Introduction
Glaucoma, one of the three major eye diseases that
lead to blindness, is caused by the depression and atrophy of optic
papilla (1). In recent years, it has
been found that approximately 60 million individuals suffer from
open angle or angle-closure glaucoma worldwide, and glaucoma is
second only to cataract in causing blindness to patients (2). Primary open angle glaucoma (POAG) is
one of the most common glaucoma types with the clinical
manifestations of pathological intraocular pressure (IOP) elevation
which causes optic nerve blood supply insufficiency in patients
(3). Previous findings showed that
elevated IOP is closely related to increased aqueous outflow
resistance in trabecular meshwork, and the main function of
trabecular meshwork is to control the aqueous flow direction
(4). Another study has shown that
the abnormal deposition of extracellular matrix (ECM) is the main
factor causing increased aqueous outflow resistance in trabecular
meshwork and elevated IOP (5).
Being a hot research field in recent years, microRNA
(miR) is a kind of non-coding short-chain RNA with a length of
approximately 22 nucleotides that functions mainly to inhibit the
translation and transcription of a target gene by binding to the 3′
untranslated region (UTR) of its cognate mRNA, thereby changing the
expression of the gene (6).
Increasing number of studies have shown that miR is closely related
to tumor and cardiovascular diseases (7,8).
Moreover, there are related studies showing that miR is related to
glaucoma. The study of Zhang et al (9) has shown that reduced miR-187 expression
in retinal cells promotes Smad7 expression and induces retinal
ganglion cells (RGCs) apoptosis in patients with glaucoma. Current
research shows that, miR-144-3p is one of the important members of
the miR family (10,11). It is differentially expressed in a
variety of tumors and cardiovascular diseases, and is closely
related to the progression of these diseases. Fibronectin-1 (FN-1),
also known as FN, is a high-molecular-weight glycoprotein that can
regulate various biological functions and widely exists in animal
tissues and tissue fluids (12). A
study has shown that, the expression of FN-1 protein is increased
in ECM of human trabecular meshwork cells (HTMCs) in POAG patients,
and FN can promote ECM deposition to block trabecular meshwork,
thereby causing an increase in pathological IOP (13). However, according to Liang et
al (14), the luciferase
reporter gene confirms that miR-144-3p can targetly regulate FN-1
expression. However, it is not clear whether miR-144-3p is
differentially expressed in glaucoma patients, or it can regulate
the function of HTMCs and the role of FN-1.
Therefore, this study explored the regulatory
effects of miR-144-3p on HTMC proliferation, invasion and FN-1,
thus providing a reference for clinicians.
Materials and methods
Cell source
HTMCs (BNCC342361; BeNa Culture Collection Company,
Beijing, China).
Main kits and instruments
TRIzol Extraction kit, RIPA, BCA Protein kit,
Lipofectamine™ 2000, ECL Luminescence kit, Transwell kit (nos.
15596018, 89900, 23225, 11668030, 35055, A1142801; Thermo Fisher
Scientific, Inc., Shanghai, China); H2O2,
FBS, DMEM, Penicillin-streptomycin double-antibody (216763, F2442,
D5796, V900929; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany);
RT-qPCR kit, TransScript Green miRNA Two-Step RT-qPCR SuperMix
(AQ202-01; Beijing TransGen Biotech Co., Ltd., Beijing, China),
FN-1 monoclonal antibody, β-actin monoclonal antibody, horseradish
peroxidase (HRP)-labeled goat anti-mouse IgG secondary antibody
(nos. MAB19182, MAB8929, HAF007; R&D Systems, Inc.,
Minneapolis, MN, USA), CCK-8 kit (C0037; Beyotime Institute of
Biotechnology, Shanghai, China), miR-144-3p primer sequence,
inhibition and over-expression sequences, and independence sequence
were designed and produced by Sangon Biotech Co., Ltd. (Shanghai,
China).
Cell culture
The HTMCs were placed in DMEM medium (10% FBS,
penicillin-streptomycin double-antibody) and transferred to an
incubator containing 5% CO2 at 37°C. The solution was
changed every 2 days and 0.25% trypsin was used for digestion and
passage when the cell fusion reached approximately 90%.
Modeling and grouping
The 3rd-4th generation cells were collected and the
density was adjusted to 4×106, then inoculated in a
6-well plate. Oxidative stress models were established 24 h later,
and the cells were cultured in a 37°C constant temperature
incubator with 300 µM H2O2 serum-free DMEM
medium for 2 h. There were 5 groups in this experiment: the
control, the blank, the independence sequence, the over-expression
and the inhibition groups. The cells in the control group were not
cultured with H2O2, while the blank, the
independence sequence, the over-expression and the inhibition
groups all used H2O2 for induction culture.
Additionally, the cells in the independence sequence, the
over-expression and the inhibition groups were transferred to
corresponding sequence plasmids respectively using Lipofectamine™
2000. The procedure was carried out according to the protocol and
the cells were cultured in an incubator for 2 h.
