Immunoglobulin A (IgA) nephropathy (IgAN) is a
common primary renal disease worldwide (1,2) and has
emerged as an important healthcare issue (3). Cell proliferation (4,5),
fibrosis (6,7), apoptosis (8,9) and
sustained inflammation (10) are
involved in the pathogenesis of IgAN. Inhibition of human mesangial
cell proliferation by targeting C3a/C5a receptors has been
demonstrated to alleviate IgAN in mice (4). Glomerular endothelial proliferation has
been reported to contribute to renal injury in IgAN (11,12).
Cytotoxin-associated antigen A may induce cellular injury in
glomerular mesangium through the proliferation and secretion of the
extracellular matrix (ECM), which may have an important role in the
pathogenesis of IgAN (13). Renal
expression of microRNA (miR)-21-5p is associated with fibrosis and
renal survival in patients with IgAN (6), and rapamycin may reduce apoptosis of
podocytes under stimulated conditions of IgAN (8,9).
However, the crucial genes involved in IgAN have remained elusive
due to limited large-scale studies, and methods for the effective
early diagnosis and treatment of IgAN remain unavaliable.
Bioinformatics analysis is a powerful research
method used to predict molecular mechanisms and associations among
genes. This approach has been used to predict novel genes and
pathways associated with tumors, including hepatocellular carcinoma
(14), non-small cell lung cancer
(15), osteosarcoma (16) and esophageal adenocarcinoma (17). Bioinformatics analysis has gradually
provided insight into the molecular mechanisms of kidney disease
(18). For instance, the gene
expression profile of macrophages was recently analyzed through a
bioinformatics analysis, indicating the induction of CCL2 and CD38
in macrophages from patients with lupus nephritis (19). To date, only few bioinformatics
analyses have been performed on IgAN. A distinct glomerular
molecular signature associated with endocapillary proliferation has
been identified in patients with IgAN through a gene expression
profiling array (12). However, to
the best of our knowledge, no integrated and in-depth data analysis
associated with IgAN has been previously performed. Therefore, it
is necessary to identify genes associated with IgAN through
integrated bioinformatics analysis. In the present study, several
gene expression profile datasets were downloaded from the Gene
Expression Omnibus (GEO) database. After data integration
processing, differentially expressed genes (DEGs) were identified,
and Gene Ontology (GO) functional analysis was performed. The top
upregulated and downregulated hub genes in five disease-associated
biological functions were further analyzed. Protein-protein
interaction (PPI) networks of the DEGs were built and the hub genes
were identified. The present results indicated that these hub genes
were involved in different pathological mechanisms in IgAN. The
DEGs co-expressed with the top hub genes were then further
analyzed. The present study aimed to discover potential novel
candidate molecular targets in IgAN.
Human IgAN microarray datasets were searched and
downloaded from the National Center for Biotechnology Information
(NCBI) GEO database (http://www.ncbi.nlm.nih.gov/geo). The keyword ‘IgA
nephropathy’ was used for accurate searching. The data selection
criteria were as follows: i) All datasets were expression profiles,
ii) all samples were kidney glomerular tissues, iii) the species
was Homo sapiens, and iv) complete microarray raw data were
available. Finally, two datasets, namely GSE93798 (20) and GSE37460 (21,22),
were finally selected on the basis of the abovementioned criteria
with exclusion of the duplicate data, for integrated analysis. The
integrated datasets included 47 IgAN and 31 normal glomerular
tissues. Original CEL files and platform probe annotation
information files were subjected to further bioinformatics
analysis.
The preprocessing and normalization of microarray
datasets with raw data (.CEL files) were performed with the RMA
function in the Affy package in the R environment (version 3.2.3)
(23) with the following parameters:
Data normalization using quantile normalization and background
correction using RMA background correction with a background
similar to the pure RMA background given in the Affy version 1.1
and above (23,24). After the gene expression value was
obtained, the ‘annotate’ software package was used to annotate the
genes and the expression matrix was merged. The batch effects from
the microarray were removed by the function Combat in the SVA
package with the ‘Empirical Bayes methods’ (25).
The DEGs between the IgAN and normal tissues were
analyzed with the Limma package in R (26). The linear fit method (using the lmFit
function with default options), Bayesian analysis (using the eBayes
function with default options) and the t-test algorithm were
utilized to calculate the P-values and fold change (FC) values. The
TopTable function in the Limma package was used to screen the DEGs
(parameters: Adjust.method=‘fdr’, coef=1, adjusted
(adj.)P-value=0.05, lfc=log(2,2), number=5,000, and
sort.by=‘logFC’). An adj.P-value <0.05 and |log2FC|≥1 were set
as the cutoff parameters to screen any significantly upregulated or
downregulated genes. The ggplot2 software package was used to
visualize the DEGs.
The ClusterProfiler version 3.5 is an R package for
the biological term classification and enrichment analysis of gene
clusters (27). The cluster profiler
package was used to perform GO functional enrichment analysis for
the DEGs. Adj.P-value <0.05 was set as the cutoff criterion for
GO enrichment analysis.
The DEGs identified were subjected to PPI analysis
by using the search functionality of STRING (http://string.embl.de/) (28) to explore the association between the
DEGs, and a network interaction matrix was built. The minimum
required interaction score of 0.7 was the cutoff threshold. The PPI
network data matrix was downloaded for further analysis and
visualization by using Cytoscape (version 6.3; http://www.cytoscape.org/) (29). CytoHubba (30) is a tool used to identify hub objects
and subnetworks from a complex interactome. ‘Degree’ is a
topological analysis method in CytoHubba. ‘Degree’ was used to
discover featured nodes and identify the hub genes from all
DEGs.
The top five hub genes in the five
disease-associated GO terms were selected by consulting the
literature in the NCBI database to find the potential KEGG pathway
associations with IgAN. The KEGG tool (http://www.genome.jp/kegg/pathway.html) was used to
further analyze the signaling pathways of the selected target
genes.
Pearson correlation coefficients between each top
hub gene and all other DEGs were calculated by using the package
‘Hmisc’ (version 4.1.1). The top five significant genes associated
with each selected top hub gene were screened. The co-expression
association between each top hub gene and other DEGs was further
analyzed using Cytoscape.
A total of 78 samples, comprising 47 IgAN samples
and 31 normal samples, in the two datasets were included for
analysis in the present study. The detailed information of all of
the samples is listed in Table SI.
