Introduction
Depression is a state of low mood and aversion to
activity that can affect a person's thoughts, behavior, feelings
and sense of well-being (1). It has
been predicted that by 2030, depression will account for the
highest level of disability among all physical and mental disorders
worldwide (2). People with
depression may feel sad, anxious, empty, hopeless, helpless,
dejected or worthless (3). Certain
life events or changes may contribute to depression, including
childbirth, menopause, financial difficulties, stress, social
isolation or relationship difficulties (4). Adolescents are especially prone to
experiencing depression following social rejection, peer pressure
and bullying (5). In addition,
several diseases (including hypothyroidism, multiple sclerosis,
Parkinson's disease and chronic pain), and drugs (including heroin,
intoxication, hallucinogens and inhalants) can cause or exacerbate
depression (6,7). Depression is also a symptom of
treatment-refractory affective disorders (8). As indicated in the aforementioned
studies, depression may be a temporary reaction to life events, a
symptom of a medical condition, or a side effect of certain drugs
or medical treatments.
N-methyl-D-aspartic acid (NMDA) receptor antagonists
are a class of anesthetics that inhibit the action of NMDA
receptors. Ketamine, phencyclidine and memantine are three common
drugs belonging to the class of NMDA receptor antagonists (9,10). They
act primarily on the nervous system, with mild stimulant effects at
subanesthetic doses, and effects of dissociation and hallucinations
at higher doses (11). Ketamine is
the primary anesthetic for burn victims and emergency patients with
unknown medical history due to its moderate inhibition of
respiration and circulation (12,13).
Certain studies have indicated that NMDA antagonists also exert
antidepressant effects, particularly ketamine (14–16).
However, the underlying molecular mechanism of this remains
unclear. Therefore, the aim of the current study was to explore the
underlying mechanism, and identify potential targets of ketamine
for antidepressant effects based on molecular profile analysis.
Materials and methods
Expression profiles
mRNA expression profiles GSE73798 and GSE73799 were
downloaded from the publicly available Gene Expression Omnibus
(GEO) database (www.ncbi.nlm.nih.gov/geo). The GSE73798 profile
contained data derived from 60 mouse hippocampus samples treated
with ketamine, phencyclidyne, memantine and physiological saline or
no treatment assessed at four time points (1, 2, 4 and 8 h after
treatment). The GSE73799 profile included data derived from 60
mouse striatum samples that underwent the same treatments and
assessments. Samples had been evaluated with the Illumina MouseWG-6
version 2.0 Expression BeadChip platform (Illumina, Inc., San
Diego, CA, USA). A preliminary study using mice failed to establish
and investigate the model of depression, and, therefore, a rat
model was used in the present study to confirm differential gene
expression.
Data processing and differentially
expressed gene (DEG) analysis
Raw data were obtained and normalized with
preprocessCore package (version 1.32.0; www.bioconductor.org/packages/3.2/bioc/html/preprocessCore.html),
and probe symbols were converted to gene symbols. Subsequently,
DEGs were identified in hippocampus or striatum samples
individually treated with the three drugs, saline or no treatment
and assessed at 1, 2, 4 and 8 h, respectively. Each sample was
analyzed three times. P<0.05 and |log(fold-change)|>0.05 were
used as the threshold criteria. A total of 48 sets of DEGs were
obtained. Overlapping DEGs in the 24 sets were screened out for
subsequent evaluation.
Functional annotation and pathway
analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG
release 82; www.genome.jp/kegg) and BioCyc
(version 21.0; biocyc.org) databases were used to
perform functional annotation and pathway analysis for the
overlapping DEGs. Gene ontology (GO) terms and pathway terms were
selected at P<0.05.
Construction of the protein-protein
interaction (PPI) network
From the results of screening DEGs, the authors
found that the three drugs had a greater effect on gene expression
in striatum samples, therefore striatum samples were selected for
further study. The search tool for the retrieval of interacting
genes/proteins (STRING; version 9.1; string-db.org)
is a biological database and web resource of known and predicted
PPIs. In the current study, PPIs with a confidence score >0.4
were selected using STRING for the DEGs in the striatum samples
treated with ketamine, phencyclidyne, memantine compared with
normal samples. The PPI network was constructed using Cytoscape
software (version 3.5.1; www.cytoscape.org/download.php).
Verification of associated genes
A total of 10 male Sprague-Dawley rats (age, 12
weeks; weight, 300–350 g) were purchased from Beijing Vital River
Laboratory Animal Technology Co., Ltd. (Beijing, China). They were
fed in specific-pathogen free (SPF) facilities under standard
conditions and had access to food and water. The rats were housed
at 18–29°C with a 12 h light/dark cycle and a relative humidity of
40–70%; the rats had access to food and water ad libitum. A
depression model was constructed using the chronic unpredictable
mild stress method (17). The rats
were randomly divided into a ketamine group and a control group,
with 5 rats in each group. Ketamine (25 mg/kg; BOC Science Co.,
Ltd., Shirley, NY, USA) was injected intraperitoneally in the
ketamine group, while a similar volume of sterile saline was
injected intraperitoneally in the control group (18). At 8 h after drug administration, the
rats were sacrificed via cervical vertebrae dislocation after
anesthesia with pentobarbital sodium (45 mg/kg; intraperitoneal)
and striatum samples were collected. All rat experiments were
approved by the Animal Use and Care Committee of the First Hospital
of Hebei Medical University (Shijiazhuang, China). Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
(19) was performed to determine the
mRNA expression levels of perilipin 4 (Plin4),
serum/glucocorticoid regulated kinase 1 (Sgk1), kruppel like
factor 2 (Klf2), and DDB1 and CUL4 associated factor 12 like
1 (Dcaf12l1) with the ABI Am1005 AgPath-ID™ One-Step RT-PCR
kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. The 2−ΔΔCt
method (20) was used to quantify
the results. The following primer sequences were used: Plin4
5′-GGGACAAGAACATGGGAAGC-3′ (forward) and 5′-CCTTGACAAGACCTTTGGCC-3′
(reverse); Sgk1 5′-GAAGCTTGCCAACAACTCCT-3′ (forward) and
5′-CGTGGGGATTTGAGGATGGA-3′ (reverse); Klf2
5′-CTATCTTGCCGTCCTTTGCC-3′ (forward) and 5′-GGCTCCGGGTAGTAGAACG-3′
(reverse); Dcaf12l1 5′-CAGCAGCAAACAGGTAGCAG-3′ (forward) and
5′-CCTACCTCCCGAACCTTCAG-3′ (reverse). β-actin was as used as an
internal reference, and the primer sequences were
5′-CTACAATGAGCTGCGTGTGG-3′ (forward) and 5′-AGGCATACAGGGACAACACA-3′
(reverse).