Clinical data of patients
A total of 40 POAG patients treated in Xuzhou No. 1
People's Hospital (Xuzhou, China) were included into the
observation group, including 25 males and 15 females with an
average age of 45.24±8.23 years, and they all underwent systematic
ophthalmological examinations, including optometry, best corrected
visual acuity (BCVA), slit lamp, fundus examination, IOP
measurement, gonioscopy, visual field and visual evoked potential
(VEP) tests. Another 40 healthy people with normal visual acuity,
heart, lung, liver and kidney laboratory tests were collected as a
normal group, including 22 males and 18 females with an average age
of 46.22±7.69 years. There was no difference in sex and age between
the two groups (P>0.05). Inclusion criteria were: i) patients
with complete clinical data; ii) patients with no family history of
glaucoma; and iii) patients conforming to Diagnosis and Treatment
of Primary Glaucoma Consensus in China. Exclusion criteria were: i)
patients complicated with malignant tumor; ii) patients with
cardio-cerebrovascular disease; iii) patients receiving glaucoma
treatment before this test; and iv) patients with no previous
operation history caused atrial morphological changes and liver
fibrosis.
The study was approved by the Ethics Committee of
Xuzhou Medical University (Xuzhou, China). Patients who
participated in this study had complete clinical data. Signed
informed consents were obtained from the patients or the
guardians.
Sample collection
Fasting venous blood samples (5 ml) of patients in
the two groups were collected in the morning, left to stand for 30
min, and centrifuged at 1,509.3 × g for 10 min at 25°C. The serum
was collected for follow-up experiments.
RT-qPCR detection
Total RNAs were extracted from the patient serum and
the cells of each group after 2 h of transfection and culture using
the TRIzol extraction reagent. The purity, concentration and
integrity of total RNAs were detected using an ultraviolet
spectrophotometer and 1% agarose gel electrophoresis. The total RNA
was reverse transcribed into cDNA using 2X TS miRNA Reaction Mix in
TransScript Green miRNA Two-Step RT-qPCR SuperMix, and the specific
operation steps were carried out according to the manufacturer's
protocol. Then PCR amplification experiment was carried out for the
PCR reaction system: 1 µl of cDNA, 0.4 µl of each upstream and
downstream primers (Table I), 10 µl
of 2X TransTaq® Tip Green qPCR SuperMix, 0.4 µl of
Passive Reference Dye (50X), made up to 20 µl with
ddH2O. PCR reaction conditions were: pre-denaturation at
94°C for 30 sec, denaturation at 94°C for 5 sec, annealing at 60°C
for 15 sec, extension at 72°C for 10 sec, for a total of 40 cycles.
Each sample was tested in 3 repeat wells, and the experiment was
carried out 3 times. In this study, U6 was used as an internal
reference and 2−ΔΔCq was used to analyze the data
(15). β-actin was used as the
internal reference in this study.
 | Table I.Primer sequence. |
Table I.
Primer sequence.
| Gene | Upstream primer | Downstream
primer |
|---|
| miR-144-3p |
5′-CCCTACAGTATAGATGATG-3′ |
5′-TGCAGGGTCCGAGGT-3′ |
| U6 |
5′-TAGGGTGCTCGCTTCGGC-3′ |
5′-CTGGTGTCGTGGAGTCG-3′ |
Western blot (WB) detection
Total protein was extracted from the collected cells
using the RIPA lysis method, and its concentration was detected
with the bicinchoninic acid (BCA) method and adjusted to 4 µg/µl.
The proteins were separated by 12% SDS-PAGE and then transferred to
a polyvinylidene difluoride membrane. The membrane was stained with
Ponceau S working solution, immersed in PBS for 5 min and then
washed, blocked with 5% skimmed milk powder for 2 h, and finally
incubated overnight at 4°C with the primary antibodies (1:1,000).
Following washing to remove primary antibodies (FN-1 and β-actin
monoclonal antibody), the HRP-labeled goat anti-mouse secondary
antibody (1:5,000) was added to the membrane for a 1 h incubation
at 37°C. After that, the membrane was rinsed 3 times with PBS, for
5 min each time. The protein bands on the membrane were developed
in the dark using the enhanced chemiluminescence (ECL) reagent, and
the excess liquid on the membrane was absorbed with a filter paper.
The luminescent protein bands were scanned and the gray value was
analyzed using Quantity One software. The relative expression level
of each protein was: the gray value of the target protein band/the
gray value of the β-actin protein band.
Cell proliferation detection
The 3rd-4th generation cells were collected and the
density was adjusted to 4×106, then inoculated in a
6-well plate. Oxidative stress models were established after 24 h
and the cells were transfected, then cultured for 48 h and 10 µl of
CCK solution and 90 µl basal medium (DMEM) were added to each well,
and cultured at 37°C for 2 h. The optical density (OD) value of
cells in each group was tested at 570 nm absorbance using a
microplate reader.