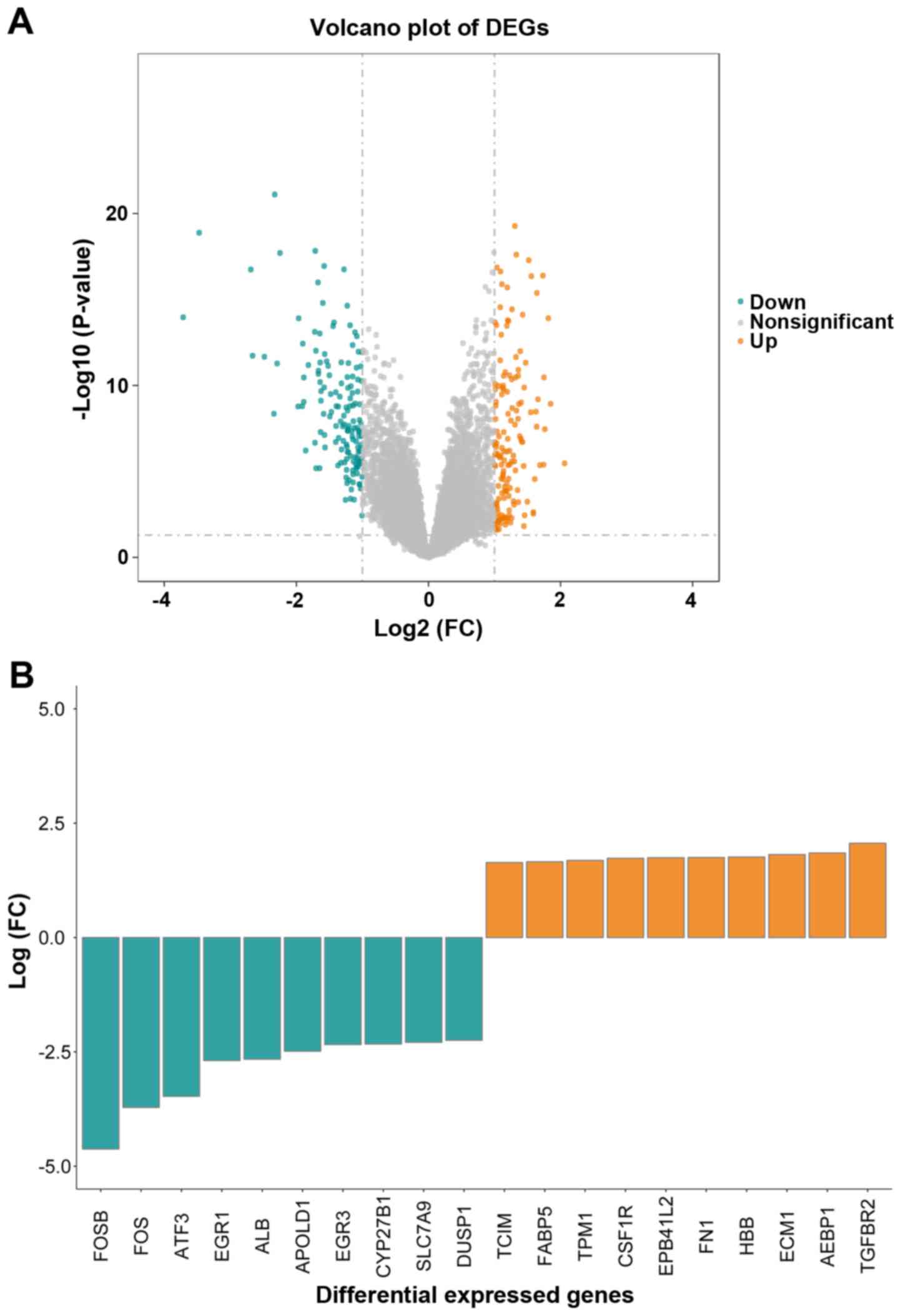
Based on the cutoff criteria of |log2 FC| ≥1.0 and adj.P-value
≤0.05, 298 DEGs in the IgAN group vs. control group were obtained.
Among those DEGs, 148 and 150 genes were upregulated and
downregulated, respectively. The results on the expression level
analysis are presented in a volcano plot in Fig. 1A. As indicated in the graph, the data
distribution of the upregulated, downregulated or insignificantly
changed gene expression levels was normal. Transcriptional and
immune response regulator, fatty acid binding protein 5,
tropomyosin 1, colony-stimulating factor 1 receptor, erythrocyte
membrane protein band 4.1 like 2 (EPB41L2), fibronectin 1 (FN1),
hemoglobin subunit β, ECM protein 1, adipocyte enhancer-binding
protein and transforming growth factor β receptor 2 (TGFBR2) were
the 10 most significantly upregulated genes in IgAN, whereas FosB
proto-oncogene, AP-1 transcription factor subunit (FOSB), FOS,
activating transcription factor 3 (ATF3), early growth response 1
(EGR1), albumin (ALB), apolipoprotein L domain containing 1, EGR3,
cytochrome P450 family 27 subfamily B member 1 (CYP27B1), solute
carrier family 7 member 9 and dual specificity phosphatase 1
(DUSP1) were the 10 most substantially downregulated genes
(Fig. 1B). Additional detailed
information on all DEGs is listed in Table SII.
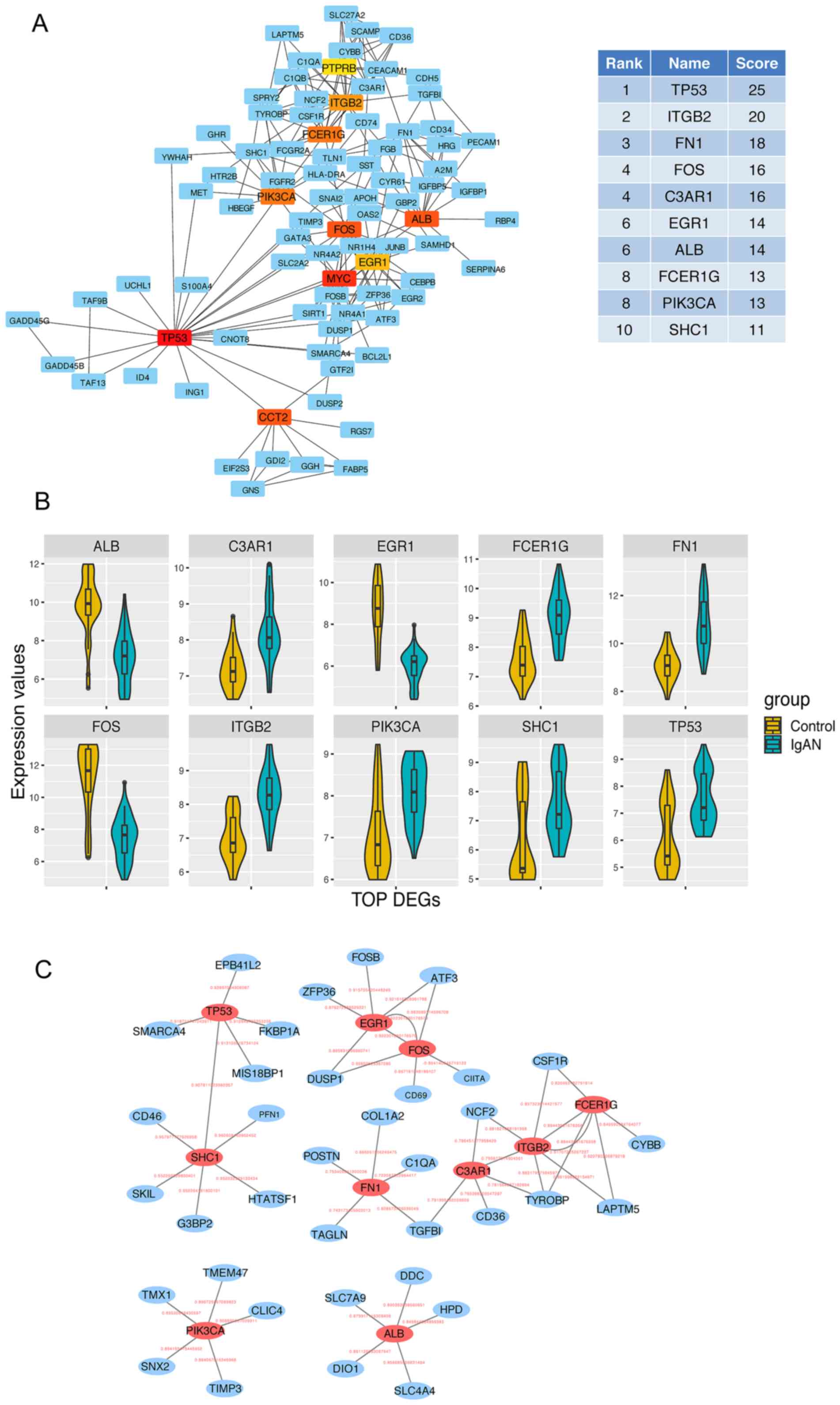
In the PPI network analysis, the average node degree
of connectivity was 2.57, the average local clustering coefficient
was 0.366 and the number of edges was 172. The P-value for clusters
of interacted proteins was <1.0×10−16. Finally, 10
top-ranked hub genes were identified from all the DEGs in the
network by Degree analysis, as presented in Fig. 2A (colored nodes). These genes were
tumor protein (TP53), integrin subunit β2 (ITGB2), FN1, FOS,
complement C3a receptor 1 (C3AR1), EGR1, ALB, Fc fragment of IgE
receptor Ig (FCER1G), phosphatidylinositol-4,5-bisphosphate
3-kinase catalytic subunit α (PIK3CA) and SHC adaptor protein 1
(SHC1). In the network, these hub genes had higher Degree scores
than the other hub genes (Fig. 2A).
The genes were closely correlated with one another. Among the 10
hub genes, seven genes, including TP53, ITGB2, FN1, C3AR1, FCER1G,
PIK3CA and SHC1, were upregulated, and three genes, including FOS,
EGR1 and ALB, were downregulated (Fig.
2B). Of note, among the top 10 hub genes, ALB (31), FOS (32) and TP53 (33) are known to be involved in the