Statistical analysis
SPSS software (version 17.0; SPSS, Inc., Chicago,
IL, USA) was used for all statistical analyses and data were
expressed as the mean ± standard error of the mean. Student's
t-test was used to compare groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
DEGs
A total of 48 sets of DEGs were identified in
hippocampus or striatum samples individually treated with the
ketamine, phencyclidine or memantine, compared with the saline and
normal groups at 1, 2, 4 and 8 h. The gene numbers of the 48 sets
of DEGs are presented in Table I.
The overlapping genes were respectively identified in the three
drug groups compared with both the saline and the normal group at
different time points, and the gene numbers are presented in
Table II. Furthermore, the
overlapping genes were screened out at the four time points, and
are presented in Table III.
Plin4 was the only overlapping gene among the aforementioned
48 sets of DEGs, and Plin4, Sgk1, Klf2 and Dcaf12l1
were overlapping in the striatum samples treated with the three
drugs at the different time points (Table III). Furthermore, Plin4,
Sgk1 and Klf2 were upregulated in the striatum samples
treated with the three drugs compared with saline or no treatment,
and Dcaf12l1 was downregulated.
 | Table I.Gene numbers of the 48 sets of DEGs
in hippocampus or striatum samples individually treated with
ketamine, phencyclidine or memantine compared with the saline or
normal group at 1, 2, 4 and 8 h, respectively. |
Table I.
Gene numbers of the 48 sets of DEGs
in hippocampus or striatum samples individually treated with
ketamine, phencyclidine or memantine compared with the saline or
normal group at 1, 2, 4 and 8 h, respectively.
|
|
| Hippocampus | Striatum |
|---|
|
|
|
|
|
|---|
| Drug | Control | 1 h | 2 h | 4 h | 8 h | 1 h | 2 h | 4 h | 8 h |
|---|
| Ketamine | Saline | 13 (2,11) | 7 (6,1) | 2 (2,0) | 0 (0,0) | 113 (23,90) | 122 (30,92) | 235 (121,114) | 103 (41,62) |
|
| Normal | 7 (6,1) | 12 (5,7) | 8 (8,0) | 2 (1,1) | 50 (36,14) | 91 (47,44) | 64 (47,17) | 26 (23,3) |
| Phencyclidine | Saline | 10 (3,7) | 4 (4,0) | 4 (1,3) | 0 (0,0) | 87 (46,41) | 106 (18,88) | 243 (119,124) | 98 (44,53) |
|
| Normal | 8 (4,4) | 14 (3,11) | 15 (10,5) | 5 (2,3) | 50 (41,9) | 43 (9,34) | 38 (21,17) | 31 (27,4) |
| Memantine | Saline | 4 (4,0) | 18 (16,2) | 11 (6,5) | 1 (0,1) | 104 (13,91) | 99 (31,68) | 520 (241,279) | 89 (44,45) |
|
| Normal | 7 (7,0) | 16 (16,0) | 7 (7,0) | 1 (1,0) | 19 (14,5) | 43 (30,13) | 134 (60,74) | 13 (11,2) |
 | Table II.Gene numbers of the overlapping genes
in three drug groups compared with both the saline and normal
groups. |
Table II.
Gene numbers of the overlapping genes
in three drug groups compared with both the saline and normal
groups.
|
| Hippocampus | Striatum |
|---|
|
|
|
|
|---|
| Drug | 1 h | 2 h | 4 h | 8 h | 1 h | 2 h | 4 h | 8 h |
|---|
| Ketamine | 3 (2,1) | 1 (0,1) | 2 (2,0) | 0 (0,0) | 10 (7,3) | 26 (19,7) | 7 (5,2) | 6 (6,0) |
| Phencyclidine | 4 (2,2) | 0 (0,0) | 4 (1,3) | 0 (0,0) | 20 (18,2) | 11 (3,8) | 2 (1,1) | 9 (7,2) |
| Memantine | 4 (4,0) | 9 (9,0) | 5 (5,0) | 0 (0,0) | 5 (4,1) | 14 (10,4) | 40 (18,22) | 2 (1,1) |
 | Table III.Overlapping genes at the four time
points. |
Table III.
Overlapping genes at the four time
points.
