Introduction
Hair graying may be classified as natural senile
canities and premature graying. Natural senile canities usually has
its onset after the age of 40 years and aggravates with the ongoing
aging process (1). Unlike senile
canities, premature graying occurs prior to the age of 25 years and
is usually progressive and permanent (2,3).
Although hair graying is considered to be genetically controlled
and inheritable, the underlying mechanisms have remained largely
elusive (4).
Hair color is determined by pigment granules in hair
follicles, wherein melanin synthesis is particularly crucial
(5). Mature melanocytes are densely
distributed in hair bulbs to sustain active melanogenesis, which is
strictly coupled to the anagen stage of the hair cycle (6,7). The
biosynthesis of melanin and its subsequent transfer from melanocyte
to hair bulb keratinocytes is a rather complex process (8). Previous molecular study has mainly
focused on identifying genes that encode characteristic markers for
pigmentation, including tyrosinase (TYR), OCA2 melanosomal
transmembrane protein, TYR-related protein 1 (TYRP1) and solute
carrier family 45 member 2 (SLC45A2) (9). Polymorphisms within these loci are
associated with a normal variation in hair color traits (10). In addition, a genome-wide association
study on the genetic basis of pigmentation in human subjects
revealed that the single nucleotide polymorphism frequency
distribution of these genes is linked to skin color and hair color
(11,12). A broader study indicated that
mutations in two pore segment channel 2, agouti signaling protein
and melanocortin 1 receptor are associated with hair color and
pigmentation (13).
Previous studies have focused on identifying genes
that encode characteristic markers for premature graying of hair. A
germline mutation in the syntaxin 17 gene of horses was recently
identified to cause premature graying of hair (14) and telomerase reverse transcriptase
mutation carriers may also present with premature hair graying
(15). Reverse transcription
polymerase chain reaction (RT-PCR) arrays on the gene expression
profiles of the hair bulge and hair bulb revealed a significant
downregulation of melanogenesis associated genes [tyrosinase (TYR),
tyrosine related protein 1 (TYRP1), melanocyte inducing
transcription factor, paired box gene 3 (PAX3) and
proopiomelanocortin] in unpigmented hair bulbs and a downregulation
of marker genes typical for melanocyte precursor cells [PAX3,
SRY-Box 10 (SOX10) and dopachrome tautomerase) in unpigmented
mid-segments compared with their pigmented analogues. Superoxide
dismutase 3 transgenic mice exhibited premature aging, including
hair graying, abnormal gait and a shortened life span (16).
Although previous studies provided candidate genes
associated with hair graying, their results are limited due to the
unavailability of related hereditary family members or an
insufficient number of subjects to provide representative results
(17). According to the central
dogma of genetics, the genotype affects the phenotype through gene
expression. Thus, it is straightforward to assume that a variance
in gene expression between the grey and black hair follicles
underlies the difference in hair color. However, to the best of our
knowledge, no previous study has assessed premature hair graying
from this genetic aspect. In the present study, an RNA sequencing
(RNA-seq) analysis was performed to reveal gene expression changes
between grey and black hair follicles from Han Chinese patients
suffering from premature hair graying. It was intended to unravel
the underlying mechanisms and potential candidate genes responsible
for hair pigmentation loss.
Materials and methods
Subjects
A total of 5 unrelated Han Chinese donors who
presented with premature graying since they were teenagers and had
a clear family history of premature hair graying were enrolled in
the present study. Their details are specified in (Table I). A total of 30 grey and 30 black
hair follicles were randomly collected from each subject and stored
in RNAlater® Stabilization Solution (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 4°C.
 | Table I.Characteristics of the subjects
affected by premature hair greying. |
Table I.
Characteristics of the subjects
affected by premature hair greying.
| Gender | Age (years) | Onset age
(years) | Head areas
affected |
|---|
| Male | 31 | 15 | Parietal, frontal
and temporal region |
| Male | 30 | 10 | Occipital
region |
| Female | 28 | 12 | Parietal and
frontal region |
| Male | 26 | 10 | Parietal
region |
| Male | 28 | 12 | Parietal and
temporal region |
RNA extraction, library preparation
and sequencing
Total RNA was extracted from each sample using the
mirVana™ miRNA Detection Kit (Thermo Fisher Scientific, Inc.) in
strict accordance with the manufacturer's protocol. The RNA-seq
library was prepared using an Ion Total RNA-seq Kit v2 (Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol.
RNA-seq libraries were sequenced on an Ion Proton™ System (Thermo
Fisher Scientific, Inc.).
Bioinformatics analysis of RNA-seq
data
Raw sequencing reads were aligned to the human
genome (GRCh38/hg38) using STAR (Spliced Transcripts Alignment to a
Reference software; http://code.google.com/p/rna-star/) (18). Ensembl gene annotation (http://www.ensembl.org/info/genome/genebuild/genome_annotation.html;
release 79) was used for evaluating gene expression using Cufflinks
(19), and differentially expressed
genes (DEGs) between grey and black hair follicles were determined
by Cuffdiff, a subpackage of Cufflinks. The ratio of Fragments per
kilobase per million of grey to black hair was calculated and
differential expression genes were then studied by log2FC. The
criteria used to define DEGs were as follows: i) Fragments per
kilobase per million mapped reads >1 in at least one sample of a
sample pair; ii) fold change of at least 2 and iii) P<0.05.
Functional classification and enrichment analysis of
the DEGs were performed using the online platform of the Database
for Annotation, Visualization and Integrated Discovery (20). Gene Ontology (GO) terms were used to
classify the DEGs. Association between microRNAs (miRs) and their
targets were predicted by Starbase v2 (21). Transcription factors (TFs) and their
target genes were predicted through Integrated Motif Activity
Response Analysis (22). The Search
Tool for the Retrieval of Interacting Genes/Proteins (STRING)
database (23) was used to construct
protein-protein interaction (PPI) networks the DEGs are involved
in.
