Introduction
Glioma is the most common malignant tumor of the
central nervous system with a high recurrence rate and low survival
rate (1,2). Despite the great advances in treatment
strategies over the last decades, the prognosis of glioma remains
unsatisfactory (3,4). Therefore, a deeper understanding of the
molecular mechanisms involved in glioma tumorigenesis is urgently
required.
Genome-wide sequencing has revealed that up to 98%
of the genome are non-coding genes with <2% of genes encoding
proteins (5,6). Among these non-transcribed genes, long
non-coding RNAs (lncRNAs), which consist of >200 nucleotides,
have been demonstrated to have critical roles in various cellular
processes (7,8). Mechanistically, lncRNAs regulate gene
expression via multiple mechanisms including gene imprinting,
epigenetic modification and microRNA (miRNA) degradation (9–11).
Recent studies have characterized a number of lncRNAs in the
initiation of glioma. For example, Feng et al (12) demonstrated that upregulation of the
lncRNA, RNA component of mitochondrial RNA processing
endoribonuclease, could promote glioma cell proliferation and
invasion. Wang et al (13)
demonstrated that the lncRNA, Prader Willi/Angelman region RNA 5,
inhibits proliferation and progression of glioma through
interaction with enhancer of zeste 2 polycomb repressive complex 2
subunit. Ma et al (14)
determined that the lncRNA, differentiation antagonizing
non-protein coding RNA, mediates cisplatin resistance in glioma
cells via activating the AXL receptor tyrosine
kinase/phosphoinositide 3-kinase/protein kinase B/nucleur factor-κB
signaling pathway. However, the roles and underlying mechanisms
through which lncRNAs modulate glioma progression remain largely
unknown.
The present study explored BLACAT1 expression both
in glioma tissues and cell lines. In vitro functional assays
were used to explore the roles of BLACAT1 on glioma progression.
Western blot analysis and reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) were used to determine the
underlying mechanism of BLACAT1 on glioma cell progression.
Patients and methods
Tissue samples
A total of 35 paired glioma tissues and adjacent
non-tumor tissues (16 female and 19 male patients; mean age,
49.61±15.51 years) were obtained from patients with glioma between
August 2016 and December 2017 at the Department of Neurosurgery,
Affiliated Hospital of Hebei University of Engineering (Handan,
China). None of the patients received preoperative treatment.
Tissues were immediately frozen in liquid nitrogen following the
surgical resection and stored at −80°C. Informed consent was
obtained from each patient. This study was approved by the Ethics
Committee of Affiliated Hospital of Hebei University of
Engineering.
Cell culture and transfection
The normal human astrocyte (NHA) cells and glioma
cell lines (U251, T98G, H4, A172 and LN229) were obtained from the
American Type Culture Collection (Manassas, VA, USA). Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) containing
10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), and incubated under standard conditions (37°C,
5% CO2).
The small interfering RNAs (siRNAs) targeting
BLACAT1 (si-BLACAT1) and scrambled negative control siRNA (si-NC)
were synthesized by Genepharma (Shanghai, China). The si-BLACAT1
sequences were as follows: si-BLACAT1-1
5′-AGGCUGGUUUCUGCCCUCAUCCUUU-3′; si-BLACAT1-2,
5′-GCCCAGCUUCUAGUCCUCUCCUUAU-3′; and si-NC,
5′-AATTCTCCGAACGTGTCACGT-3′. Cell transfection was performed by
using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according
to the manufacturer's introductions. The transfection was performed
48 h prior to the subsequent analysis.
