Introduction
Liver fibrosis is usually observed in chronic liver
diseases with different causes and its morphology is characterized
by the deposition of a large amount of extracellular matrix in
liver tissues (1). Liver fibrosis
may progress into cirrhosis that threatens the health and life of
patients (2). Chronic Hepatitis B
can cause liver inflammation and fibrosis. If not detected promptly
and treated, patients can develop cirrhosis (3). Decompensated cirrhosis has poor
prognosis and can easily develop into hepatocellular carcinoma, a
serious and life-threatening complication of cirrhosis (4,5). A
previous prospective study determined that the annual rate of
conversion from chronic Hepatitis B into liver cirrhosis is 2.1%
(6). A follow-up study of
HBeAg-negative patients with chronic Hepatitis B over an average of
9 years (1–18.4 years) demonstrated that the percentage of patients
who developed liver cirrhosis was 23% (7,8).
Interferon and nucleotide analogues are currently used in the
treatment of Hepatitis B virus (9).
In addition to antiviral therapy, the treatment of hepatic fibrosis
and cirrhosis following Hepatitis B infection needs to strengthen
hepatocyte regeneration, inhibit inflammation and reduce the risk
of further complications (10,11).
Therefore, it is important to investigate the mechanism of the
transformation from liver fibrosis to cirrhosis at the molecular
level in order to develop novel treatments for Hepatitis B.
MicroRNA (miR) molecules are a type of non-encoding
small RNA molecule that are 18–22 nucleotides long and can regulate
the expression of proteins at the mRNA level (12–14). The
processes of liver fibrosis and cirrhosis are accompanied by
alterations in the expression of different miRs and proteins,
suggesting that miR may serve important roles in regulating the
expression of proteins associated with liver fibrosis and cirrhosis
(15,16). Hepatitis B may also induce
inflammation to a certain extent (17,18),
which, in turn, may lead to the development of liver cancer
(19,20). Previous studies have demonstrated
that the inflammatory factor interleukin (IL)-6 serves a key role
in the onset of hepatitis B (21,22).
However, the regulation of liver fibrosis and cirrhosis by IL-6 and
the mechanism by which miRs regulate IL-6 remain unclear. In the
present study, the expression of IL-6 mRNA and protein in patients
with liver fibrosis and cirrhosis was assessed and the association
between miR-146a and IL-6 was evaluated.
Materials and methods
Patient samples
A total of 36 patients with Hepatitis B and liver
fibrosis and 25 patients with Hepatitis B and liver cirrhosis
admitted to the Linyi People's Hospital (Linyi, China) between June
2012 and February 2016 were included in the present study. Among
the 36 patients with liver fibrosis, 20 were male and 16 were
female, and they had a median age of 41 years (age range, 20–61
years). Among the 25 patients with liver cirrhosis, 15 were male
and 10 were female, and they had a median age of 43 years (age
range, 18–65 years). All patients were diagnosed with liver
fibrosis or cirrhosis according to the guidelines for the
prevention and treatment of chronic Hepatitis B (23). The levels of alanine aminotransferase
and aspartate aminotransferase were determined in patients with
liver fibrosis using an automatic biochemical analyzer (Beckman
Coulter, Inc., Brea, CA, USA). The normal concentrations in the
blood for alanine aminotransferase and aspartate aminotransferase
levels were 0–40 and 5–40 U/l, respectively. The levels of alanine
aminotransferase and aspartate aminotransferase in patients with
liver fibrosis were 168.6±39.6 and 146.2±28.5 U/l, respectively.
The levels of alanine aminotransferase and aspartate
aminotransferase in patients with liver cirrhosis were 182.8±51.9
and 161.2±39.3 U/l, respectively. Patients with immune-related
diseases or immune diseases, including diabetes and cancer, were
excluded from the current study. Liver biopsies (1.0–1.5 cm in
length) and peripheral blood samples (20 ml) were collected from
each patient. Blood serum was isolated from peripheral blood
following centrifugation at 400 × g for 10 min at 4°C. To obtain
peripheral blood mononuclear cells (PMBCs), a mixture of
heparin-anticoagulant venous blood and equal amount of serum-free
Iscove's modified Dulbecco's medium (1:1 v/v) was added gently to
the lymphocyte separation medium before centrifugation at 400 × g
for 30 min at 4°C. After centrifugation, the middle layer was
aspirated and mixed with 5 volumes of Hanks solution before
centrifugation at 300 × g for 10 min at 4°C, washed twice and cell
density was adjusted to 1×106 cells/ml. Cells were
seeded into six-well plates at a density of 3×106
cells/well and incubated at 37°C in a 5% CO2-humidified
incubator for 1–2 h. The cells that attached after 1–2 h incubation
were PBMCs. All procedures were approved by the Ethics Committee of
Linyi People's Hospital and written informed consent was obtained
from all patients or their families.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Prior to total RNA extraction, tissue samples (100
mg) were ground into powder using liquid nitrogen. Total RNA was
extracted using 1 ml TRIzol® (10606ES60; Shanghai Yeasen
Biotechnology, Co., Ltd., Shanghai, China). Following lysis, miRNA
was isolated using the miRcute miRNA isolation kit (Tiangen Biotech
Co., Ltd., Beijing, China). The purity of RNA was determined at
A260/A280 using an ultraviolet spectrophotometer (NanoDrop ND1000;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA). The
TIANScript II cDNA First Chain Synthesis kit (KR107; Tiangen,
Biotech, Co., Ltd., Beijing, China) was used to reverse transcribe
1 µg RNA into cDNA, which was stored at −20°C. The primer sequences
used were as follows: IL-6 forward, 5′-GGCACTGGCAGAAAACAACC-3′ and
reverse, 5′-GCAAGTCTCCTCATTGAATCC-3′; GAPDH, forward,
5′-GGGAAACTGCGGCGTGAT-3′ and reverse, 5′-AAAGGTGGAGGAGTGGGT-3′. The
PCR reaction mixture (20 µl) contained 10 µl SuperReal PreMix
SYBR-Green (Tiangen Biotech Co., Ltd.), 0.5 µl upstream primer, 0.5
µl downstream primer, 2 µl cDNA and 7 µl ddH2O. The
following thermocyclic conditions were used for the qPCR: Initial
denaturation at 95°C for 30 sec; 45 cycles of 95°C for 5 sec and
57°C for 30 sec. The qPCR was performed on an iQ5 Real-Time PCR
Detection system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The 2−ΔΔCq method was used to calculate the relative
expression of IL-6 mRNA against the reference gene GAPDH (24). For miR-146a, the primer sequences
were as follows: miR-146a forward, 5′-CGGCGGTGAGAACTGAATTCCA-3′ and
reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The following thermocyclic conditions were used for the qPCR:
Initial denaturation at 95°C for 5 min; 40 cycles of denaturation
at 95°C for 10 sec, 60°C for 20 sec and 70°C for 10 sec. The qPCR
was performed as above and the 2−ΔΔCq method was used to
calculate the relative expression of miR-146a against the reference
gene U6.