pathological process of IgAN. The top five hub genes were
considered for subsequent KEGG pathway analysis to narrow down the
analysis.
Co-expressed gene analysis. Pearson correlation
analysis revealed the major DEGs co-expressed with TP53, ITGB2,
FN1, FOS, C3AR1, EGR1, ALB, FCER1G, PIK3CA and SHC1 (Fig. 2C). The top five DEGs co-expressed
with TP53 were EPB41L2, SWI/SNF related, matrix associated,
actin-dependent regulator of chromatin, subfamily A, member 4,
MIS18 binding protein 1, FKBP prolyl isomerase 1A and SHC1. The
genes co-expressed with ITGB2 were neutrophil cytosolic factor 2,
C3AR1, FCER1G, TYRO protein tyrosine kinase binding protein and
lysosomal protein transmembrane 5. Collagen type I α2 chain, TGFB1,
periostin, transgelin and complement C1q A chain were closely
associated with FN1. Of these, ITGB2 is possibly involved in the
pathological process of IgAN through ECM remodeling and apoptosis
(34,35), whereas FN1 may be implicated in
IgAN-associated fibrosis (36).
Additional information on the top six novel hub genes is listed in
Table I (37–69).
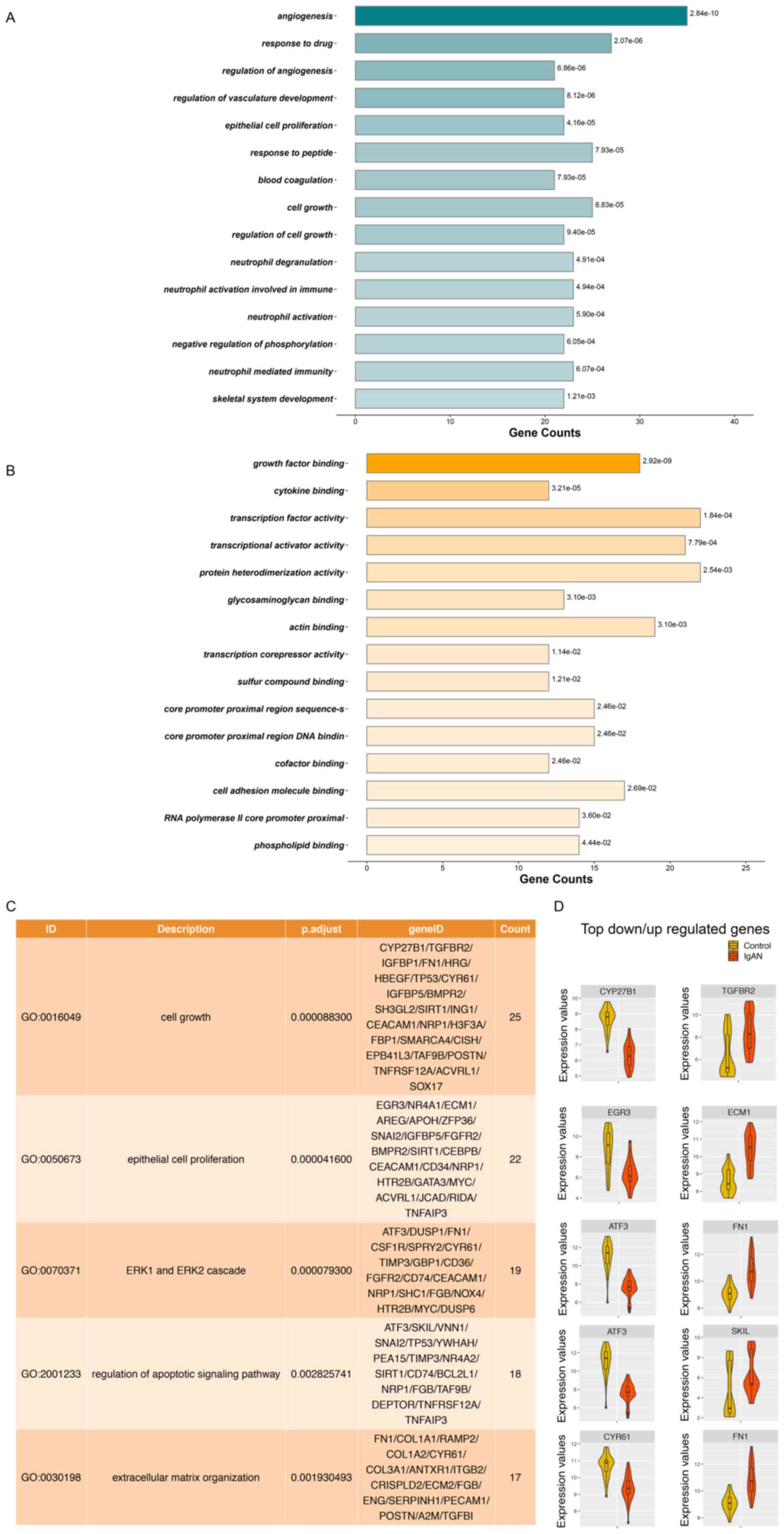
The ClusterProfiler package was used for pathway
enrichment analysis and GO analysis to reveal the biological
functions based on the DEGs. The 15 most significant GO terms in
the category biological process from the groups with adj.P<0.05
are presented in Fig. 4A and the 15
most significant GO terms in the category molecular function from
the groups with adj.P<0.05 are presented in Fig. 4B. The five GO terms closely
associated with pathological mechanisms are provided in Fig. 4C. A total of 25 DEGs were involved in
cell growth (GO:0016049), and 22 DEGs were implicated in the
regulation of cell growth and epithelial cell proliferation
(GO:0050673). Furthermore, 19 DEGs participated in the ERK1 and
ERK2 cascades (GO:0070371) and 18 DEGs functioned in the regulation
of the apoptotic signaling pathway (GO:2001233). In addition, 17
DEGs had a role in the ECM organization (GO:0030198). The top
upregulated and downregulated hub genes in these disease-associated
processes and functions are provided in Fig. 4D. CYP27B1, EGR3, ATF3, ATF3 and
cysteine-rich angiogenic inducer 61 (CYR61) were the top
downregulated hub genes in the five GO terms closely associated
with pathological mechanisms, and TGFBR2, ECM3, FN1, SKI-like
proto-oncogene and FN1 were the top upregulated hub genes in the
five GO terms. All of the significantly enriched GO terms are
listed in Table SIII.