RT-quantitative (q)PCR
Total RNA was extracted from each hair follicle
sample. The RNA (1 µg) was reverse-transcribed using a ReverTra Ace
qPCR RT Kit (Toyobo, Osaka, Japan). β-actin was used as reference
control. SYBR Green Realtime PCR Master Mix (ROX; Toyobo) was
applied for PCR amplifications on a Bio-Rad CFX96 system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) according to the
manufacturer's protocol. The thermocycling program was as follows:
95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 60°C for
40 sec and 72°C for 45 sec. The primers used are listed in Table II.
 | Table II.Primers used for polymerase chain
reaction analysis. |
Table II.
Primers used for polymerase chain
reaction analysis.
| Gene name | Primer sequence
(5′→3′) | Melting temperature
(°C) | Product length
(bp) |
|---|
| TYR | F:
CTCCCCTCTTCAGCTGATGT | 57.0 | 150 |
|
| R:
GCTGCTTTGAGAGGCATCC |
|
|
| TYRP1 | F:
CCGAAACACAGTGGAAGGTT | 58.9 | 162 |
|
| R:
TCTGTGAAGGTGTGCAGGAG |
|
|
| GJB1 | F:
CCTGCACAGACATGAGACCA | 58.9 | 192 |
|
| R:
CCACCAGCACCATGATTCTG |
|
|
| PMEL | F:
GATAGGTGCTTTGCTGGCTG | 59.26 | 159 |
|
| R:
GACACTTGACCACCTCTCCA |
|
|
| SOX10 | F:
CTGGACCGCACACCTTGG | 59.0 | 197 |
|
| R:
CTCAGCTCCACCTCCGATAG |
|
|
| SLC45A2 | F:
CTGCCGACTTCATTGATGGG | 59.0 | 175 |
|
| R:
TGCAAAGGTAGCGGTAGTGA |
|
|
| KIT | F:
GGCGGGCATCATGATCAAAA | 59.25 | 160 |
|
| R:
GCTTGCTTTGGACACAGACA |
|
|
| TRPM1 | F:
CACCCAGAGCTACCCAACAGA | 59.2 | 165 |
|
| R:
CGGATATACATGGCTTTATTGGAA |
|
|
| OCA2 | F:
ACTCTTCTTTGCCCCAGATG |
|
|
|
| R:
TCCCAAGACTCTTCAGCAGTG | 59.0 | 158 |
| β-actin | F:
GCGTGACATCAAGGAAGAAGC | 57.5 | 108 |
|
| R:
CCGTCGGGTAGTTCGTAGCT |
|
|
Results
Summary of RNA-seq data
To identify changes in the gene expression profile
associated with premature hair greying, RNA-seq libraries derived
from each grey or black hair follicle were sequenced using the Ion
Proton™ System (Thermo Fisher Scientific, Inc.). The number of raw
sequenced reads for each sample ranged from 15.9 to 27.9 million,
and the mapping percentages were 72.1–80.0%, which demonstrated an
overall good mappability.
Gene expression changes between grey
and black hair follicles
In order to identify genes with a role in premature
hair greying, the samples were classified into a grey hair group
and a black hair group, and the gene expression changes were
compared between them. With the criteria of a 2-fold change in
expression and P<0.05, a total of 127 DEGs were identified,
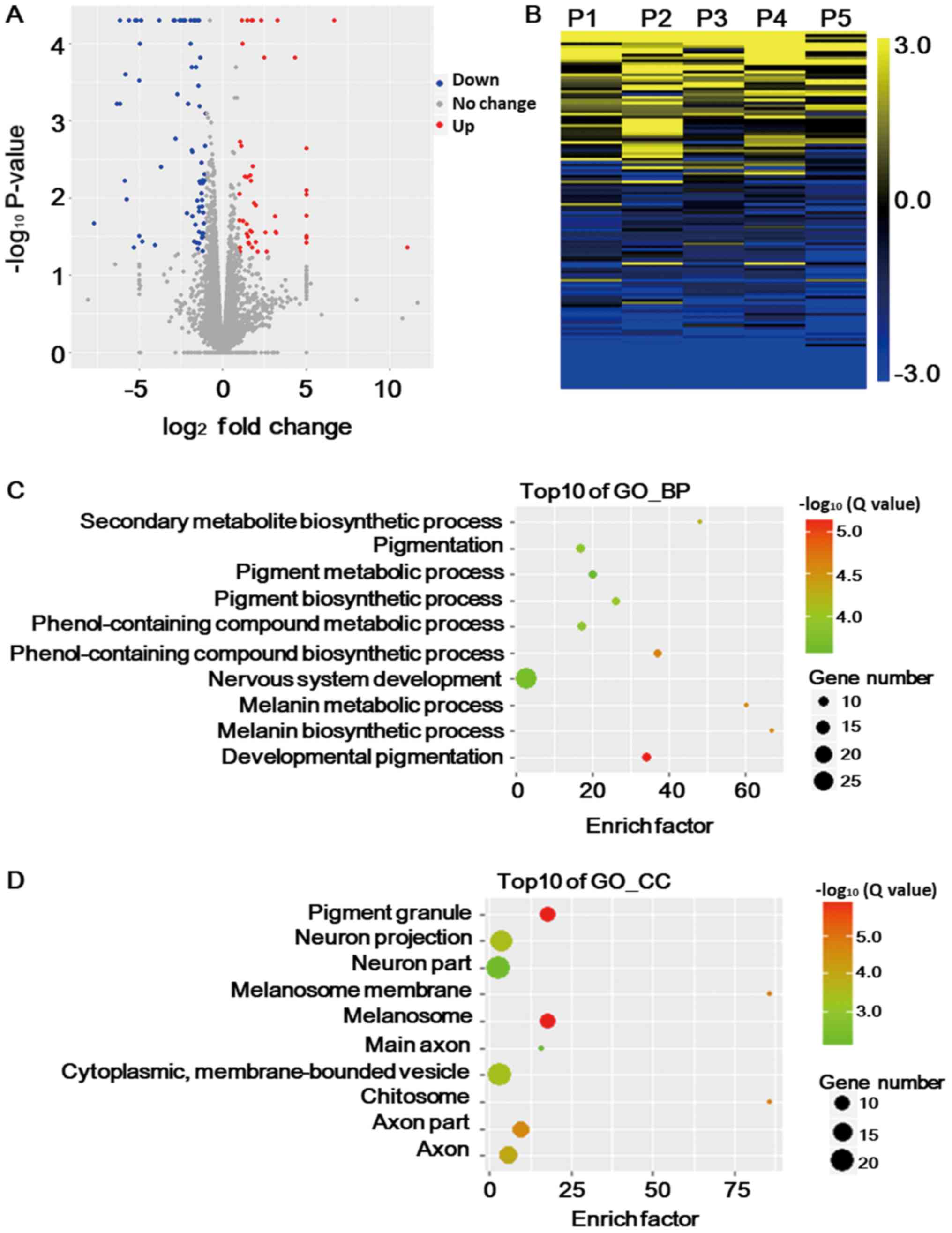
including 47 upregulated and 80 downregulated DEGs (Fig. 1A). In order to confirm these DEGs,
the changes in their expression were also assessed for each subject
individually. Most of these DEGs were changed in the same direction
among the individual subjects (Fig.