Cell invasion detection
Transwell chamber was placed in a 24-well plate, 500
µl DMEM culture solution was added to the upper chamber, and 100 µl
DMEM was added to the lower chamber, then the cells were cultured
for 1 h (37°C). The 3rd-4th generation cells were collected and the
density was adjusted to 4×106, then inoculated in a
6-well plate. Oxidative stress models were established after 24 h
and the cells were transfected, digested and resuspended with 0.25%
trypsin, then added into a serum-free DMEM medium. The cells then
were inoculated in a 24-well plate, and the density was adjusted by
1.5×106 cells. The Transwell chamber was taken out, the
lower chamber was replaced with 100% FBS, and 100 µl cell
suspension was added into the upper one, then the cells were
incubated at 37°C for 10 h. The substrates and cells that did not
penetrate the membrane surface in the upper chamber were wiped off,
washed three times with PBS, fixed with paraformaldehyde for 10
min, flushed three times with double-distilled water, dyed with
0.1% crystal violet for 10 min after drying, then rinsed three
times with double-distilled water. A microscope was used to observe
the cell invasion.
Statistical analysis
In this study, SPSS20.0 (Cabit Information
Technology Co., Ltd., Shanghai, China) software package was used to
carry out statistical analysis on the collected data. GraphPad
Prism 7 (Soft Head Technology Co., Ltd., Shenzhen, China) was used
to draw the data figure. The measurement data were expressed as
mean ± standard deviation. The comparison between the two groups
was carried out by independent sample t-test, and denoted by t. The
comparison between multiple groups was carried out by variance
analysis, and denoted by F, and the the pairwise comparison was
carried out by LSD-t-test afterwards. P<0.05 was considered to
indicate a statistically significant difference between the two
groups.
Results
Expression of miR-144-3P in serum of
patients
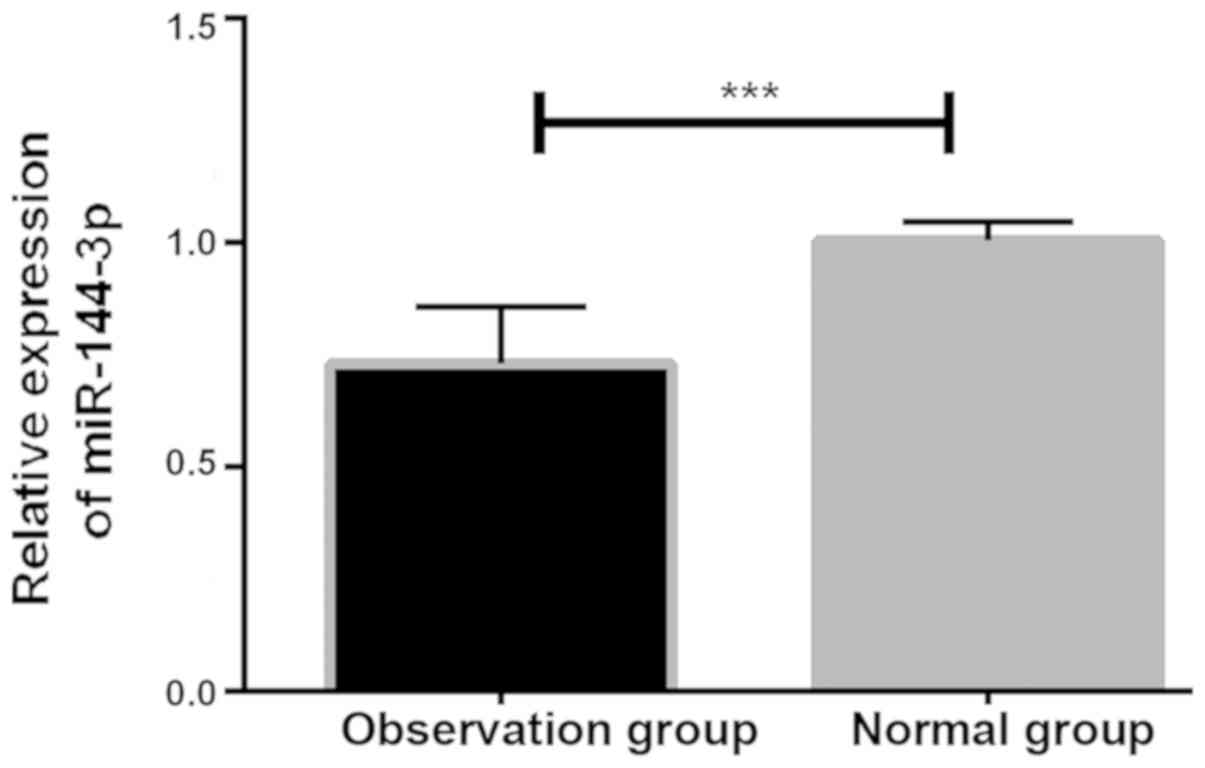
By detecting the expression of miR-144-3p in serum
of the patients, it was found that the expression in the normal
group was significantly higher than that in the observation group,
with significant differences (P<0.001; Fig. 1).