Reverse transcription-quantitative
polymerase chain reaction
Total RNA was extracted from tissues or cells using
TRIzol reagent (Invitrogen) according to the manufacturer's
instructions. Then, the reverse transcription was done with a
Reverse Transcription kit (Takara Bio, Inc., Otsu, Japan). qPCR
analysis was performed with SYBR Green (Takara Bio, Inc.). The
relative expression levels were calculated by the 2−ΔΔCq
method (15) and normalized to
GAPDH. qPCR reactions were performed using the ABI7500 system
(Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The thermocycling conditions of qPCR were as follows: 94°C
for 15 min, followed by 45 cycles at 94°C for 10 sec, 60°C for 30
sec and 72°C for 30 sec. The primer sequences were as follows:
Vimentin, 5′-AATTTCACGCAGAGCAACAG-3′ and
5′-CCACTAAGGCAGCACGTAAA-3′; E-cadherin, 5′-TTAACCTCACCAATCCTTGCT-3′
and 5′-TAGCCCATTTCTTCCCAATC-3′; and N-cadherin,
5′-AACCTAGCCTACTGGCCAAA-3′ and 5′-AACATCGAGGTCGTAAACCC-3′.
Cell proliferation assays
Cell proliferation was measured by CCK-8 (Roche
Diagnostics, Basel, Switzerland) and colony formation assay. For
CCK-8 assay, cells (1×103/well) were plated in the
96-well plates (Corning Inc., Corning, NY, USA) then 10 µl of CCK-8
was added to each well at the time of harvest. After 2 h, the
absorbance was recorded at 450 nm to determine cell viability. For
the colony formation assay, cells were seeded in the 6-well plate
at 1,000 cells per well and cultured for 10 days. Colonies were
fixed with methanol and stained with 0.1% crystal violet. The
colonies were observed and counted with microscope (magnification,
×40; Nikon Corporation, Tokyo, Japan).
Cell apoptosis analysis
Cells were harvested following transfection were
harvested and stained with propidium iodide (PI) for 30 min. The
fluorescein isothiocyanate (FITC)-Annexin V Apoptosis Detection kit
(BD Biosciences, San Jose, CA, USA) based on double staining with
FITC-Annexin V and PI was applied to detect the apoptosis level.
Then apoptotic cells were analyzed with a FC500 flow cytometer
equipped with Cell Quest 3.0 software (BD Biosciences, Franklin
Lakes, NY, USA).
Wound-healing assay
Cells were seeded in 6-well plates, cultured to ~90%
confluence, then a wound was generated by scraping the cells with a
200 µl tip. The cells were washed with PBS to remove debris and
cultured with DMEM medium containing 10% FBS. After 24 h, the wound
healing of different groups was observed using microscopy.
(magnification, ×100; Nikon Corporation).
Transwell invasion assay
The Transwell invasion assay was performed using
Matrigel-coated (BD Biosciences) 24-well Transwell chambers
(Corning Inc.). Cells were resuspended in serum-free 200 µl DMEM
medium at a density of 5×104 cells/well into the upper
chamber of Transwell cell culture chambers. The lower chambers of
the Transwell were filled with DMEM supplemented with 10% FBS.
Following incubation for 24 h, cells on the upper surface of the
filter were removed using a cotton swab. Cells migrating through
the filter to the lower surface were fixed with 4% paraformaldehyde
and stained with 0.1% crystal violet for 10 min. The cells on the
bottom of the membrane were calculated from five random light
microscopic fields using a light microscope (magnification, ×100;
Nikon Corporation).
Western blot analysis
Protein lysates were prepared using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Shanghai, China) and the concentration was measured
with a bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). Then, 40 µg (loaded per lane) protein samples were
separated by electrophoresis in 10% SDS-polyacrylamide gel and then
transferred onto a polyvinylidene difluoride membrane. The
membranes were incubated with primary antibody against N-cadherin
(cat. no. ab76011; 1:1,000), Vimentin (cat. no. ab71144; 1:1,000),
E-cadherin (cat. no. ab133597; 1:1,000), β-catenin (cat. no.
ab27798; 1:1,000), Cyclin D1 (cat. no. ab21699, 1:1,000) and GAPDH
(cat. no. b37168, 1:5,000; all Abcam, Cambridge, UK) at 4°C
overnight. The membrane was washed for three times with
Tris-buffered saline and Polysorbate 20 and incubated with
horseradish peroxidase-labeled secondary antibody goat anti-rabbit
(cat. no. ab97051; 1:2,000 dilution; Abcam) for 1 h at room
temperature. The bands were visualized using enhanced
chemiluminescent reagent (Millipore, Bedford, MA, USA) and the
bands were quantified by densitometry using ImageJ software
(V1.8.0; National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
All data were presented as mean ± standard
deviation. Statistical analysis was performed using the SPSS
software package (SPSS, Inc., Chicago, IL, USA). Statistical
significance was assessed by Student's t-test or one-way analysis
of variance was applied followed by Least Significance Difference
post hoc test. P<0.05 was considered to indicate statistical
significance.
