849–4854. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Clarimundo VS, Farinon M, Pedó RT,
Teixeira VON, Nör C, Gulko PS, Xavier RM and de Oliveira PG:
Gastrin-releasing peptide and its receptor increase arthritis
fibroblast-like synoviocytes invasiveness through activating the
PI3K/AKT pathway. Peptides. 95:57–61. 2017. View Article : Google Scholar : PubMed/NCBI
|