| Tissue | Drug | Upregulated | Downregulated |
|---|
| Hippocampus | Ketamine
Phencyclidine | Plin4, Sgk1, Gh,
Txnip Klf2, Plin4, mtDNA_ND6 | Ttr mtDNA_ND4L,
Stk32c, Hebp1, mt-Nd4l, Prr7 |
|
| Memantine | Plin4, Klf2,
Gadd45g, Mfsd2a, Ddit4, Cdkn1a, LOC240672, Dusp1, Sult1a1, Sgk1,
Fos, Egr4, Txnip, Tsc22d3 |
|
| Striatum | Ketamine | Plin4, Sgk1,
Klf2, Il22ra1, Vmn1r78, Slc13a4, D7Zem2, Txnip, Ddit4, Olfr938,
Kcne2, Star, 1500015O10Rik, Tmem26, Wfdc2, Spink8, Calml4,
Olfr247, LOC380910, Ttr, Sostdc1, Cap2, Gm4758, Cox8b, Vmn1r89,
Lbp, Col8a1, 932418N15Rik, Tmem28, Prr32, LOC672705, Folr1, Otx2,
Aqp1, Clic6, Wdr86 | Dcaf12l1, Meox1,
Olfr837, Slc6a13, Crh, LOC100046930, Fut1, Krt31, Fam171a1,
Olfr938, Olfr1168, Fam214b |
|
| Phencyclidine | Plin4, Sgk1,
Klf2, Kl, Enpp2, Olfr474, Dctn1, A030006E20Rik, D7Zem2, Aqp1,
Fgf15, Cox8b, Wfdc2, Zg16, Folr1, LOC381105, Hbb-b2, Wdr86,
Slc13a4, Ptgds, Crispld1, Osgin2, LOC383036, Igfbp2, Ces1a,
Ctgf | Dcaf12l1,
Aldh1a1, Fbxw15, Egr2, Cyp2c55, Egr4, Cldn5, Prr7, A230065H16Rik,
LOC194360, Cic, Ethe1, Olfr1317 |
|
| Memantine | Plin4, Sgk1,
Klf2, Nfkbia, E130102H24Rik, Cyp2c39, Pnpla2, Slc2a1, Gjb6,
Gadd45g, Il22ra1, Megf9, Cdkn1a, Nostrin, BAI3, Ism1, Cfp, Ramp2,
Txnip, Bcl6, Sult1a1, Tsc22d3, LOC100044968, 9930108O06Rik,
Adipor2, Thbd, Ddit4 | Dcaf12l1,
LOC331511, Cyp2c55, Phyhip, LOC386233, Mid1ip1, Rpl27a-ps1,
LOC381105, Cplx2, Calm3, Fam168b, Actb, Krt31, LOC241621,
Pimreg, |
|
|
| Megf9, Cdkn1a,
Nostrin, BAI3, Ism1, Cfp, Ramp2, Txnip, Bcl6, Sult1a1, Tsc22d3,
LOC100044968, 9930108O06Rik, Adipor2, Thbd, Ddit4 | C330049H01Rik,
LOC433476, Cldn5, LOC385068, Olfr1362, Hes5, Tmsb10, Rhbdd3,
BC037112, LOC382092, LOC382237, LOC673501, LOC381132 |
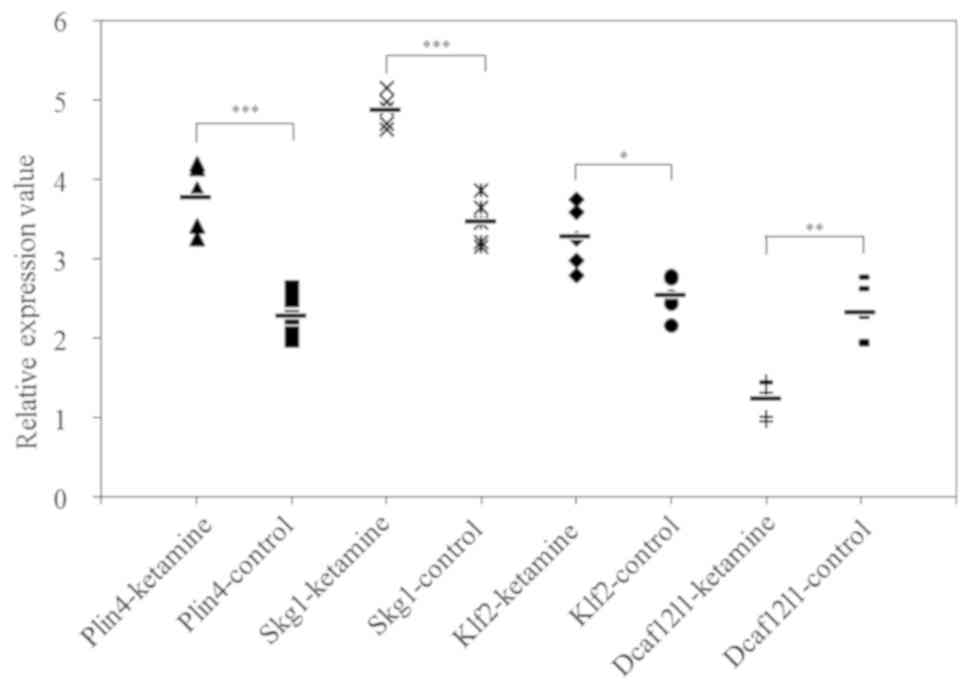
The results of RT-qPCR are presented in Fig. 1. The mRNA expression levels of