Results
LncRNA BLACAT1 is upregulated in
glioma
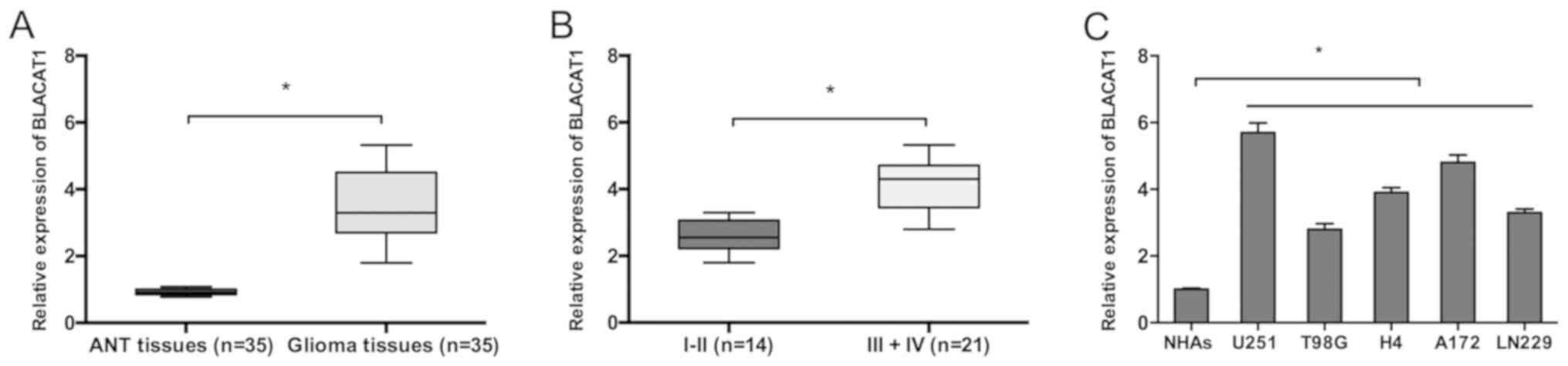
BLACAT1 mRNA expression in glioma was determined by
RT-qPCR. Results demonstrated that BLACAT1 mRNA expression was
significantly increased in glioma tissues compared to adjacent
non-tumor (ANT) tissues (Fig. 1A;
P<0.05). BLACAT1 mRNA expression was also markedly increased in
glioma tumors of high grade (III–IV) compared with low grade (I–II)
tumors (Fig. 1B; P<0.05). The
upregulation of BLACAT1 in glioma was further verified in glioma
cell lines by RT-qPCR. Results determined that BLACAT1 was highly
expressed in glioma cell lines (U251, T98G, H4, A172 and LN229)
compared with the NHA cell line. U251 and A172 cells were selected
for further assays due to displaying the highest BLACAT1 expression
(Fig. 1C; P<0.05). Taken
together, these findings demonstrated that BLACAT1 may have a
prominent role in glioma carcinogenesis.