1B), demonstrating a consistence regarding the DEGs
identified.
To determine the functional roles of these DEGs, the
functional categories they were involved in were then examined. Of
note, the DEGs were mainly enriched in functional categories
closely linked to pigment synthesis or metabolic pathways (Fig. 1C). The DEGs were also known to be
located in pigment granules and melanosomes (Fig. 1D), which provided an explanation for
the downregulation of these genes in grey hair follicles that had
lost their ability to produce melanin, which lead to hair graying.
Of note, genes associated with the nervous system accounted for the
largest number of DEGs, although the significance of their
enrichment was not as high as that in the categories associated
with pigmentation. This may imply a disturbance of the nervous
system in grey hair follicles compared with that in black ones.
Genes associated with pigmentation and
melanin synthesis are predominantly decreased in grey hair
follicles
In order to obtain a comprehensive view of genetic
changes associated with pigmentation and melanin synthesis, the
DEGs identified were classified into
pigmentation/melanin-associated GO terms (24). The results indicated that all of the
pigmentation/melanin-associated DEGs were decreased in grey hair
follicles (Fig. 2A). This suggested
that pathways involved in melanin biosynthesis are downregulated in
grey hair follicles. These DEGs comprised various genes that are
well-known to be responsible for melanin biosynthesis, including
TYR, melan-A (MLANA), premelanosome protein (PMEL), TYRP1, SLC45A2,
KIT, G protein-coupled receptor 143 (GPR143) and OCA2.
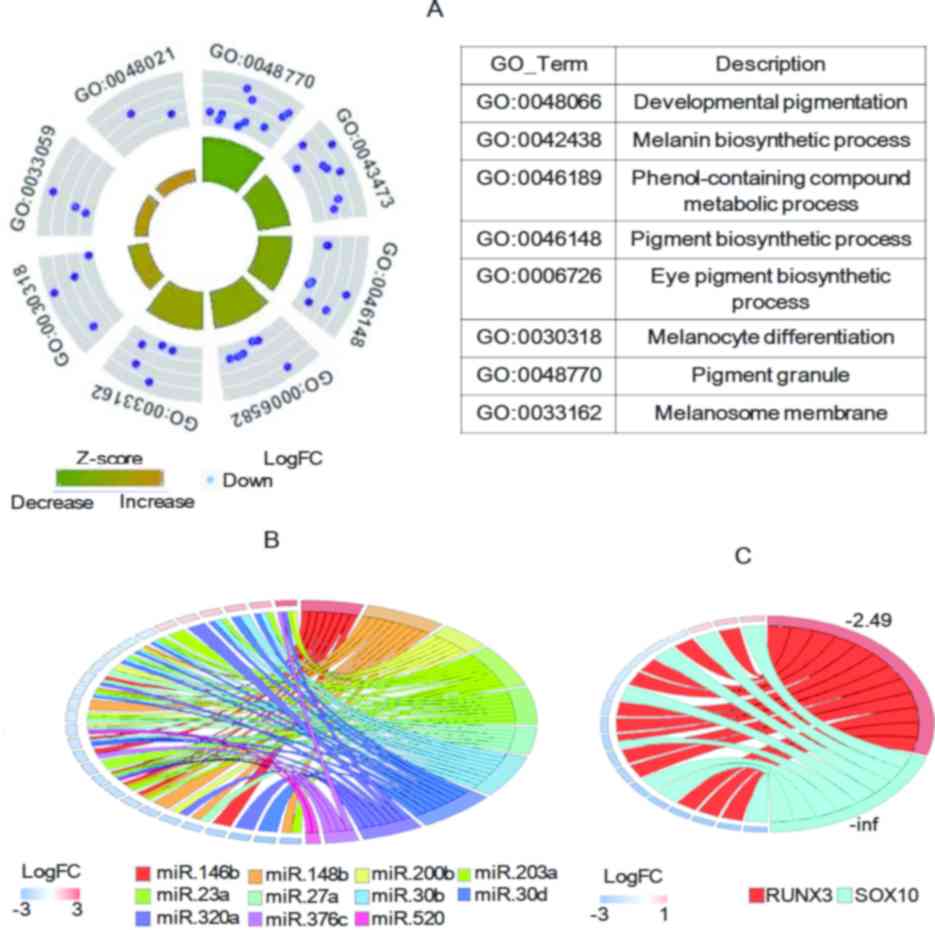 | Figure 2.Genes associated with pigmentation
and melanin synthesis are predominantly decreased in white hair
follicles. (A) GOCircle plot of DEGs associated with
pigmentation-associated GO terms. The height of the inner bars
indicates the significance of the corresponding terms
(-log10-adjusted P-value), and the color corresponds to the
enrichment Z-score (GOplot). The outer ring displays scatterplots
of the expression changes (log2|FC|) for genes in each term,
wherein blue represents decreased expression. (B and C) Expression
changes of DEGs predicted to be targeted by miRs derived from (B)
upregulated miR-encoding genes and (C) downregulated TFs. The
gradually changing green-to-pink bars denote the log2|FC| of the
target DEGs. The targeted genes are displayed on the left of the
circle, while miRs or TFs are presented on the right (red indicates
RUNX3 and blue indicates SOX10). miRs or TFs and their predicted
targets are linked via ribbons inside. DEG, differentially
expressed gene; TF, transcription factor; GO, gene ontology; FC,
fold change; miR, microRNA; SOX, sex-determining region Y box;
RUNX, runt-related TF. |
Differentially expressed miRs and TFs
regulating pigmentation/melanin-associated DEGs in grey hair
follicles
Among the DEGs identified, 13 genes encoded miRs,
all of which were significantly increased in grey hair follicles;
they accounted for 28% (13/47) of all upregulated DEGs. It has been
previously reported that miRs may be transcribed by RNA polymerase
II and have polyA tails (25), and
it was therefore likely that the miR-associated RNA-seq reads are
from precursors of miRs. Since the majority of the DEGs (80/127;
63%) were decreased in the grey hairs, the changes in the
expression of DEGs targeted by these upregulated miR-encoding genes
were examined (Fig. 2B). Of note,
most of the miR-encoding DEGs (11 out of 13) had the other DEGs as
their predicted targets (Table
III), the majority of which were significantly decreased in
grey compared with black hair follicles. Furthermore, among the
miR-targeted DEGs, two genes (syndecan binding protein and KIT) are
involved in pigment-associated pathways (26,27).