Western blotting
Total protein was extracted from tissues and PBMCs
using radioimmunoprecipitation assay lysis buffer (600 µl; 50 mM
Tris-base, 1 mM EDTA, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1%
Triton X-100, 1% sodium deoxycholate; Beyotime Institute of
Biotechnology, Shanghai, China) according to the manufacturer's
protocol. Following lysis for 50 min on ice, the mixture was
centrifuged at 13,400 × g and 4°C for 5 min. The supernatant was
used to determine protein concentration using a bicinchoninic acid
protein concentration determination kit [RTP7102; Real-Times
(Beijing) Biotechnology Co., Ltd., Beijing, China]. Protein samples
(20 µg) were then mixed with sodium dodecyl sulfate loading buffer
prior to denaturation in a boiling water bath for 5 min.
Afterwards, 50 µg protein/lane was separated via SDS-PAGE on a 10%
gel. Resolved proteins were then transferred to polyvinylidene
difluoride membranes on ice (100 V, 2 h) and blocked with 5%
skimmed milk at room temperature for 1 h. Subsequently, membranes
were incubated with rabbit anti-human IL-6 polyclonal primary
antibody (1:1,000; ab6672) and rabbit anti-human β-actin primary
antibody (1:5,000; ab129348; both Abcam, Cambridge, UK) at 4°C
overnight. Following three extensive washes with phosphate-buffered
saline with Tween 20 for 15 min each time, membranes were incubated
with goat anti-rabbit horseradish peroxidase-conjugated secondary
antibody (1:3,000; ab6721; Abcam) for 1 h at room temperature prior
to three washes with phosphate-buffered saline with Tween-20 for 15
min each time. Subsequently, the membrane was developed using an
enhanced chemiluminescence detection kit (ab65623; Abcam) for
imaging. Image lab v3.0 software (Bio-Rad Laboratories, Inc.) was
used to acquire and analyze imaging signals. The relative content
of IL-6 protein was expressed as the IL-6/β-actin ratio.
Enzyme-linked immunosorbent assay
(ELISA)
Serum samples were tested using an IL-6 ELISA kit
(ab178013; Abcam). In microplates, standards (50 µl), samples (10
µl sample liquid and 40 µl diluent) and blank were added to
predefined wells. In the wells for standards and samples,
horseradish peroxidase-labeled conjugates (100 µl) were added prior
to sealing the plates for incubation at 37°C for 1 h. Following
five washes of the plates, substrates A (50 µl) and B (50 µl) were
added to each well. Following incubation at 37°C for 15 min, stop
solution (50 µl) was added to each well, and the absorbance of each
well was measured at 450 nm within 15 min.
Dual luciferase reporter assay
Bioinformatic predictions are a useful tool to use
to evaluate miR function. To determine the regulation of IL-6 in
liver fibrosis and cirrhosis, miRanda (www.microrna.org/rnicrorna/home.do), TargetScan
(www.targetscan.org), PiTa (genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNAhybrid (bibiserv.techfak.uni-bielefeld.de/rnahybrid) and
PICTA (pictar.mdc-berlin.de) were used to
predict the miR molecules that regulate IL-6 and indicated that
miR-146a may regulate IL-6 (Fig. 1).
According to the bioinformatic results, wild-type (WT) and mutant
seed regions of miR-146a in the 3′-untranslated region (UTR) of
IL-6 were chemically synthesized in vitro, SpeI and
HindIII restriction sites were added, and the WT and mutant
miR-146a were cloned into pMIR-REPORT luciferase reporter plasmids
(Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Plasmids (0.8 µg) with WT or mutant 3′-UTR DNA sequences were
co-transfected with 100 nM agomiR-146a (Sangon Biotech, Shanghai,
China) into 293T cells were obtained from the Cell Bank of the
Chinese Academy of Sciences (Shanghai, China). Following 24 h
cultivation, cells were lysed using a dual luciferase reporter
assay kit (Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol and fluorescence intensity was measured
using a GloMax 20/20 luminometer (Promega Corporation). Renilla
fluorescence activity was used as internal reference to measure the
fluorescence values of each group of cells.