In the present study, 148 upregulated and 150
downregulated DEGs were identified from microarray data by applying
an integrated bioinformatics analysis to further elucidate the
molecular pathology of IgAN. GO analysis revealed that the DEGs
were significantly enriched in cell growth, epithelial cell
proliferation, ERK1 and ERK2 cascade, apoptotic signaling pathway
regulation and ECM organization. CYP27B1 and TGFBR2 were enriched
in the GO terms associated with cell growth. miR-195 was reported
to inhibit proliferation, invasion and metastasis by targeting
CYP27B1 in breast cancer cells (70), and CYP27B1 is involved in the
anti-proliferative effects of 25-hydroxyvitamin D (71). miR-9-5p has been demonstrated to
promote cell growth and metastasis in non-small cell lung cancer
through repression of TGFBR2 (72).
Cell proliferation is a vital factor in the pathogenesis of IgAN.
For instance, circulating galactose-deficient IgA forms immune
complexes deposited in the glomerular mesangium and causes local
proliferation in IgAN (73). The
present analysis revealed that ATF3 was enriched in GO terms
including ERK1 and ERK2 cascades and regulation of the apoptotic
signaling pathway. Cell migration and invasion may be strengthened
by ATF3 through the activation of the p53 signaling pathway
(74). Uremic toxins have been
indicated to induce ATF3/c-Jun complex-mediated cannabinoid
receptor type 1 expression by modulating the ERK1/2 and JNK
signaling pathways and reactive oxygen species (75). FN1 was enriched in GO terms including
the ERK1 and ERK2 cascades and the ECM organization. Depletion of
FN1 was reported to markedly reduce the invasive capacity of
prostate cancer cells (76).
Furthermore, increased expression of FN1 in tumors may alter the
primary tumor architecture, resulting in decreased metastasis
formation (36). Treatment of PC-3
cells with 1 µM FN1 was observed to result in a decrease in
activated ERK1/2 (77). CYR61, which
is also named cellular communication network factor 1 (CCN1), is an
ECM-associated matricellular protein and one of the six members of
the CCN family (78,79). It may impair fibroblast
responsiveness to TGF-β signaling and upregulation of matrix
metalloproteinase 1 (80). Fibrosis
is closely associated with IgAN (6,7).
Overall, the above indicates that the results of the functional
analysis of the identified DEGs in the present study are reasonable
and consistent with mechanisms identified by previous studies.
Several hub genes were identified from the PPI
network, and this result is consistent with previously described
genes, including ALB (31), FOS
(32) and TP53 (33). TP53/p53 is a known regulator of
apoptosis and macro-autophagy/autophagy (39). The coupled induction of inducible
nitric oxide synthase and upregulation of TP53 in intrinsic renal
cells of IgAN may be linked to pro- and anti-apoptotic activities
(33). The present analysis revealed
relatively few novel hub genes with a close association with the
pathological processes of IgAN, offering novel insight. ITGB2 is a
receptor of intercellular adhesion molecule (ICAM)1, ICAM2, ICAM3
and ICAM4, and it is also called CD18. ITGB2 is involved in
cellular adhesion and ECM remodeling in patients with renal cancer
(34). Furthermore, ITGB2 was
identified to be closely associated with apoptosis in patients with
Alzheimer's disease (81). However,
to the best of our knowledge, no previous study has reported on the
role of ITGB2 in IgAN. In the present study, ITGB2 was the
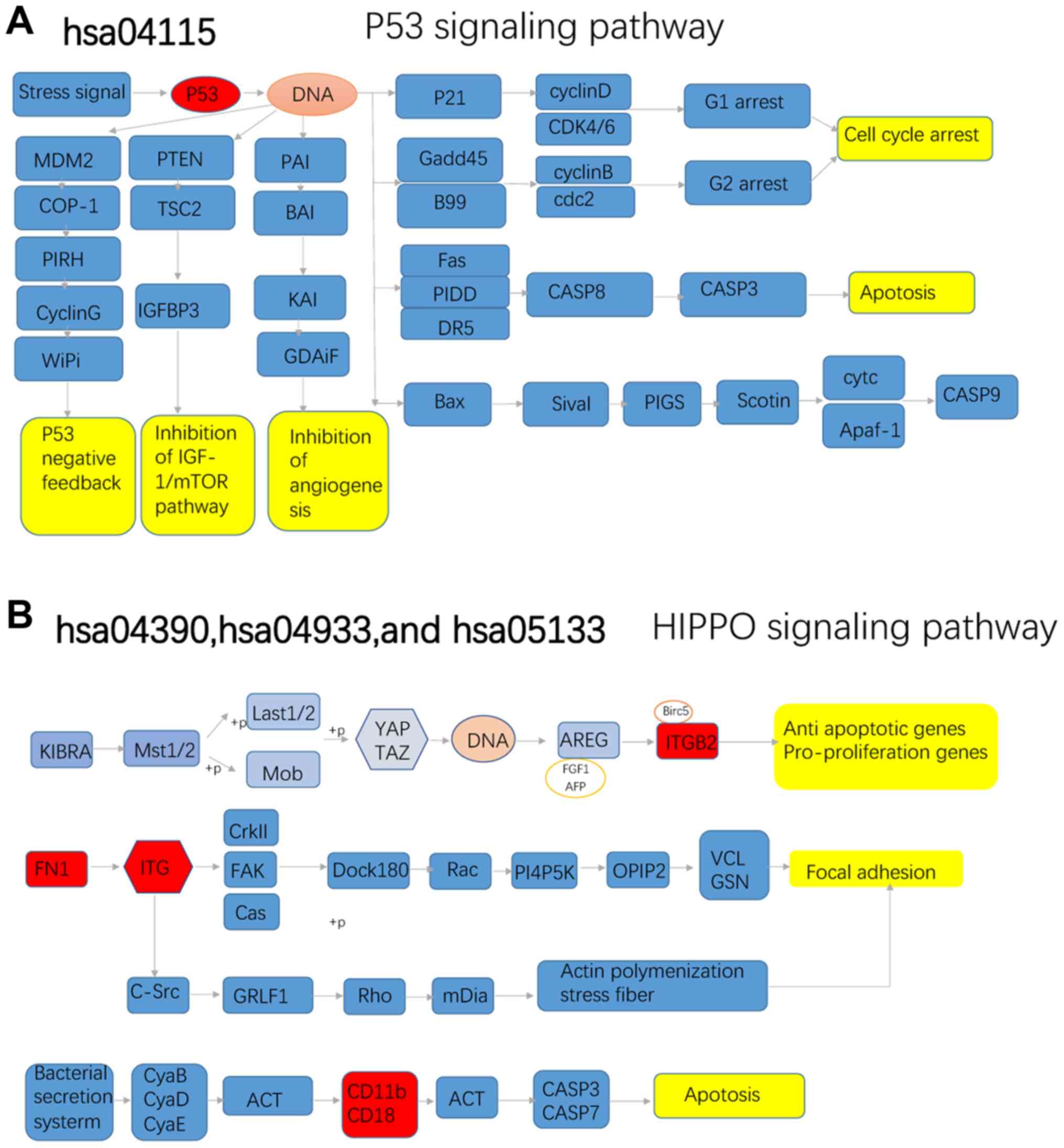
second-ranked hub gene in the PPI network. The KEGG analysis
results confirmed that ITGB2 was directly involved in apoptosis and
focal adhesion. Collectively, the novel hub gene ITGB2 was
indicated to have an important role in IgAN.
Another novel and noteworthy hub gene identified in
the present analysis is EGR1, a zinc finger transcription factor
with an essential role in cell growth and proliferation (65). EGR1 contributes to diabetic kidney
disease by enhancing epithelial-mesenchymal transition (65). Specific inhibition of EGR1 was
observed to prevent mesangial cell hypercellularity in experimental
nephritis (82). EGR1 overexpression
in rhabdomyosarcoma significantly decreases cell proliferation,
mobility and anchorage-independent growth (83). However, no previous study has
reported on the role of EGR1 in IgAN. In the present study, EGR1
was among the top 10 hub genes in the PPI network. The present
co-expression analysis indicated a close association between EGR1
and FOS. The expression levels of these two DEGs were decreased,
possibly leading to a reduction in the inhibition of cell
proliferation and resulting in the progression of IgAN.