Expression of miR-144-3p in
transfected cells
The detection of miR-144-p expression in cells of
each group after transfection showed that there was a significant
difference in the expression of miR-144-3p among groups (F=100.954,
P<0.001). The expression in the control group was significantly
higher than that in the blank, the independence sequence and the
inhibition groups, but significantly lower than that in the
over-expression group (P<0.05). While the expression in blank
and independence sequence groups was significantly higher than that
in the inhibition group but lower than that in control and the
over-expression groups (P<0.05). There was no significant
difference between the blank and the independence sequence groups
(P>0.05). However, the expression of miR-144-p in the inhibitory
group was significantly lower than that in the other four groups
(P<0.05), and the expression in the over-expression group was
significantly higher than that in other four groups (P<0.05;
Table II).
 | Table II.Expression of miR-144-3p in cells of
each group. |
Table II.
Expression of miR-144-3p in cells of
each group.
| Group | miR-144-3p | F | P-value |
|---|
| Control | 1.450±0.102 |
|
|
| Blank |
1.032±0.042a |
|
|
| Independence
sequence |
1.055±0.010a | 100.954 | <0.001 |
| Inhibition |
0.350±0.067a–c |
|
|
| Over-expression |
7.167±1.07a–d |
|
|
Cell proliferation in each group after
48 h
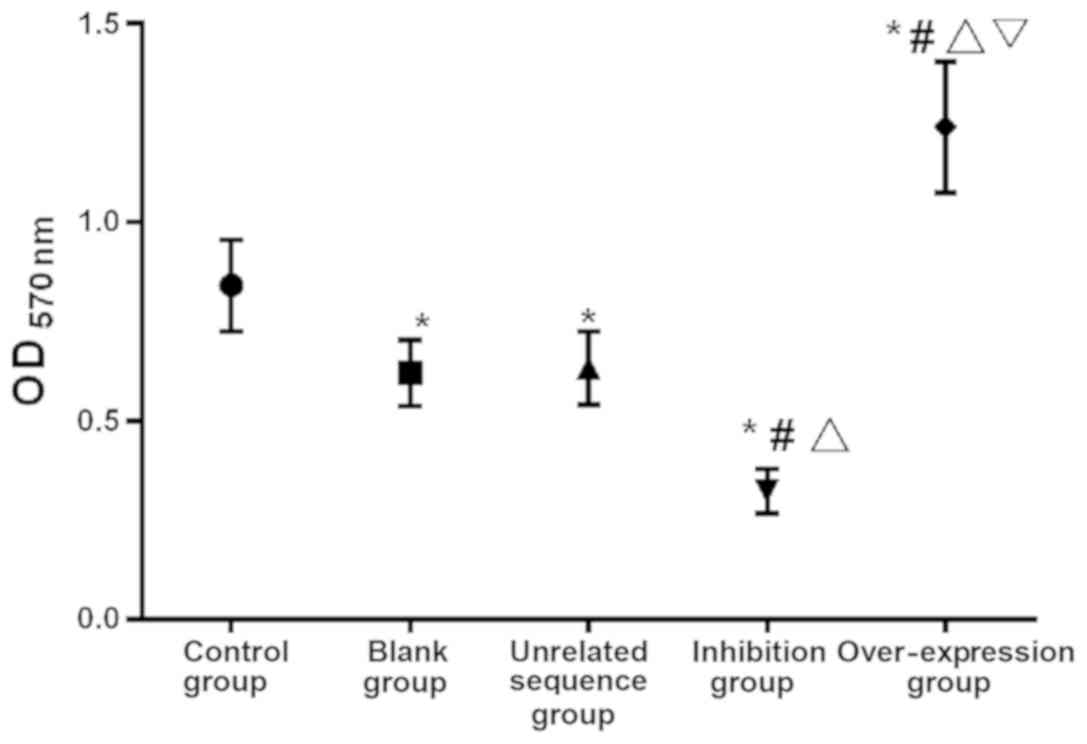
The comparison of OD value among the five groups of
cells cultured for 48 h showed that there was a significant
difference in cell proliferation among groups (F=29.146,
P<0.001). The value in the over-expression group was
significantly higher than that in the other groups (P<0.05), and
in the inhibition group was significantly lower than that in the
other groups (P<0.05). There was no difference between the blank
and the independence sequence groups (P>0.05), while the OD
value in these two groups was significantly higher than that in the
inhibition group but lower than that in the control and the
over-expression groups (P<0.05; Table III and Fig. 2).
 | Table III.Cell proliferation in each group after
48 h. |
Table III.