Statistical analysis
The results were analyzed using SPSS 18.0
statistical software (SPSS, Inc., Chicago, IL, USA). The data are
presented as the mean ± standard deviation and were tested for
normality. Multigroup measurement data were analyzed using one-way
analysis of variance. In the case of homogeneity of variance, Least
Significant Difference and Student-Newman-Keuls methods were used;
in the case of heterogeneity of variance, Tamhane's T2 or Dunnett's
T3 method were used. P<0.05 indicated a statistically
significant difference.
Results
IL-6 mRNA expression in liver tissues,
PBMCs and serum from patients with liver cirrhosis is higher than
that from patients with liver fibrosis
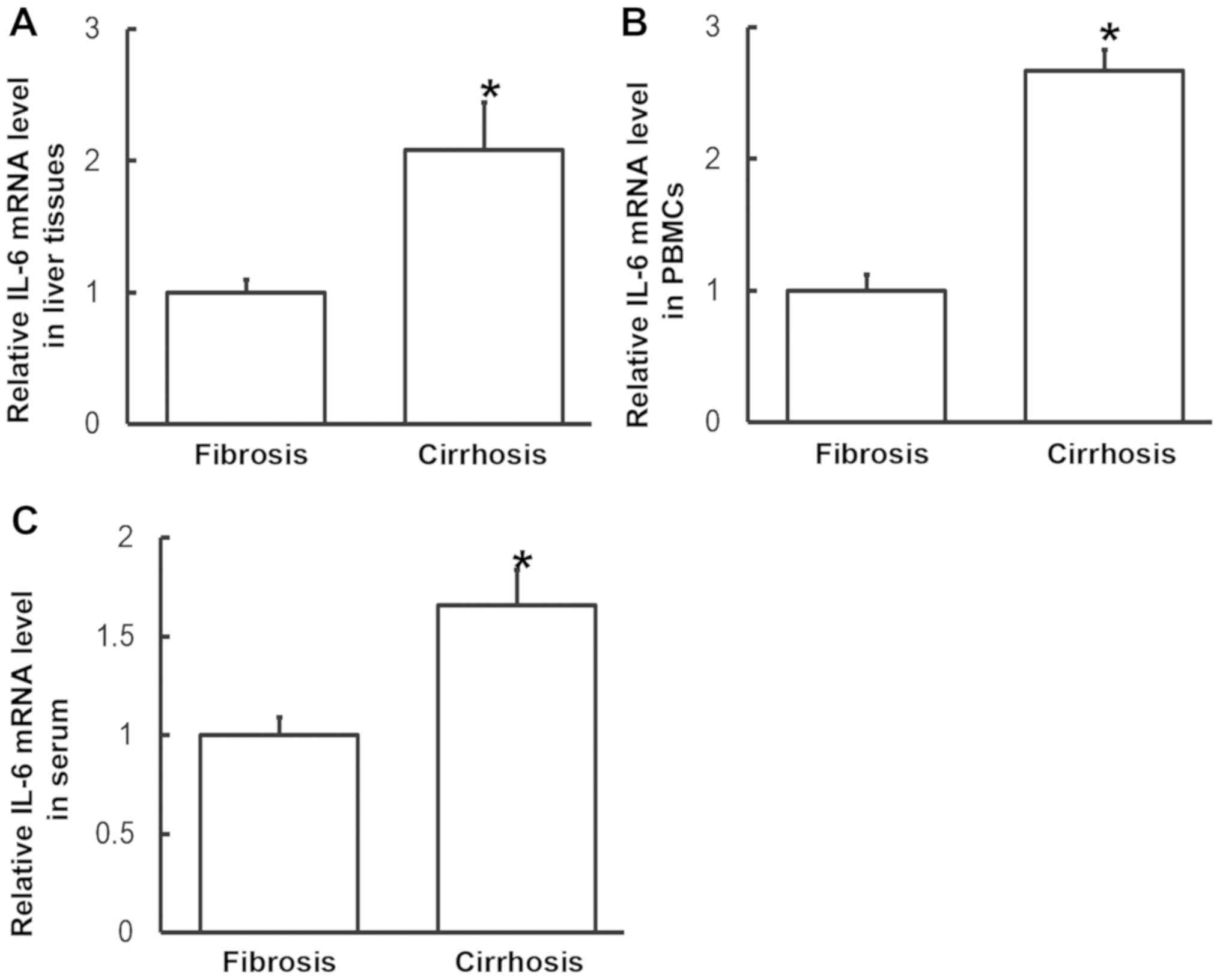
To measure the expression of IL-6 mRNA in the
different samples, RT-qPCR was performed. The data indicated that
levels of IL-6 mRNA in liver tissues, PBMCs and serum from patients
with liver cirrhosis were significantly increased compared with
those from patients with liver fibrosis (P<0.05; Fig. 2). These results suggest that the
expression of IL-6 mRNA in patients with liver cirrhosis is higher
than in patients with liver fibrosis.