LncRNA BLACAT1 inhibition decreases
glioma cell proliferation
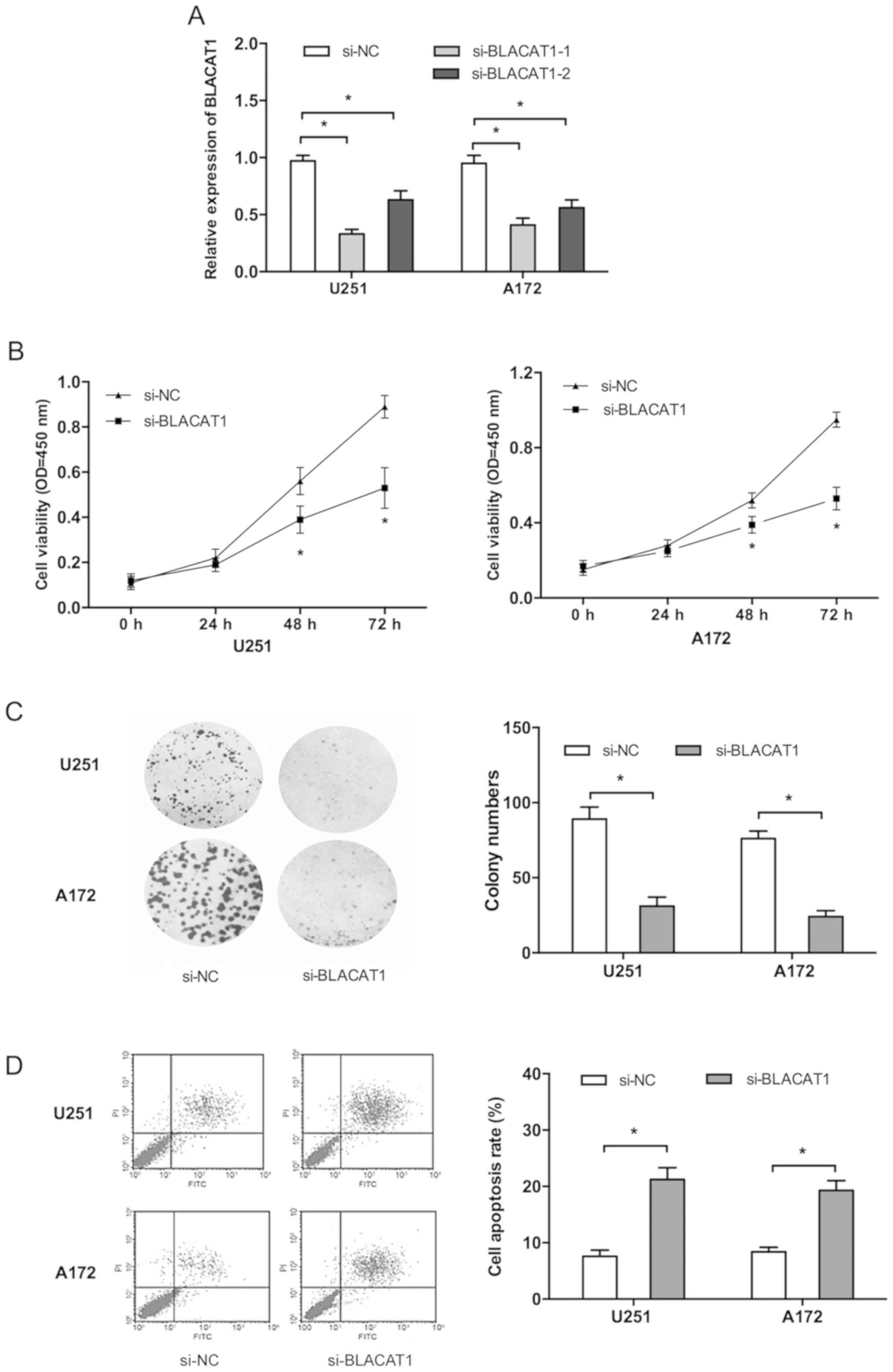
To further explore the roles of BLACAT1 in glioma,
both U251 and A172 cells were transfected with si-BLACAT1 or si-NC,
and one si-RNA (si-BLACAT1-1) with a high interference efficiency
was selected for subsequent experiments (Fig. 2A; P<0.05). CCK-8 assay
demonstrated that BLACAT1 inhibition significantly suppressed
glioma cell proliferation compared with the si-NC group (Fig. 2B; P<0.05). Colony formation assay
similarly indicated that colony numbers of glioma cells transfected
with si-BLACAT1 were significantly inhibited compared with the
si-NC group (Fig. 2C; P<0.05). In
addition, flow cytometry analysis demonstrated that BLACAT1
inhibition significantly increased glioma cell apoptosis compared
with si-NC group (Fig. 2D;
P<0.05).