Cell proliferation in each group after
48 h.
| Group | OD value | F | P-value |
|---|
| Control | 0.842±0.115 |
|
|
| Blank |
0.622±0.084a |
|
|
| Independence
sequence |
0.634±0.092a | 29.146 | <0.001 |
| Inhibition |
0.325±0.055a–c |
|
|
|
Over-expression |
1.241±0.165a–d |
|
|
Cell invasion
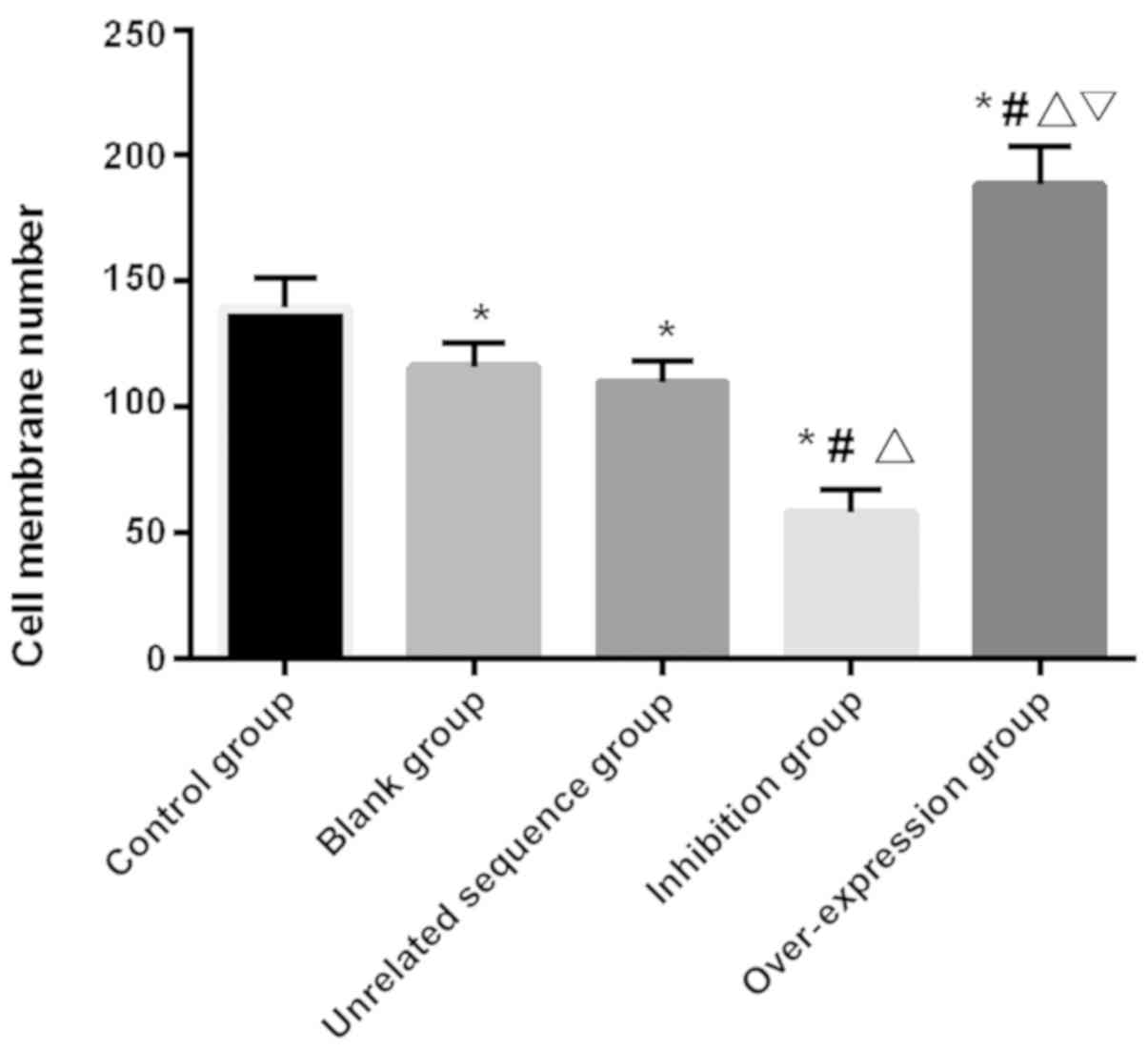
The number of cell-penetration in each group was
counted and found that there was a significant difference in the
number of cell-penetration among groups (F=52.656, P<0.001). The
number in the over-expression group was significantly higher than
that in other groups (P<0.05), and that in the inhibition group
had the least compared with the other four groups (P<0.05).
There was no significant difference in the number of
cell-penetration between the blank and the independence sequence
groups (P>0.05), and the number in these two groups
significantly decreased compared with the control and the
over-expression groups, but significantly increased compared with
the inhibition group (P<0.05; Table
IV and Fig. 3).
 | Table IV.Cell-penetration in each group. |
Table IV.