Expression of IL-6 protein in liver
tissues and PBMCs from patients with liver cirrhosis is
enhanced
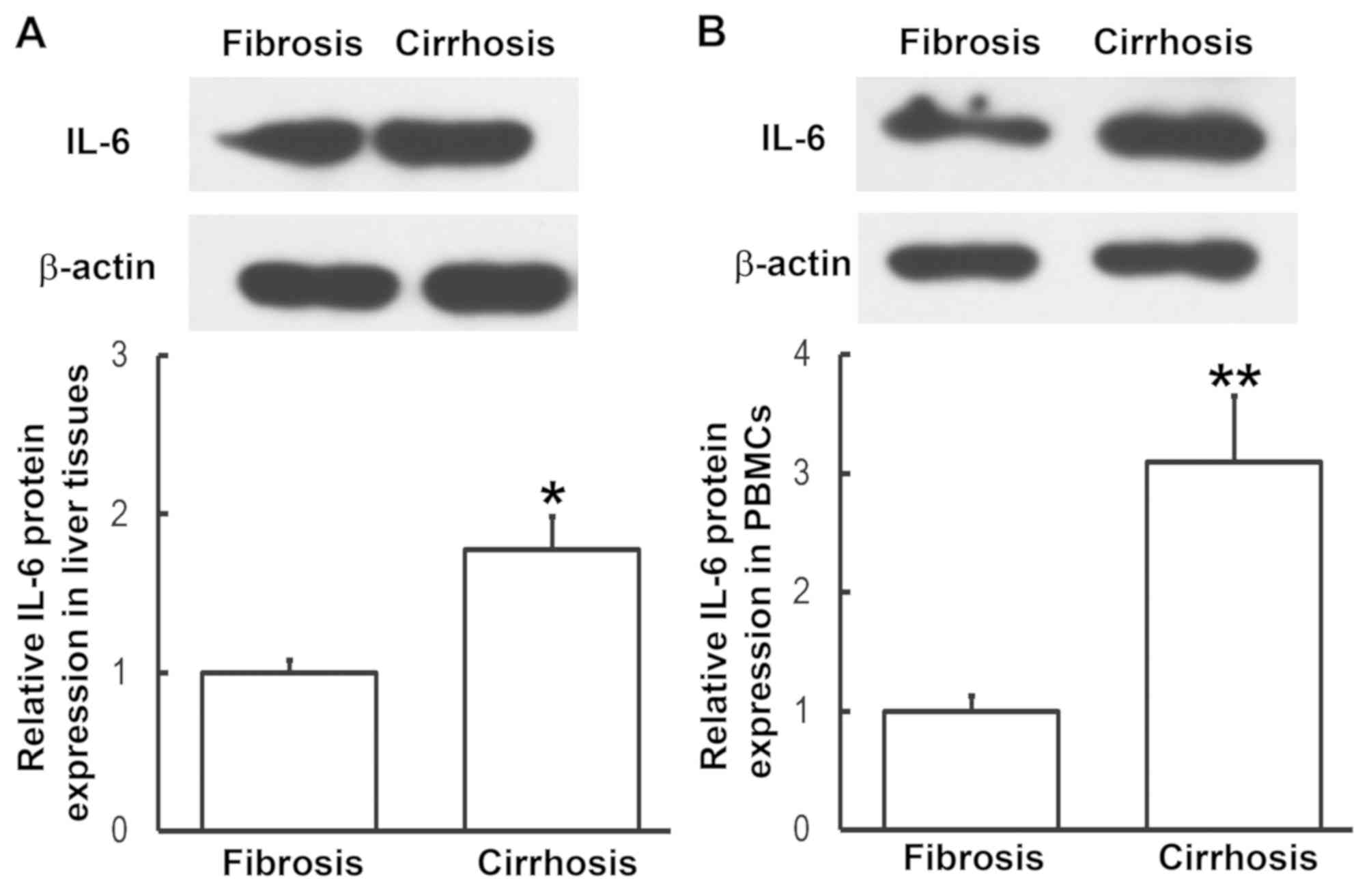
To determine the expression of IL-6 protein in liver
tissues and PBMCs, western blotting was performed. Compared with
patients with liver fibrosis, the expression of IL-6 in liver
tissues and PBMCs from patients with liver cirrhosis was
significantly enhanced (P<0.05, P<0.01, respectively;
Fig. 3). This was consistent the
results from RT-qPCR. These results indicate that IL-6 exerts its
regulatory effect on liver cirrhosis at the protein level.
IL-6 protein content in serum from
patients with liver cirrhosis is elevated compared with patients
with liver fibrosis
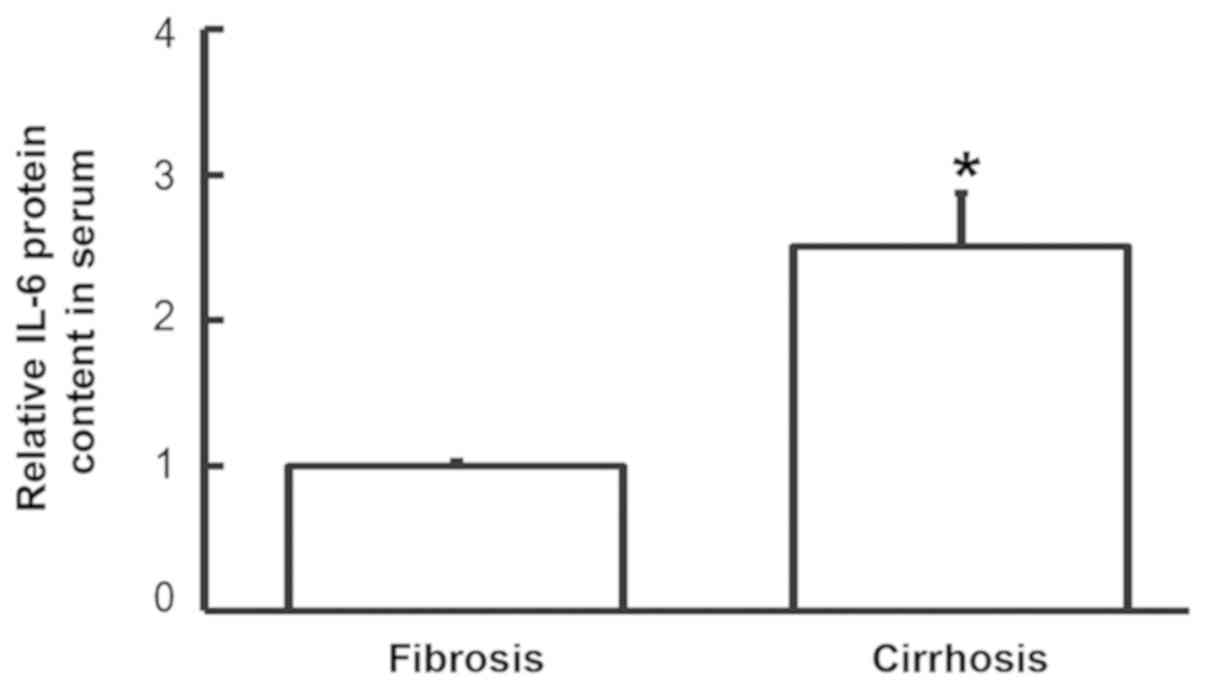
To examine the content of IL-6 protein in the serum,
ELISA was employed. The data demonstrated that IL-6 levels in the
serum of patients with liver cirrhosis were significantly higher
that in patients with liver fibrosis (P<0.05; Fig. 4). This suggests that increased IL-6
protein levels in serum may be released from PBMCs.