Plin4, Sgk1 and Klf2 were significantly higher in the
striatum samples of the ketamine group compared with the control
group (P<0.05), while the mRNA expression of Dcaf12l1 was
significantly lower compared with the control group (P<0.05). In
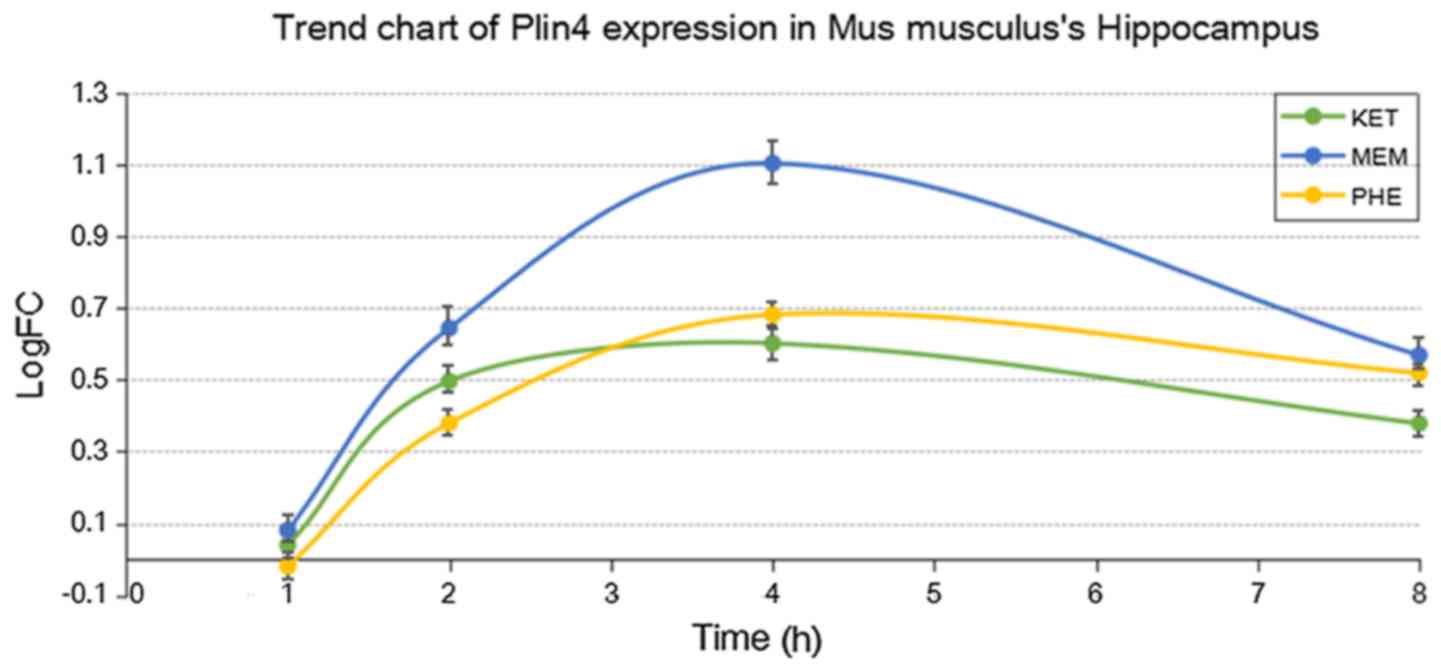
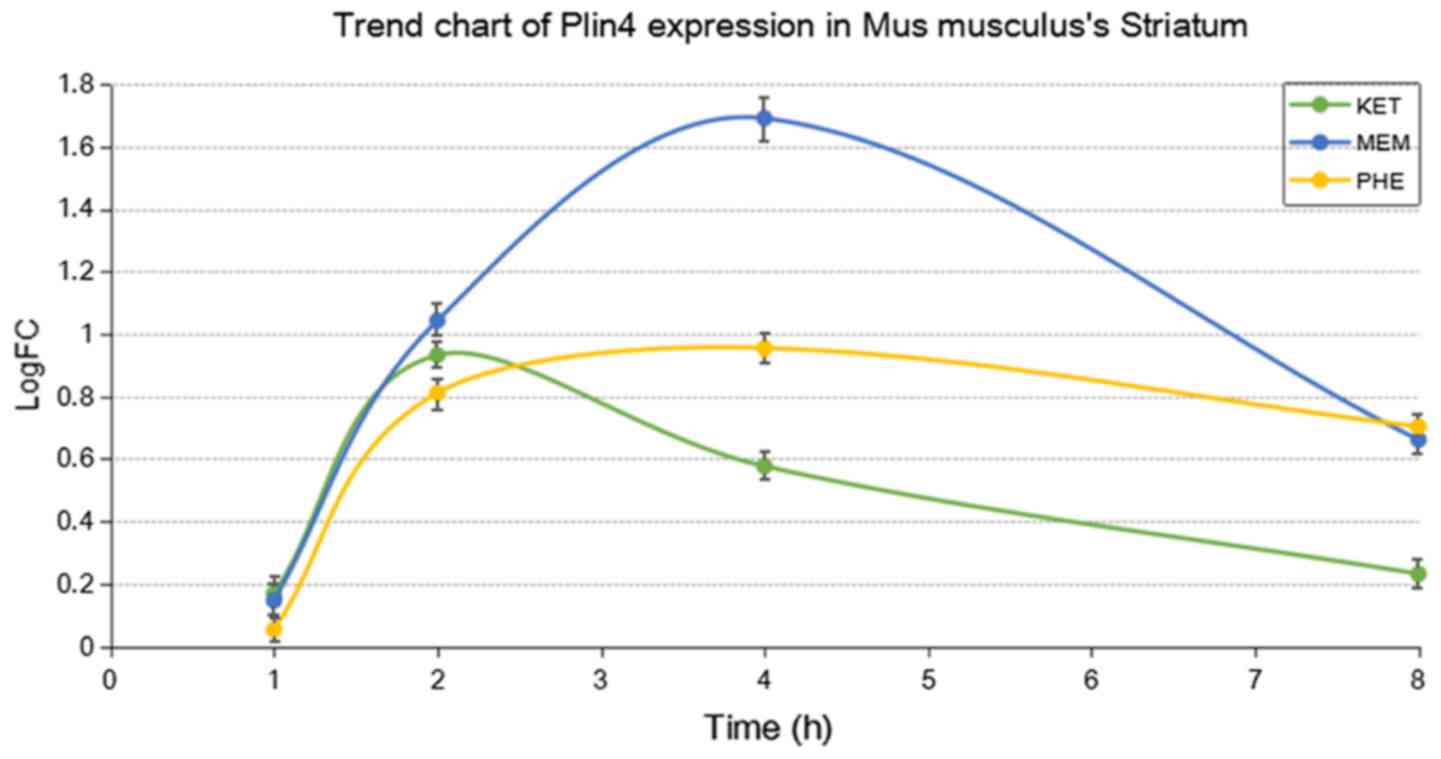
addition, the expression of Plin4 in the hippocampus and
striatum following treatment with the three drugs is presented in
Figs. 2 and 3, respectively. The results indicated that
the expression of Plin4 was highest both in the hippocampus
and striatum after treatment with ketamine compared with
phencyclidine and memantine.
GO terms and pathway analysis
The enriched GO terms and KEGG pathways of Plin4,
Sgk1, Klf2 and Dcaf12l1 were selected, and the results
are presented in Tables IV and
V. Plin4 was enriched in 1
KEGG pathway; Sgk1 was enriched in 5 GO terms and 4 KEGG
pathways; and Klf2 was enriched in 4 GO terms and 3 KEGG
pathways. Among these, Sgk1 and Klf2 were enriched in
the ‘forkhead box O (FoxO) signaling pathway’, and Sgk1 was
additionally enriched in the ‘mechanistic target of rapamycin
kinase (mTOR) signaling pathway’. Plin4 and Dcaf12l1
were enriched in no GO terms, and Dcaf12l1 was enriched in
no pathways.
 | Table IV.mRNA levels of Plin4, Sgk1,
Klf2 and Dcaf12l1 in the striatum samples. |
Table IV.
mRNA levels of Plin4, Sgk1,
Klf2 and Dcaf12l1 in the striatum samples.
|
| Relative expression
value |
|---|
|
|
|
|---|
| Group | Plin4 | Sgk1 | Klf2 |
Dcaf12l1 |
|---|
| Ketamine group | 3.77±0.38 | 4.87±0.19 | 3.27±0.36 | 1.23±0.21 |
| Control group | 2.28±0.25 | 3.46±0.27 | 2.53±0.23 | 2.31±0.34 |
| P-value | <0.0001 | <0.0001 | 0.0402 | 0.0018 |
| T-value | 8.76 | 12.81 | −3.68 | 7.87 |
 | Table V.The enriched GO terms of Sgk1
and Klf2. |
Table V.