Cell-penetration in each group.
| Group | No. of
cell-penetration | F | P-value |
|---|
| Control | 139.54±11.66 |
|
|
| Blank |
115.88±9.45a |
|
|
| Independence
sequence |
109.75±8.77a | 52.656 | <0.001 |
| Inhibition |
57.95±9.44a–c |
|
|
|
Over-expression |
188.15±15.74a–d |
|
|
Expression of FN-1 protein
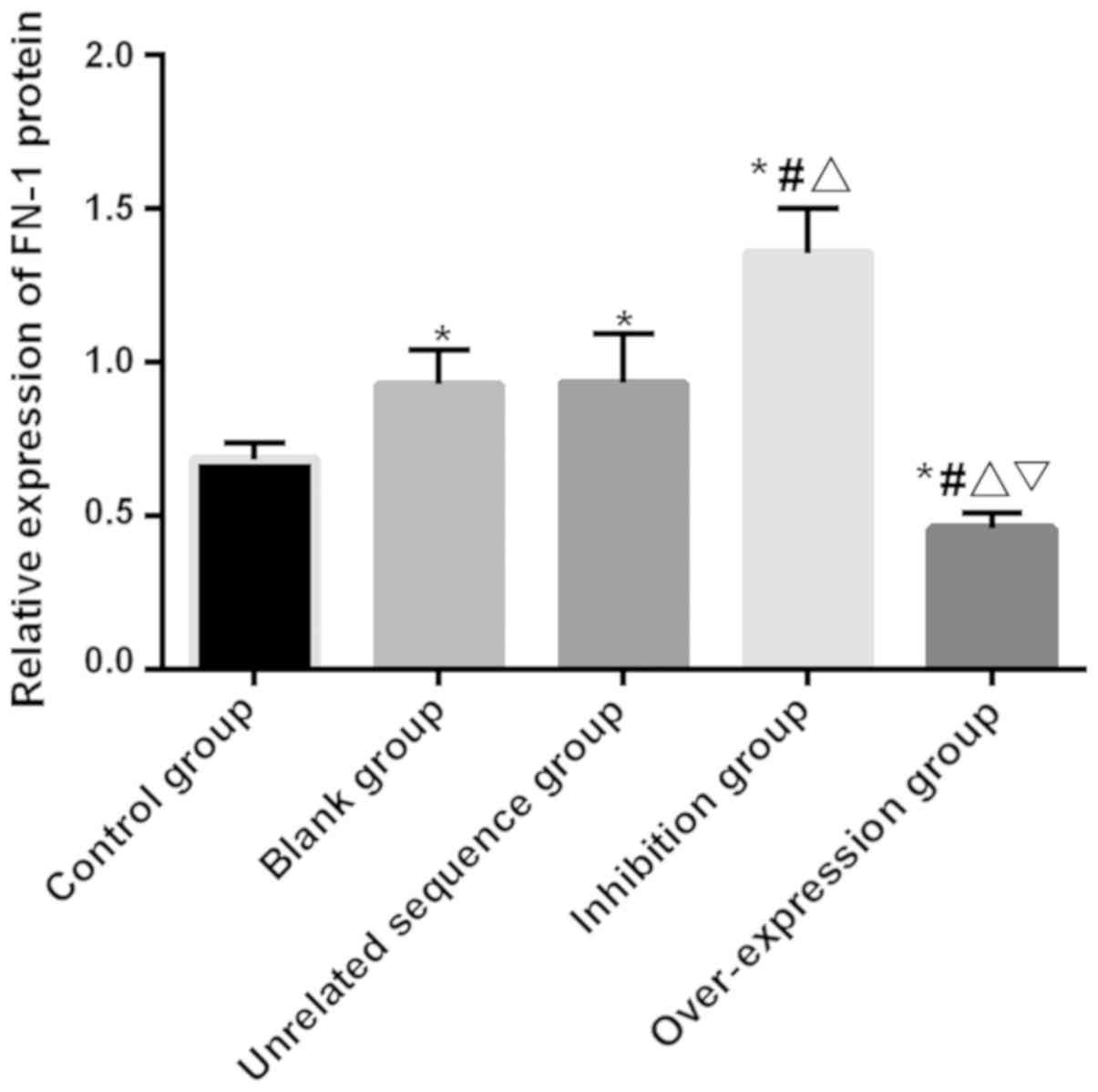
The expression of FN-1 protein in cells of each
group was detected, and the results of Western blotting showed that
there was a significant difference in the expression of FN-1 among
groups (F=25.611, P<0.001). The expression in the
over-expression group was significantly lower than that in the
other four groups (P<0.05). The expression in the blank and the
independence sequence groups had no significant difference
(P>0.05), but was significantly higher than that in the control
and the over-expression groups and significantly lower than that in
the inhibition group (P<0.05). The expression of FN-1 protein in
the inhibition group was significantly higher than that in the
other four groups (P<0.05; Table
V and Fig. 4).
 | Table V.Comparison of FN-1 protein. |
Table V.
Comparison of FN-1 protein.
| Group | Relative expression
of FN-1 protein | F | P-value |
|---|
| Control | 0.684±0.054 |
|
|
| Blank |
0.925±0.112a |
|
|
| Independence
sequence |
0.930±0.160a | 25.611 | <0.001 |
| Inhibition |
1.354±0.147a–c |
|
|
|
Over-expression |
0.459±0.049a–d |
|
|
Discussion
Glaucoma is a serious and irreversible blinding eye
disease, of which POAG is the most common type. However, each type
can cause retinal ganglion cell damage (16). The clinical pathological
manifestations of glaucoma are mainly elevated IOP caused by
excessive aqueous humor. Therefore, the main way to treat glaucoma
is to reduce aqueous humor secretion or aqueous outflow resistance
(5). Trabecular meshwork, an
important part of aqueous humor circulation, mainly regulates ECM
and aqueous outflow (17).