Altered IL-6 expression may be induced
by miR-146a levels
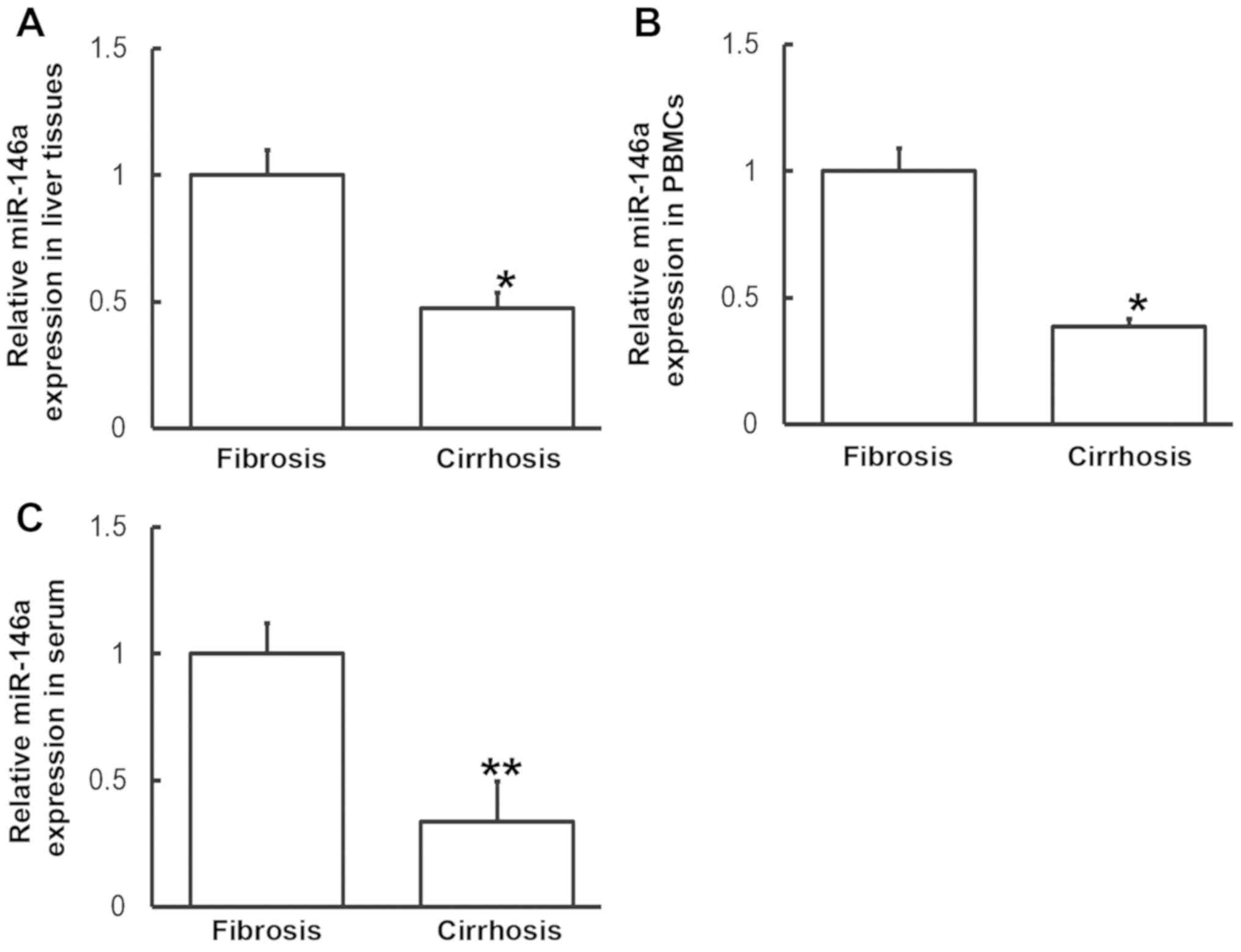
To measure miR-146a levels in various samples,
RT-qPCR was performed. Compared with patients with liver fibrosis,
miR-146a levels in the liver tissues, PBMCs and serum from patients
with liver cirrhosis were significantly downregulated (P<0.05,
P<0.05 and P<0.01, respectively; Fig. 5). These results indicate that altered
IL-6 expression may be induced by alterations in miR-146a
levels.