 | Table III.Differentially expressed genes that
are predicted targets of miRs in Table
IV. |
Table III.
Differentially expressed genes that
are predicted targets of miRs in Table
IV.
| Gene ID | Gene | FPKM in black
hair | FPKM in white
hair | Log2|FC| | P-value |
|---|
|
ENSG00000157404 | KIT | 4.22 | 0.14 | −4.88 | 0.00005 |
|
ENSG00000136040 | PLXNC1 | 6.36 | 0.46 | −3.79 | 0.00005 |
|
ENSG00000104177 | MYEF2 | 1.29 | 0.17 | −2.92 | 0.00005 |
|
ENSG00000020633 | RUNX3 | 1.48 | 0.26 | −2.49 | 0.00005 |
|
ENSG00000145335 | SNCA | 6.53 | 1.28 | −2.35 | 0.00005 |
|
ENSG00000071575 | TRIB2 | 2.72 | 0.56 | −2.28 | 0.00005 |
|
ENSG00000123095 | BHLHE41 | 5.33 | 1.2 | −2.16 | 0.01575 |
|
ENSG00000130558 | OLFM1 | 1.27 | 0.33 | −1.94 | 0.00010 |
|
ENSG00000137575 | SDCBP | 10.59 | 2.79 | −1.93 | 0.00005 |
|
ENSG00000048740 | CELF2 | 1.84 | 0.49 | −1.91 | 0.00005 |
|
ENSG00000197283 | SYNGAP1 | 20.4 | 5.71 | −1.84 | 0.00240 |
|
ENSG00000166173 | LARP6 | 1.39 | 0.4 | −1.8 | 0.01725 |
|
ENSG00000026025 | VIM | 26.35 | 7.8 | −1.76 | 0.00005 |
|
ENSG00000204764 | RANBP17 | 1.35 | 0.43 | −1.65 | 0.02195 |
|
ENSG00000115825 | PRKD3 | 6.13 | 2.31 | −1.41 | 0.01485 |
|
ENSG00000082397 | EPB41L3 | 1.85 | 0.73 | −1.34 | 0.00015 |
|
ENSG00000173482 | PTPRM | 2.16 | 0.88 | −1.29 | 0.00870 |
|
ENSG00000153823 | PID1 | 1.42 | 0.59 | −1.26 | 0.00345 |
|
ENSG00000115414 | FN1 | 1.68 | 0.72 | −1.23 | 0.01050 |
|
ENSG00000185008 | ROBO2 | 1.5 | 0.65 | −1.2 | 0.04845 |
|
ENSG00000086289 | EPDR1 | 3.04 | 1.42 | −1.09 | 0.00595 |
|
ENSG00000116641 | DOCK7 | 11.49 | 5.51 | −1.06 | 0.00210 |
|
ENSG00000249859 | PVT1 | 10.11 | 20.29 | 1 | 0.01935 |
|
ENSG00000135346 | CGA | 14.88 | 32.89 | 1.14 | 0.00005 |
|
ENSG00000137941 | TTLL7 | 1.77 | 4.44 | 1.32 | 0.00525 |
|
ENSG00000105784 | RUNDC3B | 0.44 | 1.41 | 1.7 | 0.00595 |
|
ENSG00000113532 | ST8SIA4 | 0.33 | 1.21 | 1.88 | 0.01150 |
|
ENSG00000151967 | SCHIP1 | 0.09 | 1.84 | 4.32 | 0.00015 |
Another category of regulators affecting gene
expression changes are TFs. Therefore, it was then examined whether
the DEGs included any TFs and their potential targets. A total of 4
DEGs encoding TFs were identified: AE binding protein 1, coiled
coil domain-containing 17, runt-related transcription factor 3
(RUNX3) and SOX10, all of which were significantly decreased in
grey hair follicles. Of note, two of these TFs, namely RUNX3 and
SOX10, had potential targets among the other DEGs (Table IV), which also tended to be
decreased in grey hair follicles (Fig.
2C), indicating an association between the downregulation of
the TFs and their potential target genes in grey hair follicles.
Transient receptor potential cation channel subfamily M member 1
(TRPM1), which encodes a permeable cation channel that is expressed
in melanocytes and has a role in melanin synthesis, was among the
downregulated genes regulated by these TFs (28,29). Of
note, SOX10 have been proved to have the ability to drive the
differentiation of melanocytes (30).
 | Table IV.Differentially expressed genes
targeted by differential expression of genes encoding transcription
factors (RUNX3 and sex-determining region Y box 10). |
Table IV.