As a hot research field in recent years, miR has
been shown to be closely related to diseases such as glaucoma
(18,19). Being an important member of the miR
family, miR-144-3p plays a role in the miR-451 gene cluster and can
also regulate the expression of a plurality of genes involved in
erythropoiesis (20). It was shown
that miR-144 is a potential therapeutic tool for treating ischemic
heart disease (21). By contrast,
Liu et al (22) showed that
miR-144 was under-expressed in liver fibrosis cells. It is well
known that fibrosis is an over-reaction in wound healing, which
leads to excessive ECM that can be reduced by miR-144 regulating
TGF-β1 expression. ECM deposition also occurs after HTMCs are
damaged in glaucoma. However, whether miR-144-3p has the same
expression in glaucoma patients has not yet been studied.
Therefore, this study provided reference for treatment by exploring
the effect of miR-144-3p on HTMCs and its expression in the serum
of glaucoma patients.
Research has shown that there is a close
relationship between chronic oxidative stress and the occurrence
and development of trabecular meshwork lesions (23). Elevated IOP decreased blood flow
velocity in glaucoma patients, resulting in the decrease of oxygen
flow rate, thus keeping HTMCs in a hypoxic state. Therefore, in
this study, HTMC oxidative stress models were established and the
expression of miR-144-3p in HTMCs was detected. The results showed
that the expression of miR-144-3p in the modeled cells was
significantly lower than that in the control group without
modeling, and the clinical detection showed that the expression in
serum of normal people was significantly higher than that of
glaucoma patients, indicating that miR-144-3p is expected to become
a potential diagnostic indicator for glaucoma. FN-1, as a
non-collagen a2 glycoprotein, participates in cell-to-cell and
cell-to-matrix adhesion in the body (24). Babizhayev and Brodskaya (25) showed that there was a large amount of
FN-1 deposition in the trabecular meshwork adjacent area and the
Schlemm inner wall of HTMCs in POAG patients, and the deposition
increased with the aggravation of the disease. By detecting the
expression of FN-1 mRNA and protein in the cells after modeling, it
was found that the expression in the blank group was significantly
higher than that in the control group, which was basically similar
to the expression of FN-1 in the HTMCs induced by glucocorticoid
dexamethasone as indicated by Filla et al (26). However, it is not clear whether there
is a connection between miR-144-3p and FN-1 protein. Therefore,
Targetscan, an online miR target gene prediction software, was used
in that study, and it was found that there were possible binding
targets between miR-144-3p and FN-1 (26). Moreover, Liang et al (14) confirmed their relationship using
double luciferase report. The expression detection of FN-1 protein
in cells of each group found that the expression of miR-144-3p in
cells of the over-expression group was significantly higher than
that of the blank and the independence sequence groups, while the
expression of FN-1 protein was significantly lower than that of the
two groups. In contrast, the expression of miR-144-3p in cells of
the inhibition group was significantly lower than that of the blank
and the independence sequence groups, while the expression of FN-1
protein was significantly higher than that of the two groups. These
results suggest that the over-expression of miR-144-3p reduces the
expression of FN-1 protein in cells of trabecular meshwork
oxidative stress models. Moreover, the detection showed that the
proliferation and invasion of cells after modeling in the
over-expression group were significantly enhanced compared with the
other groups, which indicates that over-expression of miR-144-3p
can upregulate the proliferation and migration of HTMCs.
This study proved the expression of miR-144-3p in
oxidative stress HTMCs and its role in regulating FN-1. However,
there were still some limitations. First, a double luciferase
experiment was not carried out. Although it has been confirmed in
other studies, it is not clear whether there is any difference
caused by the two different research directions. Second, this study
served as a basic experiment, and whether miR-144-3p can be used as
a potential target for clinical treatment of glaucoma has not been
verified. Therefore, relevant experiments are to be carried out in
clinic to confirm the potential value of miR-144-3p in the
treatment of glaucoma.
To sum up, the over-expression of miR-144-3p
promotes the proliferation and invasion of HTMCs by inhibiting the
expression of FN-1 in oxidative stress HTMCs, and is a potential
target for glaucoma treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
RY wrote the manuscript. XC performed PCR. RY was
responsible for western blot analysis and Transwell. XC contributed
to analysis of observation indexes. Both authors read and approved
the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Xuzhou Medical University (Xuzhou, China). Patients who
participated in this study had complete clinical data. Signed
informed consents were obtained from the patients or the
guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zetterberg M: Age-related eye disease and
gender. Maturitas. 83:19–26. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quigley HA and Broman AT: The number of
people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol.