LncRNA BLACAT1 inhibition decreases
glioma cell invasion
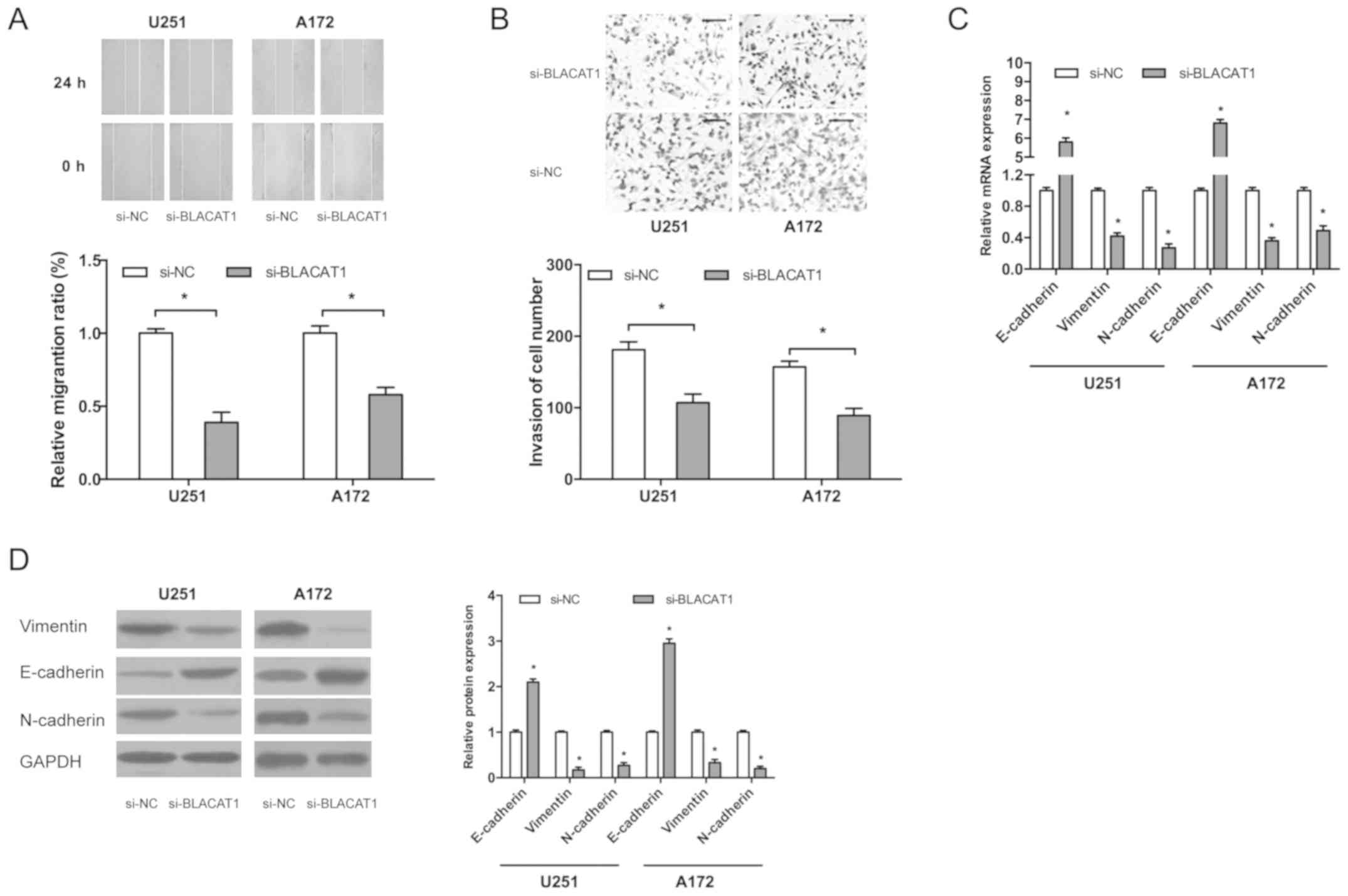
Wound-healing assay indicated that BLACAT1
inhibition significantly decreased the migration ability of glioma
cells compared with the control (Fig.