Differentially expressed genes
targeted by differential expression of genes encoding transcription
factors (RUNX3 and sex-determining region Y box 10).
| Gene ID | Gene | FPKM in black
hair | FPKM in white
hair | Log2|FC| | P-value |
|---|
|
ENSG00000020633 | RUNX3 | 1.48 | 0.26 | −2.49 | 0.00005 |
|
ENSG00000048740 | CELF2 | 1.84 | 0.49 | −1.91 | 0.00005 |
|
ENSG00000069424 | KCNAB2 | 4.88 | 0.99 | −2.31 | 0.00005 |
|
ENSG00000086289 | EPDR1 | 3.04 | 1.42 | −1.09 | 0.00595 |
|
ENSG00000089101 | CFAP61 | 1.42 | 0.25 | −2.52 | 0.00005 |
|
ENSG00000091986 | CCDC80 | 4.3 | 8.75 | 1.03 | 0.00185 |
|
ENSG00000104177 | MYEF2 | 1.29 | 0.17 | −2.92 | 0.00005 |
|
ENSG00000115414 | FN1 | 1.68 | 0.72 | −1.23 | 0.01050 |
|
ENSG00000123560 | PLP1 | 3.13 | 0.06 | −5.74 | 0.01035 |
|
ENSG00000130751 | NPAS1 | 0.55 | 1.47 | 1.42 | 0.02150 |
|
ENSG00000134160 | TRPM1 | 8.52 | 0.15 | −5.83 | 0.00025 |
|
ENSG00000136732 | GYPC | 1.43 | 0.09 | −4.05 | 0.04075 |
|
ENSG00000154277 | UCHL1 | 8.49 | 1.48 | −2.52 | 0.00005 |
|
ENSG00000173482 | PTPRM | 2.16 | 0.88 | −1.29 | 0.00870 |
|
ENSG00000184724 | KRTAP6-1 | 6.05 | 16.75 | 1.47 | 0.00005 |
|
ENSG00000187045 | TMPRSS6 | 1.19 | 0.53 | −1.16 | 0.02810 |
|
ENSG00000197415 | VEPH1 | 2.63 | 0.37 | −2.85 | 0.00170 |
|
ENSG00000204764 | RANBP17 | 1.35 | 0.43 | −1.65 | 0.02195 |
|
ENSG00000221887 | HMSD | 1.58 | 0.02 | −6.14 | 0.00060 |
PPI network centers for genes
associated with melanin synthesis
A protein usually interacts with other proteins to
function properly. To obtain a systematic view of the biological
roles of the DEGs in grey hair follicles, the PPI networks were
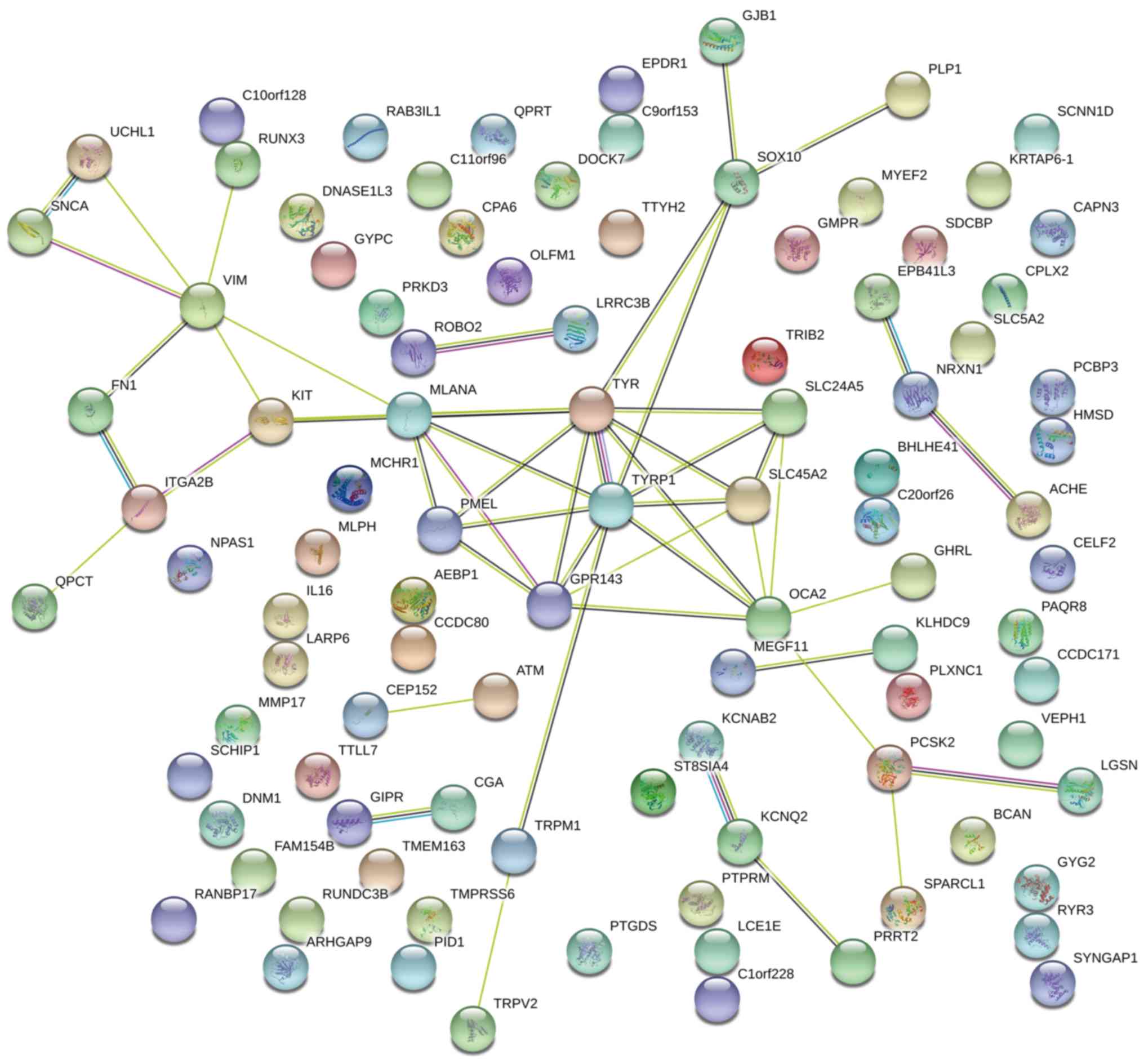
analyzed using DEGs by means of the STRING database. The result
indicated that DEGs were involved in a PPI network centering on
genes associated with melanin biosynthesis (Fig. 3), including TYR, TYRP1, MLANA,
GPR143, OCA2, PMEL, SLC24A5 and SLC45A2. Further genes that are not
involved in melanin synthesis, including KIT ligand, checkpoint
kinase 1 and integrin subunit β3, were also identified. However,
they may affect the melanin biosynthesis process and ultimately
affect hair pigmentation through interacting with genes directly
involved in melanin biosynthesis.