90:262–267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kapetanakis VV, Chan MP, Foster PJ, Cook
DG, Owen CG and Rudnicka AR: Global variations and time trends in
the prevalence of primary open angle glaucoma (POAG): A systematic
review and meta-analysis. Br J Ophthalmol. 100:86–93. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tamm ER, Braunger BM and Fuchshofer R:
Intraocular pressure and the mechanisms involved in resistance of
the aqueous humor flow in the trabecular meshwork outflow pathways.
Prog Mol Biol Transl Sci. 134:301–314. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Braunger BM, Fuchshofer R and Tamm ER: The
aqueous humor outflow pathways in glaucoma: A unifying concept of
disease mechanisms and causative treatment. Eur J Pharm Biopharm 95
(Pt B). 173–181. 2015. View Article : Google Scholar
|
|
6
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Condorelli G, Latronico MV and Cavarretta
E: microRNAs in cardiovascular diseases: Current knowledge and the
road ahead. J Am Coll Cardiol. 63:2177–2187. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang QL, Wang W, Li J, Tian SY and Zhang
TZ: Decreased miR-187 induces retinal ganglion cell apoptosis
through upregulating SMAD7 in glaucoma. Biomed Pharmacother.
75:19–25. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang SY, Lu ZM, Lin YF, Chen LS, Luo XN,
Song XH, Chen SH and Wu YL: miR-144-3p, a tumor suppressive
microRNA targeting ETS-1 in laryngeal squamous cell carcinoma.
Oncotarget. 7:11637–11650. 2016.PubMed/NCBI
|
|
11
|
Chen B, Luo L, Wei X, Gong D and Jin L:
Altered plasma miR-144 as a novel biomarker for coronary artery
disease. Ann Clin Lab Sci. 48:440–445. 2018.PubMed/NCBI
|
|
12
|
Pankov R and Yamada KM: Fibronectin at a
glance. J Cell Sci. 115:3861–3863. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Medina-Ortiz WE, Belmares R, Neubauer S,
Wordinger RJ and Clark AF: Cellular fibronectin expression in human
trabecular meshwork and induction by transforming growth factor-β2.
Invest Ophthalmol Vis Sci. 54:6779–6788. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang W, Xie Z, Cui W, Guo Y, Xu L, Wu J
and Guan H: Comprehensive gene and microRNA expression profiling
reveals a role for miRNAs in the oncogenic roles of SphK1 in
papillary thyroid cancer. J Cancer Res Clin Oncol. 143:601–611.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fry LE, Fahy E, Chrysostomou V, Hui F,
Tang J, van Wijngaarden P, Petrou S and Crowston JG: The coma in
glaucoma: Retinal ganglion cell dysfunction and recovery. Prog
Retin Eye Res. 65:77–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vranka JA, Kelley MJ, Acott TS and Keller
KE: Extracellular matrix in the trabecular meshwork: Intraocular
pressure regulation and dysregulation in glaucoma. Exp Eye Res.
133:112–125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li G, Luna C and Gonzalez P: miR-183
inhibits UV-induced DNA damage repair in human trabecular meshwork
cells by targeting of KIAA0101. Invest Ophthalmol Vis Sci.
57:2178–2186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li R, Jin Y, Li Q, Sun X, Zhu H and Cui H:
MiR-93-5p targeting PTEN regulates the NMDA-induced autophagy of
retinal ganglion cells via AKT/mTOR pathway in glaucoma. Biomed
Pharmacother. 100:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rasmussen KD, Simmini S, Abreu-Goodger C,
Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern
M, Enright AJ and O'Carroll D: The miR-144/451 locus is required
for erythroid homeostasis. J Exp Med. 207:1351–1358. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu
WT, Jegga AG and Fan GC: Synergistic effects of the GATA-4-mediated
miR-144/451 cluster in protection against simulated
ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell
Cardiol. 49:841–850. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Z, Yi J, Ye R, Liu J, Duan Q, Xiao J
and Liu F: miR-144 regulates transforming growth factor-β1 iduced
hepatic stellate cell activation in human fibrotic liver. Int J
Clin Exp Pathol. 8:3994–4000. 2015.PubMed/NCBI
|
|
23
|
Ruibin W, Zheng X, Chen J, Zhang X, Yang X
and Lin Y: Micro RNA-1298 opposes the effects of chronic oxidative
stress on human trabecular meshwork cells via targeting on EIF4E3.
Biomed Pharmacother. 100:349–357. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Singh P, Carraher C and Schwarzbauer JE:
Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev
Biol. 26:397–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Babizhayev MA and Brodskaya MW:
Fibronectin detection in drainage outflow system of human eyes in
ageing and progression of open-angle glaucoma. Mech Ageing Dev.
47:145–157. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Filla MS, Dimeo KD, Tong T and Peters DM:
Disruption of fibronectin matrix affects type IV collagen,
fibrillin and laminin deposition into extracellular matrix of human
trabecular meshwork (HTM) cells. Exp Eye Res. 165:7–19. 2017.
View Article : Google Scholar : PubMed/NCBI
|