The present study did not identify any direct
significant GO term for ‘fibrosis’. Renal biopsy in patients with
IgAN is generally performed in the early stages of the disease,
when renal fibrosis is not prominent. The GSE37460 dataset did not
provide any clinical information. However, the GSE93798 dataset
suggested that most of the patients' chronic kidney disease grades
were below 3a and the Oxford Classification scores were relatively
low, indicating that these patients were in the early stages of the
disease (20). Therefore, abnormal
expression of fibrosis-associated genes was not common in these
samples. However, DUSP1, a gene associated with fibrosis, was among
the DEGs. In chronic hypertension, angiotensin-1-7 increased DUSP1
to decrease fibrosis in resistance arterioles and attenuate
end-stage organ damage (84). In the
present study, the downregulation of DUSP1 may have acted as a
fibrotic factor and prompt the onset of fibrosis.
The present study was the first to identify novel
molecular targets by integrating all microarray datasets of IgAN in
GEO. Thereby, the sample size was expanded and further information
was obtained. The microarray matrix of the expression values was
combined and the batch effects were removed by using the empirical
Bayes method to make the data more comparable (85). The novel results may enhance the
current understanding of the molecular pathogenesis of IgAN.
However, the present study has certain limitations. First, the
clinical data from the GEO database were not available for each
sample. Furthermore, the array data came from typical IgAN in the
early stage, and therefore, the expression levels of certain genes
may not be identical to those in the later stage. For instance, the
protein levels of IGFBP1 have been reported to be upregulated in
this disease (86), while this gene
was downregulated in the present study. The cause of the
inconsistency may be that it is unrealistic to dynamically obtain
kidney tissue from a patient at different time-points. In addition,
the novel potential candidate targets should be further validated
in experimental studies. The present results were obtained using a
bioinformatics screening to identify several novel DEGs between
IgAN and healthy control samples, and suggested that part of the
top hub genes have vital roles in the pathological process of cell
proliferation in IgAN. Of note, the information provided in the
present study was not limited to the top 10 hub genes, but included
certain other representative DEGs. The present results provide a
valuable resource for future research on IgAN.
In conclusion, the present study was the first to
apply an integrated bioinformatics analysis to investigate novel
candidate genes and mechanisms involved in the pathogenesis of
IgAN. ITGB2, FN1, ATF3 and EGR1 genes may have important roles in
the development of IgAN and act as potential candidate molecular
targets for the diagnosis and treatment of IgAN.
Not applicable.
This work was supported by the National Natural
Science Foundation of China (grant no. 81270805) and the Science
and Technology Department of Sichuan Province (grant no.
2018SZ0378).
All data generated or analyzed during this study are
available from the corresponding author on reasonable request.
WT designed the experiments; WT and DZ analyzed the
data; YC, YZ, ZW and XM made substantial contributions to the
acquisition of data wrote the manuscript. All authors read and
approved the final manuscript.
This research used public microarray data from the
GEO database for bioinformatics analysis and did not involve any
human participants.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Wyatt RJ and Julian BA: IgA nephropathy. N
Engl J Med. 368:2402–2414. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rodrigues JC, Haas M and Reich HN: IgA
nephropathy. Clin J Am Soc Nephrol. 12:677–686. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schena FP: A retrospective analysis of the
natural history of primary IgA nephropathy worldwide. Am J Med.
89:209–215. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Yan X, Zhao T, Xu Q, Peng Q, Hu
R, Quan S, Zhou Y and Xing G: Targeting C3a/C5a receptors inhibits
human mesangial cell proliferation and alleviates immunoglobulin A
nephropathy in mice. Clin Exp Immunol. 189:60–70. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rops A, Jansen E, van der Schaaf A,
Pieterse E, Rother N, Hofstra J, Dijkman HBPM, van de Logt AE,
Wetzels J, van der Vlag J and van Spriel AB: Interleukin-6 is
essential for glomerular immunoglobulin A deposition and the
development of renal pathology in Cd37-deficient mice. Kidney Int.