Validation of RNA-seq data by
RT-qPCR
In order to verify gene expression differences
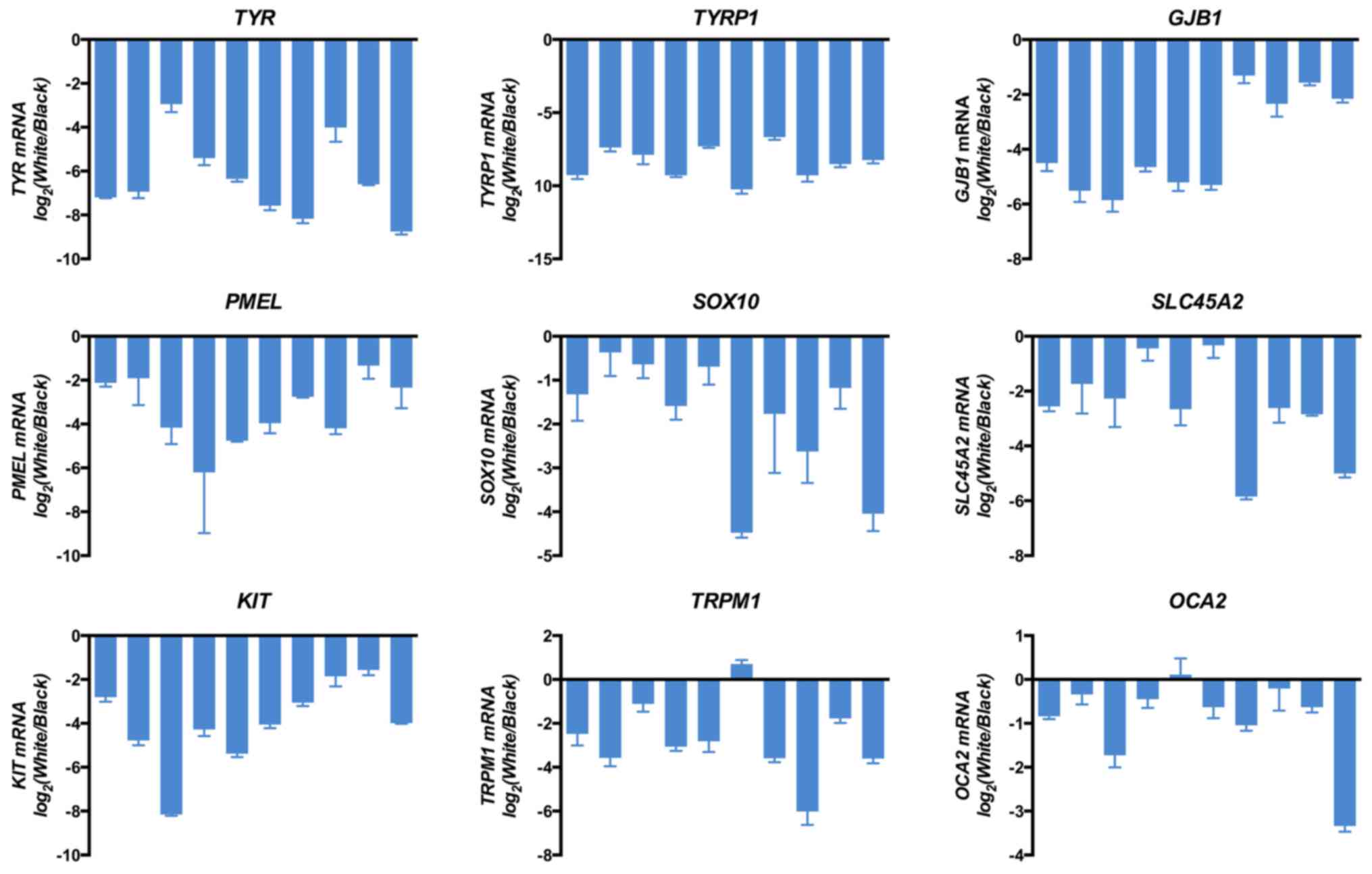
determined via RNA-seq analysis, 9 randomly selected genes [TYR,
PMEL, TYRP1, gap junction β1 (GJB1), SOX10, SLC45A2, KIT, TRPM1 and
OCA2] were detected by RT-qPCR. As presented in Fig. 4, the abscissa represents different
samples, and ordinates are the relative expression value of grey
vs. black hair. The results indicated that the expressional changes
of all of these genes were consistent with those detected by
RNA-seq analysis, thus confirming that all genes were consistently
downregulated in grey vs. black hair follicles.
Discussion
Hair graying, particularly premature hair graying,
changes the appearance of affected individuals in a mostly
undesired manner and has attracted the attention of researchers. To
date, the underlying causes have remained largely elusive. In the
present study, a genome-wide RNA-seq profiling analysis was
performed using grey and black hair follicles from the same
individuals with premature hair graying. It was revealed that
pigment synthesis pathways were significantly impaired in grey vs.
black hair, with a significantly decreased expression of multiple
key genes crucial for the stability, trafficking and proliferation
of melanocytes (31). The present
results support the theory that premature graying may occur due to
exhaustion of the melanocytes' capability to produce hair
pigmentation.
Previous studies have suggested that premature hair
graying is associated with factors affecting melanogenesis,
including nutritional deficiencies (32), insufficient neuroendocrine
stimulation (33) and ionic
signaling across melanosomes (34).
The identified DEGs included those with similar functions with this
regard, e.g. MCHR1, TRPM1 and SLC45A2. MCHR1 acts as a receptor for
melanin-concentrating hormone (35),
TRPM1 regulates pigmentation at the plasma membrane level (29) and SLC45A2 modulates the melanosomal
pH for optimal TYR activity required for melanogenesis (36). Of note, the present study identified
that GJB1 was downregulated in grey vs. black hair follicles, which
known to contribute to pigment transfer between melanocytes and
neighboring keratinocytes (37).