The enriched GO terms of Sgk1
and Klf2.
| Gene | GO term | Function |
|---|
| Sgk1 | GO:0004672 | ‘Protein kinase
activity’ |
|
| GO:0004674 | ‘Protein
serine/threonine kinase activity’ |
|
| GO:0005524 | ‘ATP binding’ |
|
| GO:0016301 | ‘Kinase
activity′’ |
|
| GO:0016740 | ‘Transferase
activity’ |
| Klf2 | GO:0003677 | ‘DNA binding’ |
|
| GO:0003700 | ‘Transcription
factor activity, sequence-specific DNA binding’ |
|
| GO:0008270 | ‘Zinc ion
binding’ |
|
| GO:0046872 | ‘Metal ion
binding’ |
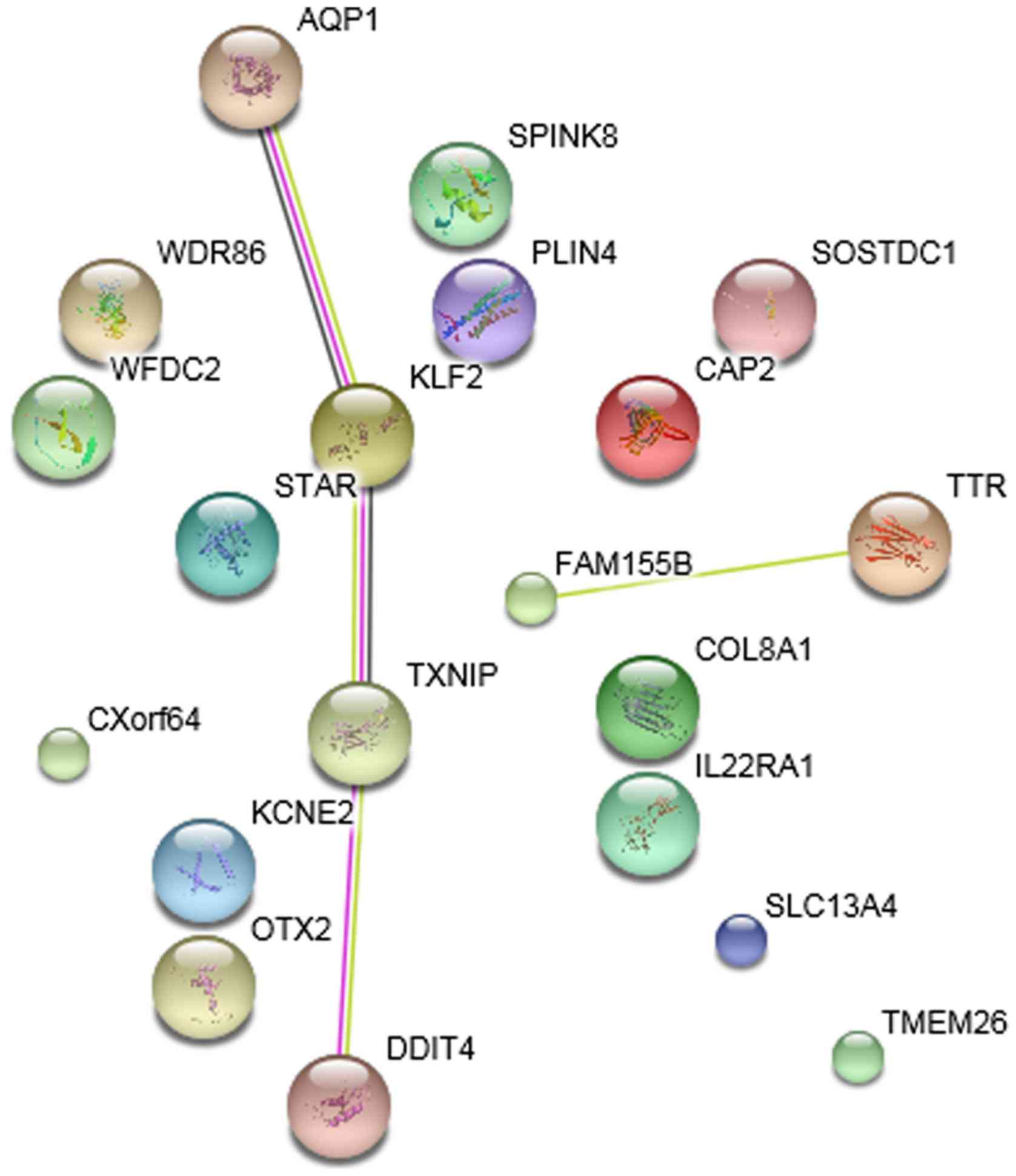
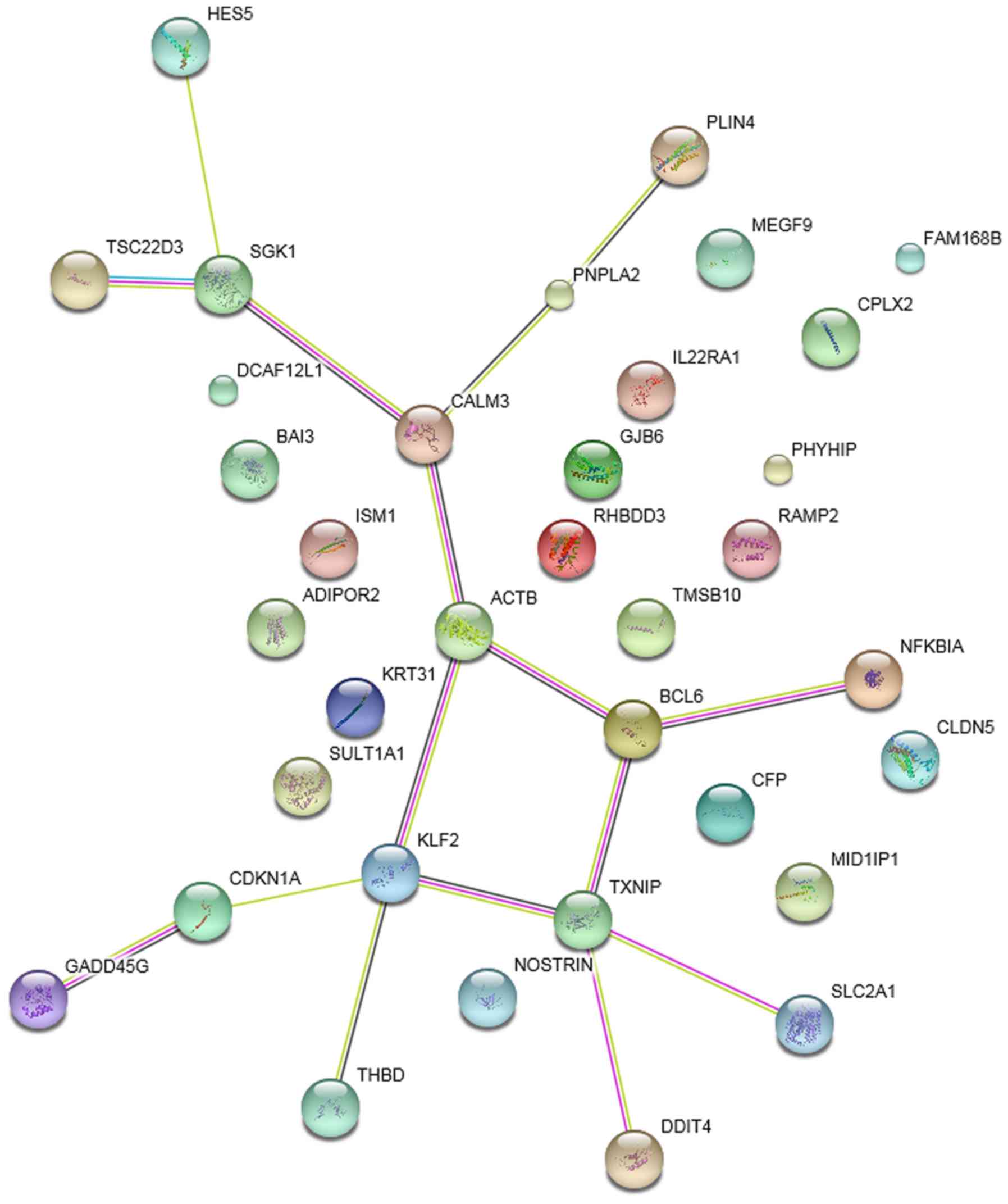
PPI network
The PPI networks of DEGs in the striatum samples
treated with ketamine, phencyclidine and memantine compared with
normal samples are presented in Figs.
4–6, respectively. The results
indicated that Klf2 was involved in more PPI pairs in these
three PPI networks and was therefore the hub gene.
Discussion
Genetic factors may promote or even cause the
occurrence of depression (21). A
systematic review found that major histocompatibility complex,
class I-related gene polymorphisms, and glutamate decarboxylase
(GAD) genes (GAD1 and GAD2) contributed to the
development of depression (22). One
study reported that 5-hydroxytryptamine receptor 2A functional
rs6311 polymorphism may modulate the severity of depression
symptoms in children with autism spectrum disorder (23). Another study reported that glutamate
ionotropic receptor kainate type subunit 4 variants were involved
in treatment-resistant depression (24). Apoptosis regulator BCL2 was
considered to serve a role in mediating the outcome of
antidepressant treatment (24). In
the current study, rat models of depression were selected to
investigate the underlying mechanism of treatment with ketamine.
The present study was primarily based on bioinformatics and a
comparison between the ketamine and control groups in vivo
further demonstrated the effect of ketamine during depression, and
provided an additional method for exploring the mechanism of action
of ketamine. In addition, the underlying mechanism requires further
verification in animal models and human trials, however the current
results provide a preliminary basis for these.
In the present study, Plin4, Sgk1, Klf2 and
Dcaf12l1 were the overlapping DEGs in the striatum samples
treated with three NMDA receptor antagonists at different time