93:1356–1366. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hennino MF, Buob D, Van der Hauwaert C,
Gnemmi V, Jomaa Z, Pottier N, Savary G, Drumez E, Noël C, Cauffiez
C and Glowacki F: miR-21-5p renal expression is associated with
fibrosis and renal survival in patients with IgA nephropathy. Sci
Rep. 6:272092016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tanaka K, Sugiyama H, Yamanari T, Mise K,
Morinaga H, Kitagawa M, Onishi A, Ogawa-Akiyama A, Tanabe K, Eguchi
J, et al: Renal expression of trefoil factor 3 mRNA in association
with tubulointerstitial fibrosis in IgA nephropathy. Nephrology
(Carlton). 23:855–862. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang S, Jin J, Lin B, Gong J, Li Y and He
Q: Rapamycin induces autophagy and reduces the apoptosis of
podocytes under a stimulated condition of immunoglobulin a
nephropathy. Kidney Blood Press Res. 42:177–187. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Leung JC, Chan LY, Saleem MA, Mathieson
PW, Tang SC and Lai KN: Combined blockade of angiotensin II and
prorenin receptors ameliorates podocytic apoptosis induced by
IgA-activated mesangial cells. Apoptosis. 20:907–920. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rauen T and Floege J: Inflammation in IgA
nephropathy. Pediatr Nephrol. 32:2215–2224. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bao H, Chen H, Zhu X, Zhang M, Yao G, Yu
Y, Qin W, Zeng C, Zen K and Liu Z: MiR-223 downregulation promotes
glomerular endothelial cell activation by upregulating importin α4
and α5 in IgA nephropathy. Kidney Int. 85:624–635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hodgin JB, Berthier CC, John R, Grone E,
Porubsky S, Gröne HJ, Herzenberg AM, Scholey JW, Hladunewich M,
Cattran DC, et al: The molecular phenotype of endocapillary
proliferation: Novel therapeutic targets for IgA nephropathy. PLoS
One. 9:e1034132014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Tan RZ, Chen Y, Wang HL, Liu YH,
Wen D and Fan JM: CagA promotes proliferation and secretion of
extracellular matrix by inhibiting signaling pathway of apoptosis
in rat glomerular mesangial cells. Ren Fail. 38:458–464. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui H, Zhang Y, Zhang Q, Chen W, Zhao H
and Liang J: A comprehensive genome-wide analysis of long noncoding
RNA expression profile in hepatocellular carcinoma. Cancer Med.
6:2932–2941. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian W, Liu J, Pei B, Wang X, Guo Y and
Yuan L: Identification of miRNAs and differentially expressed genes
in early phase non-small cell lung cancer. Oncol Rep. 35:2171–2176.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong B, Wang G, Yao J, Yuan P, Kang W, Zhi
L and He X: Predicting novel genes and pathways associated with
osteosarcoma by using bioinformatics analysis. Gene. 628:32–37.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu D: LYN, a key gene from bioinformatics
analysis, contributes to development and progression of esophageal
adenocarcinoma. Med Sci Monit Basic Res. 21:253–261. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou LT, Qiu S, Lv LL, Li ZL, Liu H, Tang
RN, Ma KL and Liu BC: Integrative bioinformatics analysis provides
insight into the molecular mechanisms of chronic kidney disease.
Kidney Blood Press Res. 43:568–581. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shu B, Fang Y, He W, Yang J and Dai C:
Identification of macrophage-related candidate genes in lupus
nephritis using bioinformatics analysis. Cell Signal. 46:43–51.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu P, Lassen E, Nair V, Berthier CC,
Suguro M, Sihlbom C, Kretzler M, Betsholtz C, Haraldsson B, Ju W,
et al: Transcriptomic and proteomic profiling provides insight into
mesangial cell function in IgA nephropathy. J Am Soc Nephrol.
28:2961–2972. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Berthier CC, Bethunaickan R,
Gonzalez-Rivera T, Nair V, Ramanujam M, Zhang W, Bottinger EP,
Segerer S, Lindenmeyer M, Cohen CD, et al: Cross-species
transcriptional network analysis defines shared inflammatory
responses in murine and human lupus nephritis. J Immunol.
189:988–1001. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shved N, Warsow G, Eichinger F, Hoogewijs
D, Brandt S, Wild P, Kretzler M, Cohen CD and Lindenmeyer MT:
Transcriptome-based network analysis reveals renal cell
type-specific dysregulation of hypoxia-associated transcripts. Sci
Rep. 7:85762017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Forero DA, Guio-Vega GP and
Gonzalez-Giraldo Y: A comprehensive regional analysis of
genome-wide expression profiles for major depressive disorder. J
Affect Disord. 218:86–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Leek JT, Johnson WE, Parker HS, Jaffe AE
and Storey JD: The sva package for removing batch effects and other
unwanted variation in high-throughput experiments. Bioinformatics.
28:882–883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kawai Y, Masutani K, Torisu K, Katafuchi
R, Tanaka S, Tsuchimoto A, Mitsuiki K, Tsuruya K and Kitazono T:
Association between serum albumin level and incidence of end-stage
renal disease in patients with immunoglobulin A nephropathy: A
possible role of albumin as an antioxidant agent. PLoS One.
13:e01966552018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Park HJ, Kim JW, Cho BS and Chung JH:
Association of FOS-like antigen 1 promoter polymorphism with
podocyte foot process effacement in immunoglobulin A nephropathy
patients. J Clin Lab Anal. 28:391–397. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiu LQ, Sinniah R and Hsu SI: Coupled
induction of iNOS and p53 upregulation in renal resident cells may
be linked with apoptotic activity in the pathogenesis of
progressive IgA nephropathy. J Am Soc Nephrol. 15:2066–2078. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Boguslawska J, Kedzierska H, Poplawski P,
Rybicka B, Tanski Z and Piekielko-Witkowska A: Expression of genes
involved in cellular adhesion and extracellular matrix remodeling
correlates with poor survival of patients with renal cancer. J
Urol. 195:1892–1902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livingston MJ, Ding HF, Huang S, Hill JA,
Yin XM and Dong Z: Persistent activation of autophagy in kidney
tubular cells promotes renal interstitial fibrosis during
unilateral ureteral obstruction. Autophagy. 12:976–998. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Glasner A, Levi A, Enk J, Isaacson B,
Viukov S, Orlanski S, Scope A, Neuman T, Enk CD, Hanna JH, et al:
NKp46 receptor-mediated interferon-γ production by natural killer
cells increases fibronectin 1 to alter tumor architecture and
control metastasis. Immunity. 48:107–119.e4. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jovanovic KK, Escure G, Demonchy J,
Willaume A, Van de Wyngaert Z, Farhat M, Chauvet P, Facon T,
Quesnel B and Manier S: Deregulation and targeting of TP53 pathway
in multiple myeloma. Front Oncol. 8:6652019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yogosawa S and Yoshida K: Tumor
suppressive role for kinases phosphorylating p53 in DNA
damage-induced apoptosis. Cancer Sci. 109:3376–3382. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang SZ, Qiu XJ, Dong SS, Zhou LN, Zhu Y,
Wang MD and Jin LW: MicroRNA-770-5p is involved in the development
of diabetic nephropathy through regulating podocyte apoptosis by
targeting TP53 regulated inhibitor of apoptosis 1. Eur Rev Med
Pharmacol Sci. 23:1248–1256. 2019.PubMed/NCBI
|
|
40
|
Chen SN, Lombardi R, Karmouch J, Tsai JY,
Czernuszewicz G, Taylor MRG, Mestroni L, Coarfa C, Gurha P and
Marian AJ: DNA damage Response/TP53 pathway is activated and
contributes to the pathogenesis of dilated cardiomyopathy
associated With LMNA (Lamin A/C) mutations. Circ Res. 124:856–873.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu GC, Zhou YF, Su XC and Zhang J:
Interaction between TP53 and XRCC1 increases susceptibility to
cervical cancer development: A case control study. BMC Cancer.