miR-146a regulates the expression of
IL-6 by binding to its 3′-UTR
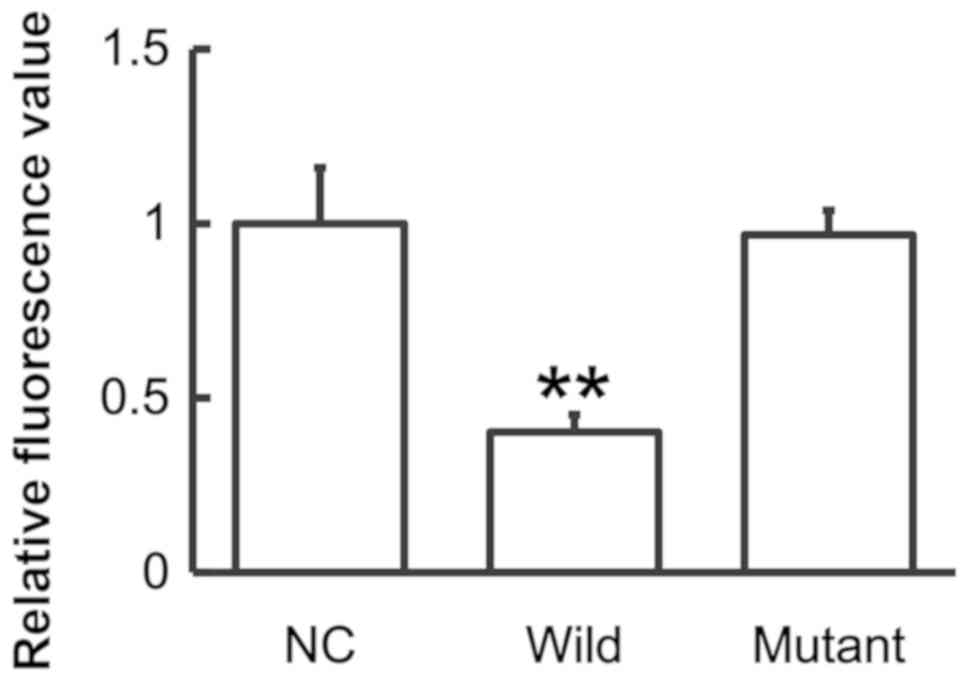
To identify the interaction between miR-146a and
IL-6, a dual luciferase reporter assay was performed. The data
demonstrated that the fluorescence value of the WT group was
significantly lower than that of the NC (P<0.01), whereas that
of the mutant group did not differ significantly from the control
(P>0.05; Fig. 6). The results
suggest that miR-146a regulates the expression of IL-6 by binding
to its 3′-UTR.
Discussion
IL-6 is an interleukin that serves a variety of
functions and is released during immune responses. Bacteria,
endotoxin and dust particles may stimulate the production of IL-6
in the body (25). IL-6 serves
important roles in immune responses, inflammation, cell
differentiation, coagulation and the occurrence and development of
tumors (26). The expression of IL-6
is markedly elevated in inflammatory responses caused by injury,
trauma, stress and infection (27).
In inflammation, IL-6 induces the production of C-reactive protein
and fibrinogen in the body, and promotes thrombogenesis (28). Additionally, elevated IL-6 levels
induce the onset of inflammatory diseases including rheumatoid
arthritis and Crohn's disease by binding to IL-6 receptors
(29). In rheumatoid arthritis, IL-6
stimulates the secretion of inflammatory mediators by T and B
lymphocytes, promotes the maturation and differentiation of B
lymphocytes and enhances the effects of IL-1β and TNF-α (30). In inflammatory responses, IL-6 has
chemotaxis on other inflammatory cells including neutral
lymphocytes and mononuclear macrophages (31). In the present study, the expression
of IL-6 mRNA and protein in liver tissues, PBMCs and serum from
patients with liver cirrhosis was higher than in patients with
liver fibrosis, suggesting that inflammation may be aggravated
during the transformation from liver fibrosis to cirrhosis.
Upregulation of IL-6 may be caused by the activation of monocytes
and lymphocytes, which can secrete abundant IL-6 factors and
produce antigen immune responses (32).
As an important gene regulator, miR molecules
participate in various pathophysiological processes, including the
proliferation, invasion and metastasis of tumor cells,
hypertension, diabetes or atherosclerosis (33,34). The
general function of miR molecules is to regulate the expression of
their target genes (33,34), however the expression and mechanism
of action of miR in Hepatitis B remains unclear. miR-146 was the
first miR identified to induce a regulatory effect in the immune
response and is associated with a number of diseases. For example,
the abnormal expression of miR-146a has been detected in autoimmune
diseases, such as rheumatoid arthritis (35,36).
Furthermore, clinical and animal models of osteoarthritis suggest
that miR-146a is associated with pain in osteoarthritis (37–39).