While pigmentation-associated pathways are impaired
in grey hair follicles, the underlying mechanisms of their
inhibition/damage during hair greying remain to be elucidated. Of
note, all of the 13 DEGs encoding miRs identified in the present
study were significantly upregulated in grey hair follicles,
indicating a substantial increase in the abundance of the
corresponding mature miRs. Furthermore, the majority of the DEGs
predicted to be targets of these miRs were significantly decreased
in grey hair follicles, which is in line with the known mechanism
that miRs reduce the expression of their target genes.
Another reason could be the decreased TFs in grey
hair follicles. Among the four decreased TFs expressed in gray
hair, two TFs (RUNX3 and SOX10) had potential target genes in other
DEGs, which were also downregulated in gray hair, indicating that
the downregulation of TFs were associated with their potential
target genes in gray hair follicles. In addition, GJB1 was
co-expressed with a crucial node, SOX10, in the PPI network. It has
been reported that SOX10, in synergy with early growth response 2,
may activate GJB1 in melanocytes, which may cause and alteration in
melanogenesis (38,39).
Another point worth mentioning is that a relatively
high fraction of DEGs was associated with the nervous system,
including potassium voltage-gated channel subfamily Q member 2,
basic helix-loop-helix family member e41, prostaglandin D2 synthase
and centrosomal protein of 152 kDa, which are thought to be
involved in controlling the circadian rhythm. Defects in these
genes have been reported to be associated with a short sleep
phenotype (40,41). It is widely accepted that nerve
signaling defects, including a disturbed sleeping ability, may lead
to hair graying, which probably functions through its further
effects on the nervous system and downstream pigmentation pathways
(42). While it remains elusive
whether and how the nervous system contributes to premature hair
graying, the present results may indicate a novel aspect regarding
the causes of hair graying. Since hair graying has been considered
to be associated with aging of the hair follicle pigmentation
system (43), the JenAge Ageing
Factor Database (44) was searched
for the DEGs in grey hair identified in the present study. A total
of 5 genes were included in the online database, namely
acetylcholine esterase, ATM, proprotein convertase subtilisin/kexin
type 2, ubiquitin C-terminal hydrolase L1 and ventricular zone
expressed PH domain containing 1, all of which had a decreased
expression in white hair follicles, implying the decline of
melanocyte-associated processes.
In conclusion, the present study was the first, to
the best of our knowledge, to perform a genome-wide transcriptome
profiling of human hair follicles affected by premature hair
graying, which uncovered that damage of the melanin biosynthesis
pathway was the direct cause of the decline hair pigmentation and
the resulting hair graying. Furthermore, it was indicated that
deregulated miRs and TFs, as well as neural disturbances, may be
underlying causes. The present study provided multiple clues worthy
of subsequent study to elucidate the mechanisms underlying canities
and human premature hair graying. However, the present study is
limited as only the processes occurring in subjects with premature
hair greying were assessed, while the genetic predisposition to
hair greying was not be determined. The genetic differences between
subjects with premature hair greying and normal individuals should
be assessed to identify the genes that are the primary cause of
this condition.
Acknowledgements
The authors would like to thank Dr Shiming Wang, Dr
Cong Huai and Dr Hexige Saiyin (School of Life Sciences, Fudan
University, Shandhai, China), as well as Dr Xiaotian Wang (Thermo
Fisher Scientific, Inc.) for their assistance in sample collection
and RNA-seq library construction.
Funding
The present study was supported by the National
Natural Science Foundation (grant no. 31571371) and the National
Key Research and Development Plan (grant no. 2017YFC090750).
Availability of data and materials
The data used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
DL, TN and HC conceived and designed the
experiments. YB, XS and LY collected hair follicles, prepared RNA
samples and constructed RNA-Seq libraries. GW analyzed
transcriptome data. YB and LY performed qRT-PCR. YB and GW wrote
the paper. All authors read and approved the final version of the
manuscript.
Ethical approval and consent to
participate
The present study was performed according to the
declaration of Helsinki and was approved by the Research Ethics
Committee at Fudan University (Shanghai, China). Informed consent
was obtained from each participant prior to enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Panhard S, Lozano I and Loussouarn G:
Greying of the human hair: A worldwide survey, revisiting the ‘50’
rule of thumb. Br J Dermatol. 167:865–873. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tobin DJ: Human hair
pigmentation-biological aspects. Int J Cosmet Sci. 30:233–257.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tobin DJ and Paus R: Graying:
Gerontobiology of the hair follicle pigmentary unit. Exp Gerontol.
36:29–54. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pandhi D and Khanna D: Premature graying
of hair. Indian J Dermatol Venereol Leprol. 79:641–653. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Slominski A, Wortsman J, Plonka PM,
Schallreuter KU, Paus R and Tobin DJ: Hair follicle pigmentation. J
Invest Dermatol. 124:13–21. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Slominski A, Paus R and Costantino R:
Differential expression and activity of melanogenesis-related
proteins during induced hair growth in mice. J Invest Dermatol.
96:172–179. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Slominski A, Paus R, Plonka P, Chakraborty
A, Maurer M, Pruski D and Lukiewicz S: Melanogenesis during the
anagen-catagen-telogen transformation of the murine hair cycle. J
Invest Dermatol. 102:862–869. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tobin DJ: The cell biology of human hair
follicle pigmentation. Pigment Cell Melanoma Res. 24:75–88. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rees JL: Genetics of hair and skin color.
Annu Rev Genet. 37:67–90. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sturm RA: Molecular genetics of human
pigmentation diversity. Hum Mol Genet. 18:R9–R17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sabeti PC, Varilly P, Fry B, Lohmueller J,
Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R,
et al: Genome-wide detection and characterization of positive
selection in human populations. Nature. 449:913–918. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han J, Kraft P, Nan H, Guo Q, Chen C,
Qureshi A, Hankinson SE, Hu FB, Duffy DL, Zhao ZZ, et al: A
genome-wide association study identifies novel alleles associated
with hair color and skin pigmentation. PLoS Genet. 4:e10000742008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sulem P, Gudbjartsson DF, Stacey SN,
Helgason A, Rafnar T, Jakobsdottir M, Steinberg S, Gudjonsson SA,
Palsson A, Thorleifsson G, et al: Two newly identified genetic
determinants of pigmentation in Europeans. Nat Genet. 40:835–837.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Z, Duffy D, Thomas S, Martin NG,
Hayward NK and Montgomery GW: Polymorphisms in the syntaxin 17 gene
are not associated with human cutaneous malignant melanoma.