19:242019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Robin M, Issa AR, Santos CC, Napoletano F,
Petitgas C, Chatelain G, Ruby M, Walter L, Birman S, Domingos PM,
et al: Drosophila p53 integrates the antagonism between autophagy
and apoptosis in response to stress. Autophagy. 15:771–784. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dai J, Xu LJ, Han GD, Jiang HT, Sun HL,
Zhu GT and Tang XM: Down-regulation of long non-coding RNA
ITGB2-AS1 inhibits osteosarcoma proliferation and metastasis by
repressing Wnt/β-catenin signalling and predicts favourable
prognosis. Artif Cells Nanomed Biotechnol. 46 (Suppl 3):S783–S790.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu H, Dai X, Cao X, Yan H, Ji X, Zhang H,
Shen S, Si Y, Zhang H, Chen J, et al: PRDM4 mediates YAP-induced
cell invasion by activating leukocyte-specific integrin β2
expression. EMBO Rep. 19(pii): e451802018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Aziz MH, Cui K, Das M, Brown KE, Ardell
CL, Febbraio M, Pluskota E, Han J, Wu H, Ballantyne CM, et al: The
upregulation of integrin αDβ2 (CD11d/CD18) on inflammatory
macrophages promotes macrophage retention in vascular lesions and
development of atherosclerosis. J Immunol. 198:4855–4867. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kijas JM, Bauer TR Jr, Gafvert S, Marklund
S, Trowald-Wigh G, Johannisson A, Hedhammar A, Binns M, Juneja RK,
Hickstein DD and Andersson L: A missense mutation in the beta-2
integrin gene (ITGB2) causes canine leukocyte adhesion deficiency.
Genomics. 61:101–107. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yee NK and Hamerman JA: β(2) integrins
inhibit TLR responses by regulating NF-κB pathway and p38 MAPK
activation. Eur J Immunol. 43:779–792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Duan J, Zhang X, Zhang S, Hua S and Feng
Z: miR-206 inhibits FN1 expression and proliferation and promotes
apoptosis of rat type II alveolar epithelial cells. Exp Ther Med.
13:3203–3208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang J, Deng L, Huang J, Cai R, Zhu X, Liu
F, Wang Q, Zhang J and Zheng Y: High expression of Fibronectin 1
suppresses apoptosis through the NF-κB pathway and is associated
with migration in nasopharyngeal carcinoma. Am J Transl Res.
9:4502–4511. 2017.PubMed/NCBI
|
|
50
|
Liu C, Feng Z, Chen T, Lv J, Liu P, Jia L,
Zhu J, Chen F, Yang C and Deng Z: Downregulation of NEAT1 reverses
the radioactive iodine resistance of papillary thyroid carcinoma
cell via miR-101-3p/FN1/PI3K-AKT signaling pathway. Cell Cycle.
18:167–203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liao YX, Zhang ZP, Zhao J and Liu JP:
Effects of Fibronectin 1 on cell proliferation, senescence and
apoptosis of human glioma cells through the PI3K/AKT signaling
pathway. Cell Physiol Biochem. 48:1382–1396. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bakiri L, Hamacher R, Grana O,
Guío-Carrión A, Campos-Olivas R, Martinez L, Dienes HP, Thomsen MK,
Hasenfuss SC and Wagner EF: Liver carcinogenesis by FOS-dependent
inflammation and cholesterol dysregulation. J Exp Med.
214:1387–1409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xu X, Kwon OK, Shin IS, Mali JR, Harmalkar
DS, Lim Y, Lee G, Lu Q, Oh SR, Ahn KS, et al: Novel benzofuran
derivative DK-1014 attenuates lung inflammation via blocking of
MAPK/AP-1 and AKT/mTOR signaling in vitro and in vivo. Sci Rep.
9:8622019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen M, Li X, Shi Q, Zhang Z and Xu S:
Hydrogen sulfide exposure triggers chicken trachea inflammatory
injury through oxidative stress-mediated FOS/IL8 signaling. J
Hazard Mater. 368:243–254. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nagesh R, Kiran Kumar KM, Naveen Kumar M,
Patil RH and Sharma SC: Stress activated p38 MAPK regulates cell
cycle via AP-1 factors in areca extract exposed human lung
epithelial cells. Cytotechnology. 71:507–520. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wu DM, Zhang YT, Lu J and Zheng YL:
Effects of microRNA-129 and its target gene c-Fos on proliferation
and apoptosis of hippocampal neurons in rats with epilepsy via the
MAPK signaling pathway. J Cell Physiol. 233:6632–6643. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu C, Ding L, Bai L, Chen X, Kang H, Hou
L and Wang J: Folate receptor alpha is associated with cervical
carcinogenesis and regulates cervical cancer cells growth by
activating ERK1/2/c-Fos/c-Jun. Biochem Biophys Res Commun.
491:1083–1091. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hwang MS, Strainic MG, Pohlmann E, Kim H,
Pluskota E, Ramirez-Bergeron DL, Plow EF and Medof ME: VEGFR2
survival and mitotic signaling depends on joint-activation of
associated C3ar1/C5ar1 and IL-6R-gp130. J Cell Sci. 132(pii):
jcs2193522019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Mathern DR, K Horwitz J and Heeger PS:
Absence of recipient C3aR1 signaling limits expansion and
differentiation of alloreactive CD8+ T cell immunity and
prolongs murine cardiac allograft survival. Am J Transplant.