3A; P<0.05). Transwell invasion assay indicated that BLACAT1
inhibition reduced the invasion ability of glioma cells (Fig. 3B; P<0.05).
Epithelial-mesenchymal-transition (EMT) is an important process for
tumor metastasis therefore in the present study the effects of
BLACAT1 on EMT processes were investigated. Results demonstrated
that BLACAT1 inhibition decreased mRNA and protein levels of
N-cadherin and vimentin whilst E-cadherin mRNA and protein levels
increased (Fig. 3C and D;
P<0.05). These findings demonstrated that BLACAT1 knockdown
inhibited glioma cell proliferation and invasion in
vitro.
LncRNA BLACAT1 activates Wnt/β-catenin
signaling in glioma
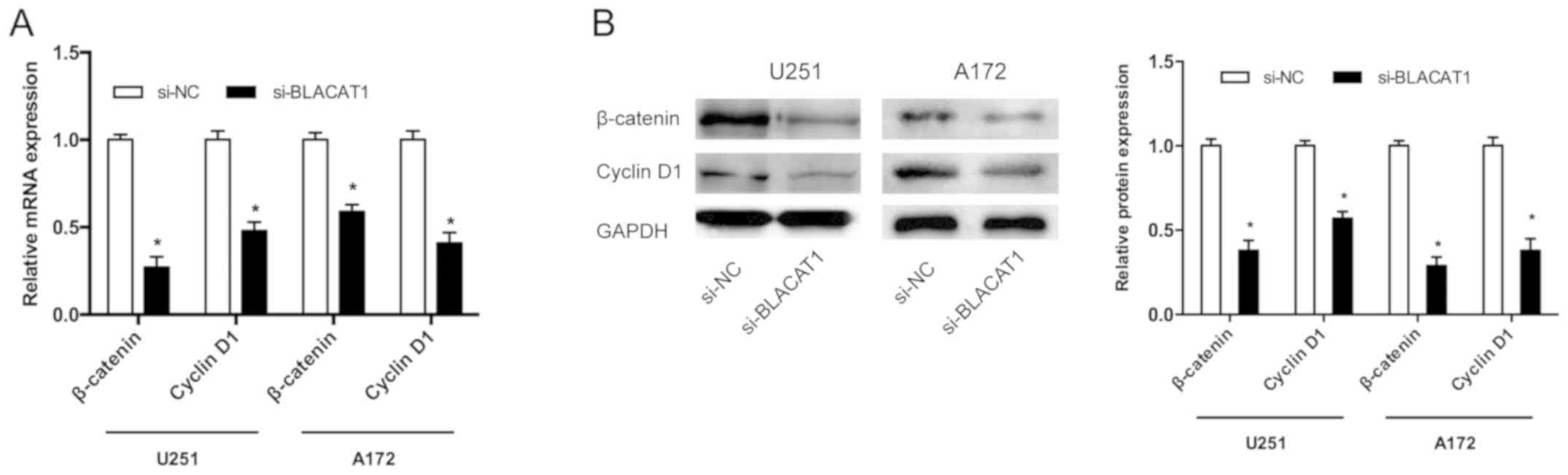
To further explore the BLACAT1-mediated molecular
mechanism, effects of BLACAT1 on the Wnt/β-catenin pathway were
investigated. Results demonstrated that BLACAT1 knockdown
significantly decreased the β-catenin and cyclin D1 mRNA and
protein levels in both U251 and A172 cells (Fig. 4A and B; P<0.05). These findings
indicated that BLACAT1 may serve as a novel oncogenic lncRNA by
regulating the Wnt/β-catenin signaling pathway in glioma.