points. Furthermore, the expression levels of Plin4, Sgk1
and Klf2 increased in the striatum samples of rat models of
depression following administration of ketamine, and the expression
of Dcaf12l1 decreased compared with the control group. Plin4
protein coats lipid droplets in adipocytes to protect them from
lipases (25,26). Plin4 is associated with
insulin resistance and obesity risk (27,28). In
the current study, Plin4 was identified as the only
overlapping DEG (upregulated) in hippocampus or striatum samples
individually treated with ketamine, phencyclidine or memantine
compared with the saline or normal groups at 1, 2, 4 and 8 h. SGK1
protein contributes to the regulation of discrete developmental
stages and pathological conditions including hypertension, diabetic
neuropathy, ischemia, trauma and neurodegenerative diseases
(29). Anacker et al
(30) identified SGK1 as a mediator
of the effects of cortisol on neurogenesis and glucocorticoid
receptor function, with particular relevance to stress and
depression. KLF2 protein is implicated in lung development,
embryonic erythropoiesis, epithelial integrity, T-cell viability
and adipogenesis (31). Another
study by Miller et al (32)
was performed using ribosome-bound mRNA footprinting and deep
sequencing, and confirmed that initiation of protein synthesis is a
defining feature of antidepressant dose ketamine in mice; with the
use of GO analysis, vasoactive intestinal peptide receptor 2 gene
was identified as a potential target for antidepressant action. In
the current study, Klf2 was involved in more pairs in the PPI
network of DEGs in the striatum samples treated with ketamine,
phencyclidine or memantine compared with normal samples. That was
to say, Klf2 was differentially expressed between the groups
compared with the control, and that Klf2 exhibited the
highest degree among all proteins in the PPI networks. However, to
the best of the authors' knowledge, the association between
Klf2 and depression or NMDA receptor antagonists has not
been previously reported. Dcaf12l1 protein is associated with
embryonic development and idiopathic nonobstructive azoospermia
(33,34). The current study indicated that
Plin4, Sgk1, Klf2 and Dcaf12l1 were differentially
expressed in depression models treated with ketamine, phencyclidine
and memantine, which suggested that these genes may be the targets
of the NMDA receptor antagonist treatment.