198:1628–1640. 2019. View Article : Google Scholar
|
|
60
|
Matsumoto N, Satyam A, Geha M, Lapchak PH,
Dalle Lucca JJ, Tsokos MG and Tsokos GC: C3a enhances the formation
of intestinal organoids through C3aR1. Front Immunol. 8:10462017.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lokman FE, Seman NA, Ismail AA, Yaacob NA,
Mustafa N, Khir AS, Hussein Z and Wan Mohamud WN: Gene expression
profiling in ethnic Malays with type 2 diabetes mellitus, with and
without diabetic nephropathy. J Nephrol. 24:778–789. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang D, Guan MP, Zheng ZJ, Li WQ, Lyv FP,
Pang RY and Xue YM: Transcription factor Egr1 is involved in high
glucose-induced proliferation and fibrosis in rat glomerular
mesangial cells. Cell Physiol Biochem. 36:2093–2107. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu F, Zhang ZP, Xin GD, Guo LH, Jiang Q
and Wang ZX: miR-192 prevents renal tubulointerstitial fibrosis in
diabetic nephropathy by targeting Egr1. Eur Rev Med Pharmacol Sci.
22:4252–4260. 2018.PubMed/NCBI
|
|
64
|
Wu C, Ma X, Zhou Y, Liu Y, Shao Y and Wang
Q: Klotho restraining Egr1/TLR4/mTOR axis to reducing the
expression of fibrosis and inflammatory cytokines in high glucose
cultured rat mesangial cells. Exp Clin Endocrinol Diabetes. Jun
11–2018.(Epub ahead of print). doi: 10.1055/s-0044-101601.
|
|
65
|
Hu F, Xue M, Li Y, Jia YJ, Zheng ZJ, Yang
YL, Guan MP, Sun L and Xue YM: Early growth response 1 (Egr1) is a
transcriptional activator of NOX4 in oxidative stress of diabetic
kidney disease. J Diabetes Res. 2018:34056952018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li Y, Hu F, Xue M, Jia YJ, Zheng ZJ, Wang
L, Guan MP and Xue YM: Klotho down-regulates Egr-1 by inhibiting
TGF-β1/Smad3 signaling in high glucose treated human mesangial
cells. Biochem Biophys Res Commun. 487:216–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wu SY, Rupaimoole R, Shen F, Pradeep S,
Pecot CV, Ivan C, Nagaraja AS, Gharpure KM, Pham E, Hatakeyama H,
et al: A miR-192-EGR1-HOXB9 regulatory network controls the
angiogenic switch in cancer. Nat Commun. 7:111692016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sun S, Ning X, Zhai Y, Du R, Lu Y, He L,
Li R, Wu W, Sun W and Wang H: Egr-1 mediates chronic
hypoxia-induced renal interstitial fibrosis via the PKC/ERK
pathway. Am J Nephrol. 39:436–448. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Lin CY, Lin TY, Lee MC, Chen SC and Chang
JS: Hyperglycemia: GDNF-EGR1 pathway target renal epithelial cell
migration and apoptosis in diabetic renal embryopathy. PLoS One.
8:e567312013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Singh R, Yadav V, Kumar S and Saini N:
MicroRNA-195 inhibits proliferation, invasion and metastasis in
breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1.
Sci Rep. 5:174542015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Banks M and Holick MF: Molecular
mechanism(s) involved in 25-hydroxyvitamin D's antiproliferative
effects in CYP27B1-transfected LNCaP cells. Anticancer Res.
35:3773–3779. 2015.PubMed/NCBI
|
|
72
|
Li G, Wu F, Yang H, Deng X and Yuan Y:
MiR-9-5p promotes cell growth and metastasis in non-small cell lung
cancer through the repression of TGFBR2. Biomed Pharmacother.
96:1170–1178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ebefors K, Liu P, Lassén E, Elvin J,
Candemark E, Levan K, Haraldsson B and Nyström J: Mesangial cells
from patients with IgA nephropathy have increased susceptibility to
galactose-deficient IgA1. BMC Nephrol. 17:402016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
You Z, Xu J, Li B, Ye H, Chen L, Liu Y and
Xiong X: The mechanism of ATF3 repression of epithelial-mesenchymal
transition and suppression of cell viability in cholangiocarcinoma
via p53 signal pathway. J Cell Mol Med. 23:2184–2193. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Hsu YJ, Hsu SC, Chang YL, Huang SM, Shih
CC, Tsai CS and Lin CY: Indoxyl sulfate upregulates the cannabinoid
type 1 receptor gene via an ATF3/c-Jun complex-mediated signaling
pathway in the model of uremic cardiomyopathy. Int J Cardiol.
252:128–135. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Armstrong HK, Gillis JL, Johnson IRD,
Nassar ZD, Moldovan M, Levrier C, Sadowski MC, Chin MY, Tomlinson
Guns ES, Tarulli G, et al: Dysregulated fibronectin trafficking by
Hsp90 inhibition restricts prostate cancer cell invasion. Sci Rep.
8:20902018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Wei X, Zhou D, Wang H, Ding N, Cui XX,
Wang H, Verano M, Zhang K, Conney AH, Zheng X and DU ZY: Effects of
pyridine analogs of curcumin on growth, apoptosis and NF-κB
activity in prostate cancer PC-3 cells. Anticancer Res.
33:1343–1350. 2013.PubMed/NCBI
|
|
78
|
Bork P: The modular architecture of a new
family of growth regulators related to connective tissue growth
factor. FEBS Lett. 327:125–130. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chen CC and Lau LF: Functions and
mechanisms of action of CCN matricellular proteins. Int J Biochem
Cell Biol. 41:771–783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Quan T, He T, Shao Y, Lin L, Kang S,
Voorhees JJ and Fisher GJ: Elevated cysteine-rich 61 mediates
aberrant collagen homeostasis in chronologically aged and photoaged
human skin. Am J Pathol. 169:482–490. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Mizwicki MT, Liu G, Fiala M, Magpantay L,
Sayre J, Siani A, Mahanian M, Weitzman R, Hayden EY, Rosenthal MJ,
et al: 1α,25-dihydroxyvitamin D3 and resolvin D1 retune the balance
between amyloid-β phagocytosis and inflammation in Alzheimer's
disease patients. J Alzheimers Dis. 34:155–170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Carl M, Akagi Y, Weidner S, Isaka Y, Imai
E and Rupprecht HD: Specific inhibition of Egr-1 prevents mesangial
cell hypercellularity in experimental nephritis. Kidney Int.
63:1302–1312. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Mohamad T, Kazim N, Adhikari A and Davie
JK: EGR1 interacts with TBX2 and functions as a tumor suppressor in
rhabdomyosarcoma. Oncotarget. 9:18084–18098. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Carver KA, Smith TL, Gallagher PE and
Tallant EA: Angiotensin-(1–7) prevents angiotensin II-induced
fibrosis in cremaster microvessels. Microcirculation. 22:19–27.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Heider A and Alt R: virtualArray: A
R/bioconductor package to merge raw data from different microarray
platforms. BMC Bioinformatics. 14:752013. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Tokunaga K, Uto H, Takami Y, Mera K,
Nishida C, Yoshimine Y, Fukumoto M, Oku M, Sogabe A, Nosaki T, et
al: Insulin-like growth factor binding protein-1 levels are
increased in patients with IgA nephropathy. Biochem Biophys Res
Commun. 399:144–149. 2010. View Article : Google Scholar : PubMed/NCBI
|