Discussion
Initiation and development of glioma involves both
genetic and epigenetic alterations (16). LncRNAs are a class of non-coding
RNAs, which have important roles under both physiological and
pathological conditions (7). BLACAT1
is present on human chromosome 1q32.1 and has a transcript of 2,616
kb (17). Recent studies
demonstrated that BLACAT1 may have important roles in tumor
progression. For example, Shan et al (18) determined that lncRNA BLACAT1 promotes
cell proliferation and invasion in human cervical cancer. Ye et
al (19) identified that lncRNA
BLACAT1 promotes the proliferation, migration, and invasion of
non-small cell lung cancer through sponging miR-144. Wu et
al (20) revealed that lncRNA
BLACAT1 modulates ABCB1 to promote oxaliplatin resistance of
gastric cancer via sponging miR-361. However, Liao et al
(21) demonstrated that lncRNA
BLACAT1 is downregulated in papillary thyroid carcinoma and could
act as a suppressor gene in tumor progression. The expression and
underlying mechanisms of lncRNA BLACAT1 in glioma progression
remains unclear. The present study demonstrated that lncRNA BLACAT1
was highly expressed in glioma tissues and cell lines. In addition,
lncRNA BLACAT1 inhibition reduced proliferation, migration, and
invasion of glioma cells.
EMT has been proposed as a key process in the
induction of tumor cell invasion and metastasis (22). For example, Li et al (23) determined that lncRNA SLC25A25
antisense RNA 1 inhibition promotes proliferation, chemoresistance,
and EMT in colorectal cancer cells. He et al (24) demonstrated that lncRNA FEZF1
antisense RNA 1 enhances EMT processes through suppressing
E-cadherin expression and regulating the WNT pathway in non-small
cell lung cancer. The present study demonstrated that BLACAT1
inhibition suppressed EMT processes by decreasing expression of the
mesenchymal markers N-cadherin and vimentin whilst increasing
expression of the epithelial marker E-cadherin. The mechanism
involved in the regulative effects of BLACAT1 on EMT processes was
then investigated.
The Wnt/β-catenin pathway has been reported to have
an important role in EMT (25). For
example, Tian et al (26)
demonstrated that miR-361-5p inhibits the mobility of gastric
cancer cells through suppressing EMT via the Wnt/β-catenin pathway.
Yuan et al (27) identified
that rhomboid 5 homolog 1 regulates the APC-mediated stimulation of
EMT and also proliferation of colorectal cancer cells via the
Wnt/β-catenin pathway. The present study investigated whether
BLACAT1 modulated the Wnt/β-catenin pathway in glioma cells.
Results demonstrated that BLACAT1 inhibition reduced β-catenin and
cyclin D1 mRNA and protein expression levels. Based on these
findings, BLACAT1 might promote EMT processes, at least partly, via
activating the Wnt/β-catenin pathway in glioma.
In conclusion, these results determined that lncRNA
BLACAT1 was upregulated in glioma, and highly expressed BLACAT1
promoted cellular proliferation and invasion. These effects may be
related to increased EMT-related gene expression via promotion of
the Wnt/β-catenin pathway. The present findings revealed that
BLACAT1 might be a novel therapeutic target for glioma
treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XL and JW conceived and designed the study, and
analyzed and interpreted the results. XL, SQ, DM and JF performed
experiments and wrote this manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
For the use of human samples, the protocol for the
present study was approved by the Institutional Ethics Committee of
Affiliated Hospital of Hebei University of Engineering, and all
enrolled patients signed a written informed consent document.
Patient consent for publication
All patients within the present study provided
consent for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schwartzbaum JA, Fisher JL, Aldape KD and
Wrensch M: Epidemiology and molecular pathology of glioma. Nat Clin
Pract Neurol. 2:494–503. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trabelsi S, Brahim DH, Ladib M, Mama N,
Harrabi I, Tlili K, Yacoubi MT, Krifa H, Hmissa S, Saad A and Mokni
M: Glioma epidemiology in the central tunisian population:
1993–2012. Asian Pac J Cancer Prev. 15:8753–8757. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wrensch M, Fisher JL, Schwartzbaum JA,
Bondy M, Berger M and Aldape KD: The molecular epidemiology of
gliomas in adults. Neurosurg Focus. 19:E52005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Reich DE, Cargill M, Bolk S, Ireland J,
Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward
R and Lander ES: Linkage disequilibrium in the human genome.