Studies have demonstrated that a number of binding sites of NF-κB
exist in the promoter region of miR-146a gene and that
lipopolysaccharides, IL-1 and TNF-α all promote miR-146a expression
in a NF-κB-dependent manner (40–42). In
the current study, bioinformatics predicted that IL-6 may be a
direct target gene of miR-146a. The results demonstrated that
miR-146a was downregulated and IL-6 was upregulated in liver
tissues and PBMCs from patients with liver cirrhosis compared with
patients with liver fibrosis, suggesting that the immune system may
negatively regulate the targeting of miR-146a on IL-6 by
downregulating miR-146a expression. This suggests that miR-146a is
not directly correlated with platelet count or serum albumin. Thus,
the expression of IL-6 is enhanced and immune responses are
generated. Similarly, it was demonstrated that miR-146a is
downregulated and IL-6 is upregulated in the serum of patients with
liver cirrhosis. This observation indicates that increased levels
of IL-6 induced by elevated miR-146a in PBMCs may be released into
the serum. Therefore, the levels of miR-146a and IL-6 in the serum
indicate the degrees of inflammatory responses and tissue lesions
during the transformation from liver fibrosis to cirrhosis. In
conclusion, the current study suggests that the decrease of
miR-146a expression in the liver tissues, peripheral blood and
serum regulates the expression of IL-6, leading to changes in the
levels of associated proteins that serve biological roles in the
occurrence and development of the disease. The balance between
miR-146a and IL-6 expression may determine the occurrence and
development of immunity. In addition, miR-146a is stable in the
blood, meaning that it may be used for diagnostic purposes.
Acknowledgements
The authors would like to thank Dr Tonglong Xu, the
president of the hospital, for his encouragement and advice. In
addition, the authors would like to thank Dr Yongli Wei for his
contribution to the writing of the manuscript.
Funding
The present study was supported by the Linyi
People's Hospital.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author's contributions
ZY and SY designed the study. ZY and YP performed
the experiments. ZY and SY analyzed the data. All authors
interpreted the results and prepared the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures performed in the current study were
approved by the Ethics Committee of Linyi People's Hospital (Linyi,
China). Written informed consent was obtained from all patients or
their parents, guardians or next of kin.
Patient consent for publication
Written informed consent for publication of any
associated data and accompanying images were obtained from all
patients or their parents, guardians or next of kin.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cabre A, Babio N, Lazaro I, Lázaro I,
Bulló M, Garcia-Arellano A, Masana L and Salas-Salvadó J: FABP4
predicts atherogenic dyslipidemia development. The PREDIMED study.
Atherosclerosis. 222:229–234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tawada A, Kanda T, Imazeki F and Yokosuka
O: Prevention of Hepatitis B virus-associated liver diseases by
antiviral therapy. Hepatol Int. 10:574–593. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Franchis R, Hadengue A, Lau G, Lavanchy
D, Lok A, McIntyre N, Mele A, Paumgartner G, Pietrangelo A, Rodés
J, et al: EASL international consensus conference on Hepatitis B.
13–14 September, 2002 Geneva, Switzerland. Consensus statement
(long version). J Hepatol. 39 (Suppl 1):S3–S25. 2003.PubMed/NCBI
|
|
4
|
Chen YC, Chu CM, Yeh CT and Liaw YF:
Natural course following the onset of cirrhosis in patients with
chronic Hepatitis B: A long-term follow-up study. Hepatol Int.
1:267–273. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chu CM and Liaw YF: Hepatitis B
virus-related cirrhosis: Natural history and treatment. Semin Liver
Dis. 26:142–152. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bataller R and Brenner DA: Liver fibrosis.
J Clin Invest. 115:209–218. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pinzani M: Liver fibrosis. Springer Semin
Immunopathol. 21:475–490. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Safadi R and Friedman SL: Hepatic
fibrosis-role of hepatic stellate cell activation. MedGenMed.
4:272002.PubMed/NCBI
|
|
9
|
Trepo C, Chan HL and Lok A: Hepatitis B
virus infection. Lancet. 384:2053–2063. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun M and Kisseleva T: Reversibility of
liver fibrosis. Clin Res Hepatol Gastroenterol. 39 (Suppl
1):S60–S63. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calvaruso V and Craxi A: Regression of
fibrosis after HBV antiviral therapy. Is cirrhosis reversible?
Liver Int. 34 (Suppl 1):S85–S90. 2014. View Article : Google Scholar
|
|
12
|
Jiang XI, Luo Y, Zhao S, Chen Q, Jiang C,
Dai Y, Chen Y and Cao Z: Clinical significance and expression of
microRNA in diabetic patients with erectile dysfunction. Exp Ther
Med. 10:213–218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jia W, Wu Y, Zhang Q, Gao GE, Zhang C and
Xiang Y: Expression profile of circulating microRNAs as a promising
fingerprint for cervical cancer diagnosis and monitoring. Mol Clin
Oncol. 3:851–858. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Graziano A, Lo Monte G, Piva I, Caserta D,
Karner M, Engl B and Marci R: Diagnostic findings in adenomyosis: A
pictorial review on the major concerns. Eur Rev Med Pharmacol Sci.
19:1146–1154. 2015.PubMed/NCBI
|
|
15
|
Hayes CN and Chayama K: MicroRNAs as
biomarkers for liver disease and hepatocellular carcinoma. Int J
Mol Sci. 17:2802016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ichikawa Y, Joshita S, Umemura T,
Shobugawa Y, Usami Y, Shibata S, Yamazaki T, Fujimori N, Komatsu M,
Matsumoto A and Tanaka E: Serum Wisteria floribunda
agglutinin-positive human Mac-2 binding protein may predict liver
fibrosis and progression to hepatocellular carcinoma in patients
with chronic hepatitis B virus infection. Hepatol Res. 47:226–233.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Cheng LS, Wu SD, Wang SQ, Li L, She
WM, Li J, Wang JY and Jiang W: IL-10-producing regulatory B-cells
suppressed effector T-cells but enhanced regulatory T-cells in
chronic HBV infection. Clin Sci (Lond). 130:907–919. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
MacParland SA, Ma XZ, Chen L, Khattar R,
Cherepanov V, Selzner M, Feld JJ, Selzner N and McGilvray ID:
Lipopolysaccharide and tumor necrosis factor alpha inhibit
interferon signaling in hepatocytes by increasing ubiquitin-like
protease 18 (USP18) expression. J Virol. 90:5549–5560. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang TS, Chen CL, Wu YC, Liu JJ, Kuo YC,
Lee KF, Lin SY, Lin SE, Tung SY, Kuo LM, et al: Inflammation
promotes expression of stemness-related properties in HBV-related
hepatocellular carcinoma. PLoS One. 11:e01498972016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li C, Deng M, Hu J, Li X, Chen L, Ju Y,
Hao J and Meng S: Chronic inflammation contributes to the
development of hepatocellular carcinoma by decreasing miR-122
levels. Oncotarget. 7:17021–17034. 2016.PubMed/NCBI
|
|
21
|
Chang L, Lan T, Wu L, Li C, Yuan Y and Liu
Z: The association between three IL-6 polymorphisms and HBV-related
liver diseases: A meta-analysis. Int J Clin Exp Med. 8:17036–17045.