Duman et al (35) investigated the signaling pathway
underlying the rapid antidepressant effects of ketamine, and the
results demonstrated that the effects were associated with the
stimulation of mTOR and increased expression levels of synaptic
proteins. Li et al (36)
observed that ketamine rapidly activated the mTOR pathway, leading
to increased expression levels of synaptic signaling proteins and
increased number and function of new spine synapses in the
prefrontal cortex of rats, while inhibition of mTOR signaling
completely blocked ketamine-mediated induction of synaptogenesis
and behavioral responses in models of depression. The above results
indicated that the effects of ketamine are opposite to the synaptic
deficits that result from exposure to stress and could contribute
to the rapid antidepressant effects of ketamine. The current study
also found that Sgk1, one of the key DEGs underlying the
rapid antidepressant effects of ketamine (37), was enriched in the ‘mTOR signaling
pathway’. In addition, the present results indicated that both
Sgk1 and Klf2 were enriched in the ‘FoxO signaling
pathway’. FoxO is a subfamily of the fork head transcription factor
family, serving roles in cell fate decisions (38). Polter et al (39) observed that FoxO may be a
transcriptional target for treatment of anxiety and mood disorders,
and serves a potential role in regulating mood-associated behavior.
This study further indicated that mice displayed reduced anxiety
when FoxO1 was knocked down in the brain (39). Hence, FoxO3a-deficient mice presented
with an antidepressant-like behavior (39). The FoxO signaling pathway is
considered to be a therapeutic target in cancer (40), and mediates stress responses
(41,42). Therefore, it is hypothesized that the
mTOR and FoxO signaling pathways may be involved in the underlying
mechanism of antidepressant effects of ketamine.
In conclusion, the present study suggested that the
mTOR and FoxO signaling pathways may serve roles in the underlying
mechanism of antidepressant effects of ketamine, and Plin4,
Sgk1, Klf2 and Dcaf12l1 may be potential biomarkers for
depression and targets for NMDA receptor antagonist treatment of
depression.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
JQ and YS made substantial contributions to the
conception and design of the study, and analysis of the data. JW
interpreted data and drafted the manuscript. LW designed the study,
drafted and revised the manuscript, gave final approval of the
version to be published and agreed to be accountable for all
aspects of the work.
Ethics approval and consent to
participate
All rat experiments were approved by the Animal Use
and Care Committee of the First Hospital of Hebei Medical
University (Shijiazhuang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Saadat M, Behboodi ZM and Saadat E:
Comparison of depression, anxiety, stress, and related factors
among women and men with human immunodeficiency virus infection. J
Hum Reprod Sci. 8:48–51. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
HM, Vega WA, Williams DR, Tarraf W, West
BT and Neighbors HW: Depression care in the United States: too
little for too few. Arch Gen Psychiatry. 67:37–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krishnan V and Nestler EJ: The molecular
neurobiology of depression. Nature. 455:894–902. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mata DA, Ramos MA, Bansal N, Khan R,
Guille C, Di Angelantonio E and Sen S: Prevalence of depression and
depressive symptoms among resident physicians: A systematic review
and meta-analysis. JAMA. 314:2373–2383. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davey CG, Yücel M and Allen NB: The
emergence of depression in adolescence: Development of the
prefrontal cortex and the representation of reward. Neurosci
Biobehav Rev. 32:1–19. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saravane D, Feve B, Frances Y, Corruble E,
Lancon C, Chanson P, Maison P, Terra JL and Azorin JM; avec le
soutien institutionnel du laboratoire Lilly, : Drawing up
guidelines for the attendance of physical health of patients with
severe mental illness. Encéphale. 35:330–339. 2009.(In French).
View Article : Google Scholar
|
|
7
|
Rogers D and Pies R: General medical with
depression drugs associated. Psychiatry (Edgmont). 5:28–41.
2008.PubMed/NCBI
|
|
8
|
Al-Harbi KS: Treatment-resistant
depression: therapeutic trends, challenges, and future directions.
Patient Prefer Adherence. 6:369–388. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aiyer R, Mehta N, Gungor S and Gulati A: A
systematic review of NMDA receptor antagonists for treatment of
neuropathic pain in clinical practice. Clin J Pain. 34:450–467.
2018.PubMed/NCBI
|
|
10
|
Leung LS and Ma J: Medial septum modulates
hippocampal gamma activity and prepulse inhibition in an,
N-methyl-d-aspartate receptor antagonist model of schizophrenia.
Schizophr Res. 198:36–44. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rasmussen KG: Psychiatric side effects of
ketamine in hospitalized medical patients administered
subanesthetic doses for pain control. Acta Neuropsychiatr.
26:230–233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ceber M and Salihoglu T: Ketamine may be
the first choice for anesthesia in burn patients. J Burn Care Res.
27:760–762. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heshmati F, Zeinali MB, Noroozinia H,
Abbacivash R and Mahoori A: Use of ketamine in severe status
asthmaticus in intensive care unit. Iran J Allergy Asthma Immunol.
2:175–180. 2003.PubMed/NCBI
|
|
14
|
Maeng S, Zarate CA Jr, Du J, Schloesser
RJ, McCammon J, Chen G and Manji HK: Cellular mechanisms underlying
the antidepressant effects of ketamine: Role of
alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors.
Biol Psychiatry. 63:349–352. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murrough JW, Perez AM, Pillemer S, Stern
J, Parides MK, aan het Rot M, Collins KA, Mathew SJ, Charney DS and
Iosifescu DV: Rapid and longer-term antidepressant effects of
repeated ketamine infusions in treatment-resistant major
depression. Biol Psychiatry. 74:250–256. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ardalan M, Rafati AH, Nyengaard JR and
Wegener G: Rapid antidepressant effect of ketamine correlates with
astroglial plasticity in the hippocampus. Br J Pharmacol.