Nature. 411:199–204. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lander ES: Initial impact of the
sequencing of the human genome. Nature. 470:187–197. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Monnier P, Martinet C, Pontis J, Stancheva
I, Ait-Si-Ali S and Dandolo L: H19 lncRNA controls gene expression
of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad
Sci U S A. 110:20693–20698. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016.PubMed/NCBI
|
|
12
|
Feng W, Li L, Xu X, Jiao Y and Du W:
Up-regulation of the long non-coding RNA RMRP contributes to glioma
progression and promotes glioma cell proliferation and invasion.
Arch Med Sci. 13:1315–1321. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang XP, Shan C, Deng XL, Li LY and Ma W:
Long non-coding RNA PAR5 inhibits the proliferation and progression
of glioma through interaction with EZH2. Oncol Rep. 38:3177–3186.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P
and Lin K: Long noncoding RNA DANCR mediates cisplatin resistance
in glioma cells via activating AXL/PI3K/Akt/NF-κB signaling
pathway. Neurochem Int. 118:233–241. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alonso MM, Diez-Valle R, Manterola L,
Rubio A, Liu D, Cortes-Santiago N, Urquiza L, Jauregi P, Lopez de
Munain A, Sampron N, et al: Genetic and epigenetic modifications of
Sox2 contribute to the invasive phenotype of malignant gliomas.
PLoS One. 6:e267402011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Dai M, Zhu H, Li J, Huang Z, Liu
X, Huang Y, Chen J and Dai S: Evaluation on the diagnostic and
prognostic values of long non-coding RNA BLACAT1 in common types of
human cancer. Mol Cancer. 16:1602017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shan D, Shang Y and Hu T: Long noncoding
RNA BLACAT1 promotes cell proliferation and invasion in human
cervical cancer. Oncol Lett. 15:3490–3495. 2018.PubMed/NCBI
|
|
19
|
Ye JR, Liu L and Zheng F: Long noncoding
RNA bladder cancer associated transcript 1 promotes the
proliferation, migration, and invasion of nonsmall cell lung cancer
through sponging miR-144. DNA Cell Biol. 36:845–852. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu X, Zheng Y, Han B and Dong X: Long
noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin
resistance of gastric cancer via sponging miR-361. Biomed
Pharmacother. 99:832–838. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liao D, Lv G, Wang T, Min J, Wang Y and
Liu S: Prognostic value of long non-coding RNA BLACAT1 in patients
with papillary thyroid carcinoma. Cancer Cell Int. 18:472018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Huang S, Li Y, Zhang W, He K, Zhao
M, Lin H, Li D, Zhang H, Zheng Z and Huang C: Decreased expression
of LncRNA SLC25A25-AS1 promotes proliferation, chemoresistance, and
EMT in colorectal cancer cells. Tumor Biol. 37:14205–14215. 2016.
View Article : Google Scholar
|
|
24
|
He R, Zhang FH and Shen N: LncRNA
FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through
suppressing E-cadherin and regulating WNT pathway in non-small cell
lung cancer (NSCLC). Biomed Pharmacother. 95:331–338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tian L, Zhao Z, Xie L and Zhu J:
MiR-361-5p inhibits the mobility of gastric cancer cells through
suppressing epithelial-mesenchymal transition via the Wnt/β-catenin
pathway. Gene. 675:102–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yuan H, Wei R, Xiao Y, Song Y, Wang J, Yu
H, Fang T, Xu W and Mao S: RHBDF1 regulates APC-mediated
stimulation of the epithelial-to-mesenchymal transition and
proliferation of colorectal cancer cells in part via the
Wnt/β-catenin signalling pathway. Exp Cell Res. 368:24–36. 2018.
View Article : Google Scholar : PubMed/NCBI
|