Melanoma Res. 19:80–85. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Diaz de Leon A, Cronkhite JT, Yilmaz C,
Brewington C, Wang R, Xing C, Hsia CCW and Garcia CK: Subclinical
lung disease, macrocytosis, and premature graying in kindreds with
telomerase (TERT) mutations. Chest. 140:753–763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kwon MJ, Lee KY, Lee HW, Kim JH and Kim
TY: SOD3 variant, R213G, altered SOD3 function, leading to
ROS-mediated inflammation and damage in multiple organs of
premature aging mice. Antioxid Redox Signal. 23:985–999. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin BD, Mbarek H, Willemsen G, Dolan CV,
Fedko IO, Abdellaoui A, de Geus EJ, Boomsma DI and Hottenga JJ:
Heritability and genome-wide association studies for hair color in
a dutch twin family based sample. Genes (Basel). 6:559–576. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dobin A, Davis CA, Schlesinger F, Drenkow
J, Zaleski C, Jha S, Batut P, Chaisson M and Gingeras TR: STAR:
Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate-a practical and powerful approach to
multiple testing. J Royal Stat Soc Series B-Methodol. 57:289–300.
1995.
|
|
21
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res 42 (Database issue). D92–D97. 2014. View Article : Google Scholar
|
|
22
|
Balwierz PJ, Pachkov M, Arnold P, Gruber
AJ, Zavolan M and van Nimwegen E: ISMARA: Automated modeling of
genomic signals as a democracy of regulatory motifs. Genome Res.
24:869–884. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res 39 (Database issue). D561–D568. 2011. View Article : Google Scholar
|
|
24
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Aissaoui H, Prévost C, Boucharaba A,
Sanhadji K, Bordet JC, Négrier C and Boukerche H: MDA-9/syntenin is
essential for factor VIIa-induced signaling, migration, and
metastasis in melanoma cells. J Biol Chem. 290:3333–3348. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang S, Yu X and Dong C: MiR-137 affects
melanin synthesis in mouse melanocyte by repressing the expression
of c-Kit and Tyrp2 in SCF/c-Kit signaling pathway. Biosci
Biotechnol Biochem. 80:2115–2121. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gees M, Owsianik G, Nilius B and Voets T:
TRP channels. Compr Physiol. 2:563–608. 2012.PubMed/NCBI
|
|
29
|
Oancea E, Vriens J, Brauchi S, Jun J,
Splawski I and Clapham DE: TRPM1 forms ion channels associated with
melanin content in melanocytes. Sci Signal. 2:ra212009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Harris ML, Buac K, Shakhova O, Hakami RM,
Wegner M, Sommer L and Pavan WJ: A dual role for SOX10 in the
maintenance of the postnatal melanocyte lineage and the
differentiation of melanocyte stem cell progenitors. PLoS Genet.
9:e10036442013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
D'Mello SA, Finlay GJ, Baguley BC and
Askarian-Amiri ME: Signaling pathways in melanogenesis. Int J Mol
Sci. 17:E11442016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fatemi Naieni F, Ebrahimi B, Vakilian HR
and Shahmoradi Z: Serum iron, zinc, and copper concentration in
premature graying of hair. Biol Trace Elem Res. 146:30–34. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Paus R: A neuroendocrinological
perspective on human hair follicle pigmentation. Pigment Cell
Melanoma Res. 24:89–106. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bellono NW and Oancea EV: Ion transport in
pigmentation. Arch Biochem Biophys. 563:35–41. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kemp EH, Waterman EA, Hawes BE, O'Neill K,
Gottumukkala RV, Gawkrodger DJ, Weetman AP and Watson PF: The
melanin-concentrating hormone receptor 1, a novel target of
autoantibody responses in vitiligo. J Clin Invest. 109:923–930.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dooley CM, Schwarz H, Mueller KP, Mongera
A, Konantz M, Neuhauss SC, Nüsslein-Volhard C and Geisler R:
Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis
in zebrafish, providing a mechanism for human pigment evolution and
disease. Pigment Cell Melanoma Res. 26:205–217. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Joshi PG, Nair N, Begum G, Joshi NB,
Sinkar VP and Vora S: Melanocyte-keratinocyte interaction induces
calcium signalling and melanin transfer to keratinocytes. Pigment
Cell Res. 20:380–384. 2007.PubMed/NCBI
|
|
38
|
LeBlanc SE, Ward RM and Svaren J:
Neuropathy-associated Egr2 mutants disrupt cooperative activation
of myelin protein zero by Egr2 and Sox10. Mol Cell Biol.
27:3521–3529. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ludwig A, Rehberg S and Wegner M:
Melanocyte-specific expression of dopachrome tautomerase is
dependent on synergistic gene activation by the Sox10 and Mitf
transcription factors. FEBS Letters. 556:236–244. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lee IC, Yang JJ and Li SY: A KCNQ2 E515D
mutation associated with benign familial neonatal seizures and
continuous spike and waves during slow-wave sleep syndrome in
Taiwan. J Formos Med Assoc. 116:711–719. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pellegrino R, Kavakli IH, Goel N,
Cardinale CJ, Dinges DF, Kuna ST, Maislin G, Van Dongen HP, Tufik
S, Hogenesch JB, et al: A novel BHLHE41 variant is associated with
short sleep and resistance to sleep deprivation in humans. Sleep.
37:1327–1336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Anderson NE and Haas LF: Neurological
complications of Werner's syndrome. J Neurol. 250:1174–1178. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tobin DJ: Aging of the hair follicle
pigmentation system. Int J Trichology. 1:83–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Huhne R, Thalheim T and Sühnel J:
AgeFactDB-the JenAge ageing factor database-towards data
integration in ageing research. Nucleic Acids Research (Database
issue). 42:D892–D896. 2014. View Article : Google Scholar
|