174:483–492. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jing XU: The establishment and evaluation
of chronic unpredictable mild stress depression model. Chin J
Behavioral Med Sci. 12:14–17. 2003.(In Chinese).
|
|
18
|
Wang J, Goffer Y, Xu D, Tukey DS, Shamir
DB, Eberle SE, Zou AH, Blanck TJ and Ziff EB: A single
subanesthetic dose of ketamine relieves depression-like behaviors
induced by neuropathic pain in rats. Anesthesiology. 115:812–821.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meijerink J, Mandigers C, van de Locht L,
Tönnissen E, Goodsaid F and Raemaekers J: A novel method to
compensate for different amplification efficiencies between patient
DNA samples in quantitative Real-Time PCR. J Mol Diagn. 3:55–61.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Corrales E, Navarro A, Cuenca P and Campos
D: Candidate gene study reveals DRD1 and DRD2 as putative
interacting risk factors for youth depression. Psychiatry Res.
244:71–77. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lacerda-Pinheiro SF, Pinheiro Junior RF,
Pereira de Lima MA, Lima da Silva CG, Vieira dos Santos Mdo S,
Teixeira Júnior AG, Lima de Oliveira PN, Ribeiro KD, Rolim-Neto ML
and Bianco BA: Are there depression and anxiety genetic markers and
mutations? A systematic review. J Affect Disord. 168:387–398. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gadow KD, Smith RM and Pinsonneault JK:
Serotonin 2A receptor gene (HTR2A) regulatory variants: Possible
association with severity of depression symptoms in children with
autism spectrum disorder. Cogn Behav Neurol. 27:107–116. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Milanesi E, Bonvicini C, Congiu C,
Bortolomasi M, Gainelli G, Gennarelli M and Minelli A: The role of
GRIK4 gene in treatment-resistant depression. Genet Res (Camb).
97:e142015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wolins NE, Quaynor BK, Skinner JR,
Schoenfish MJ, Tzekov A and Bickel PE: S3-12, Adipophilin, and
TIP47 package lipid in adipocytes. J Biol Chem. 280:19146–19155.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen W, Chang B, Wu X, Li L, Sleeman M and
Chan L: Inactivation of Plin4 downregulates Plin5 and reduces
cardiac lipid accumulation in mice. Am J Physiol Endocrinol Metab.
304:E770–E779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peters SJ, Samjoo IA, Devries MC, Stevic
I, Robertshaw HA and Tarnopolsky MA: Perilipin family (PLIN)
proteins in human skeletal muscle: The effect of sex, obesity, and
endurance training. Appl Physiol Nutr Metab. 37:724–735. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Soenen S, Mariman EC, Vogels N, Bouwman
FG, den Hoed M, Brown L and Westerterp-Plantenga MS: Relationship
between perilipin gene polymorphisms and body weight and body
composition during weight loss and weight maintenance. Physiol
Behav. 96:723–728. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schoenebeck B, Bader V, Zhu XR, Schmitz B,
Lübbert H and Stichel CC: Sgk1, a cell survival response in
neurodegenerative diseases. Mol Cell Neurosci. 30:249–264. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Anacker C, Cattaneo A, Musaelyan K,
Zunszain PA, Horowitz M, Molteni R, Luoni A, Calabrese F, Tansey K,
Gennarelli M, et al: Role for the kinase SGK1 in stress,
depression, and glucocorticoid effects on hippocampal neurogenesis.
Proc Natl Acad Sci USA. 110:8708–8713. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pearson R, Fleetwood J, Eaton S, Crossley
M and Bao S: Kruppel-like transcription factors: A functional
family. Int J Biochem Cell Biol. 40:1996–2001. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Miller OH, Grabole N, Wells I and Hall J:
Genome-wide translating mRNA analysis following ketamine reveals
novel targets for antidepressant treatment. BioRxiv. 2018.
|
|
33
|
Ramasamy R, Ridgeway A, Lipshultz LI and
Lamb DJ: Integrative DNA methylation and gene expression analysis
identifies discoidin domain receptor 1 association with idiopathic
nonobstructive azoospermia. Fertil Steril. 102:968–973.e3. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gerovska D and Arauzo-Bravo MJ: Does mouse
embryo primordial germ cell activation start before implantation as
suggested by single-cell transcriptomics dynamics? Mol Hum Reprod.
22:208–225. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Duman RS, Li N, Liu RJ, Duric V and
Aghajanian G: Signaling pathways underlying the rapid
antidepressant actions of ketamine. Neuropharmacology. 62:35–41.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li N, Lee B, Liu RJ, Banasr M, Dwyer JM,
Iwata M, Li XY, Aghajanian G and Duman RS: mTOR-dependent synapse
formation underlies the rapid antidepressant effects of NMDA
antagonists. Science. 329:959–964. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ficek J, Zygmunt M, Piechota M, Hoinkis D,
Rodriguez Parkitna J, Przewlocki R and Korostynski M: Molecular
profile of dissociative drug ketamine in relation to its rapid
antidepressant action. BMC Genomics. 17:3622016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Farhan M, Wang H, Gaur U, Little PJ, Xu J
and Zheng W: FOXO signaling pathways as therapeutic targets in
cancer. Int J Biol Sci. 13:815–827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Polter A, Yang S, Zmijewska AA, van Groen
T, Paik JH, Depinho RA, Peng SL, Jope RS and Li X: Forkhead box,
class O transcription factors in brain: Regulation and behavioral
manifestation. Biol Psychiatry. 65:150–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Farhan M, Wang H, Gaur U, Little PJ, Xu J
and Zheng W: FOXO signaling pathways as therapeutic targets in
cancer. Int J Biol Sci. 13:815–827. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huang P, Zhou Z, Shi F, Shao G, Wang R,
Wang J, Wang K and Ding W: Effects of the IGF-1/PTEN/Akt/FoxO
signaling pathway on male reproduction in rats subjected to water
immersion and restraint stress. Mol Med Rep. 14:5116–5124. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liang B, Moussaif M, Kuan CJ, Gargus JJ
and Sze JY: Serotonin targets the DAF-16/FOXO signaling pathway to
modulate stress responses. Cell Metab. 4:429–440. 2006. View Article : Google Scholar : PubMed/NCBI
|