2015.PubMed/NCBI
|
|
22
|
Lan T, Chang L, Wu L and Yuan YF: IL-6
plays a crucial role in HBV infection. J Clin Transl Hepatol.
3:271–276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chinese Society of Hepatology, Chinese
Medical Association: Chinese Society of Infectious Diseases,
Chinese Medical Association, . The guidelines of prevention and
treatment for chronic Hepatitis B. Zhonghua Gan Zang Bing Za Zhi.
13:881–891. 2005.(In Chinese). PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Badding MA, Schwegler-Berry D, Park JH,
Fix NR, Cummings KJ and Leonard SS: Sintered indium-tin oxide
particles induce pro-inflammatory responses in vitro, in part
through inflammasome activation. PLoS One. 10:e01243682015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hunter CA and Jones SA: IL-6 as a keystone
cytokine in health and disease. Nat Immunol. 16:448–457. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tanaka T, Narazaki M and Kishimoto T: IL-6
in inflammation, immunity, and disease. Cold Spring Harb Perspect
Biol. 6:a0162952014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Baeuerle PA and Henkel T: Function and
activation of NF-kappa B in the immune system. Annu Rev Immunol.
12:141–179. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tone M, Powell MJ, Tone Y, Thompson SA and
Waldmann H: IL-10 gene expression is controlled by the
transcription factors Sp1 and Sp3. J Immunol. 165:286–291. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yao X, Huang J, Zhong H, Shen N, Faggioni
R, Fung M and Yao Y: Targeting interleukin-6 in inflammatory
autoimmune diseases and cancers. Pharmacol Ther. 141:125–139. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo Q, Ma X, Wahl SM, Bieker JJ, Crossley
M and Montaner LJ: Activation and repression of interleukin-12 p40
transcription by erythroid Kruppel-like factor in macrophages. J
Biol Chem. 279:18451–18456. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aarstad HH, Vintermyr OK, Ulvestad E,
Kross K, Heimdal JH and Aarstad HJ: In vitro-stimulated IL-6
monocyte secretion and in vivo peripheral blood T lymphocyte
activation uniquely predicted 15-year survival in patients with
head and neck squamous cell carcinoma. PLoS One. 10:e01297242015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Varshney J and Subramanian S: MicroRNAs as
potential target in human bone and soft tissue sarcoma
therapeutics. Front Mol Biosci. 2:312015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pauley KM, Satoh M, Chan AL, Bubb MR,
Reeves WH and Chan EK: Upregulated miR-146a expression in
peripheral blood mononuclear cells from rheumatoid arthritis
patients. Arthritis Res Ther. 10:R1012008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Abou-Zeid A, Saad M and Soliman E:
MicroRNA 146a expression in rheumatoid arthritis: Association with
tumor necrosis factor-alpha and disease activity. Genet Test Mol
Biomarkers. 15:807–812. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamasaki K, Nakasa T, Miyaki S, Yamasaki
T, Yasunaga Y and Ochi M: Angiogenic microRNA-210 is present in
cells surrounding osteonecrosis. J Orthop Res. 30:1263–1270. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li X, Gibson G, Kim JS, Kroin J, Xu S, van
Wijnen AJ and Im HJ: MicroRNA-146a is linked to pain-related
pathophysiology of osteoarthritis. Gene. 480:34–41. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li X, Kroin JS, Kc R, Gibson G, Chen D,
Corbett GT, Pahan K, Fayyaz S, Kim JS, van Wijnen AJ, et al:
Altered spinal microRNA-146a and the microRNA-183 cluster
contribute to osteoarthritic pain in knee joints. J Bone Miner Res.
28:2512–2522. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Curtale G, Citarella F, Carissimi C,
Goldoni M, Carucci N, Fulci V, Franceschini D, Meloni F, Barnaba V
and Macino G: An emerging player in the adaptive immune response:
MicroRNA-146a is a modulator of IL-2 expression and
activation-induced cell death in T lymphocytes. Blood. 115:265–273.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Taganov KD, Boldin MP, Chang KJ and
Baltimore D: NF-kappaB-dependent induction of microRNA miR-146, an
inhibitor targeted to signaling proteins of innate immune
responses. Proc Natl Acad Sci USA. 103:12481–12486. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Larner-Svensson HM, Williams AE, Tsitsiou
E, Perry MM, Jiang X, Chung KF and Lindsay MA: Pharmacological
studies of the mechanism and function of interleukin-1beta-induced
miRNA-146a expression in primary human airway smooth muscle. Respir
Res. 11:682010. View Article : Google Scholar : PubMed/NCBI
|