Introduction
Gastric cancer (GC) is one of the most common
malignant tumors of the digestive tract in the world (1). Despite the continuous development of
novel treatment options for GC, the prognosis of the disease
remains poor (1–3). Many patients with GC exhibit tumor
metastasis upon diagnosis, making treatment very difficult
(4,5). Therefore, further elucidating the
molecular mechanism of GC to determine early diagnostic markers,
therapeutic targets, novel molecular targeted therapeutic drugs and
indicators of metastasis and recurrence is of great importance in
GC.
MicroRNAs (miRNAs) are a type of non-coding small
RNA containing 20–24 nucleotides that regulate the expression of
their target genes primarily at the post-transcriptional level
(6–8). Thereby, miRNAs participate in a variety
of cell functions including cell proliferation, apoptosis,
differentiation, metabolism and endocrine system regulation
(6,7,9). The
abnormal expression of miRNA is associated with a variety of
diseases including tumors (7). miRNA
has been demonstrated to be involved in the regulation of tumor
cell proliferation, apoptosis, differentiation, drug resistance,
invasion and metastasis, among which miR-150 also ion, drug
resistance, invasion and metastasis, among which miR-150 also
serves an important role in the development of tumors (10–15). In
colorectal cancer, breast cancer and melanoma, miR-150 is
significantly downregulated and exerts a tumor suppressive role
(11–13). However, in cervical cancer, miR-150
is highly expressed and promotes tumor progression (14). Therefore, miR-150 serves different
roles in different tumor types (10–14).
miR-150-5p, a member of the miR-150 family, has been determined to
inhibit cancer cell aggressiveness by targeting cwcv and kazal like
domains proteoglycan 1 in head and neck squamous cell carcinoma
(15). miR-150-5p affects cell
proliferation, apoptosis and epithelial-mesenchymal transition
(EMT) by regulating the BRAFV600E mutation in papillary thyroid
cancer cells (16). miR-150-5p has
also been identified as a novel prognostic biomarker in non-small
cell lung cancer (17). Yu et
al (18) hypothesized that the
aryl hydrocarbon receptor enhanced the expression of miR-150-5p to
suppress cell proliferation and invasion in prostate cancer by
regulating MAP3K12. Furthermore, Suetsugu et al (19) revealed that miR-150-5p inhibits the
aggressiveness of lung squamous cell carcinoma cells. These results
indicate the important roles of miR-150-5p in tumor progression.
However, to the best of our knowledge, the role of miR-150-5p in
the pathological development of GC remains largely unknown.
EMT is an important component of wound healing and
stem cell behavior (20), which may
develop fibrosis and tumor progression under pathological
conditions (20–22). In addition, tumor cells can obtain
transfer force and invasiveness via certain signaling pathways,
which include the TGF-β/Smad pathway (23). For example, the reorganization of the
cytoskeleton and prominent membrane behavior are all key features
of tumor invasion (23,24). In order to leave the primary tumor
and invade surrounding tissue, tumor cells must destroy contact
between cells, recombine the attachment site of the cell-matrix and
generate chemotaxis under the guidance of the extracellular matrix
(23,24). In EMT, polarized epithelial cells,
which originally exhibit no activity, dissolve their own
intercellular junctions into independent, depolarizing and active
metastatic mesenchymal cells (22,24). For
example, the expression and function of E-cadherin, which supports
epithelial cell junctions, disappear, while N-cadherin, which
supports mesenchymal cell-cell attachment, is induced (21). The expression of N-cadherin triggers
cell metastasis and invasion. Vimentin contributes to EMT cancer
cell mechanics by mediating cytoskeletal organization and focal
adhesion maturation (25). Highly
and abnormally expressed β-catenin, a multifunctional protein that
serves an important role in physiological homeostasis, results in
various diseases including cancer, which can serve as
transcriptional co-regulators and as adaptor proteins for
intracellular adhesion (26). Zonula
occluden-1 (ZO-1) is a tight junction-associated protein involved
in the maintenance and regulation of epithelial barrier function
and is often utilized as an indicator of tight junctional barrier
function and permeability of various tissues (27). To the best of our knowledge, the
effect of miR-150-5p on GC EMT, particularly on the expression of
interstitial cell markers (vimentin, N-cadherin and β-catenin) and
epithelial cell markers (E-cadherin and ZO-1) in GC cells remain
unclear.
The current study aimed to assess the role and
regulation mechanism of miR-150-5p in the pathological process of
GC.
Materials and methods
Clinical samples
A total of 36 GC tissue and paired normal tissue
samples (2 cm from the tumor lesion) were collected from patients
with GC (age range, 43–67 years; male:female, 1:1; Stage I, 12
cases; Stage II, 12 cases; Stage III–IV, 12 cases; lymph node
metastases, 18 cases; no lymph node metastasis, 18 cases) who had
undergone surgical resection at Zhuzhou Central Hospital (Zhuzhou,
China) from March 2015 to March 2017. None of the patients received
any radiotherapy or chemotherapy prior to the surgery. The present
study was approved by the Ethical Committee of the Zhuzhou Central
Hospital (Zhuzhou, China) and written informed consent was obtained
from each patient.
Cell culture
The GC cell lines MGC-803, SGC-7901, BGC-823, and
the normal gastric epithelial cell line GES-1 (all Shanghai Guandao
Bio-engineering Co., Ltd., Shanghai, China) were cultured in
Dulbecco's modified Eagle medium or RPMI-1640 (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Sigma-Aldrich; Merck KGaA; Darmstadt,
Germany). Cells were incubated in a humidified incubator at 37°C
with 5% CO2.
Dual luciferase reporter assay
TargetScan bioinformatics software (www.targetscan.org/vert_71) was used to predict
the targets of miR-150-5p. It was revealed that SRC kinase
signaling inhibitor 1 (SRCIN1) was a potential target of
miR-150-5p. To confirm this prediction, wild type (WT-SRCIN1) and
mutant (MUT-SRCIN1) 3′-untranslated regions of SRCIN1 were cloned
into a pmiR-RB-ReportTM dual luciferase reporter gene plasmid
vector (Guangzhou RiboBio Co., Ltd., Guangzhou, China). BGC-823
cells were co-transfected with WT-SRCIN1 or MUT-SRCIN1 and
miR-150-5p mimic (5′-UCUCCCAACCCUUGUACCAGUG-3′) or mimic control
(5′-UUCUCCGAACGUGUCACGUTT-3′) using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. miR-150-5p mimic and mimic control were
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China).
Following 48-h incubation, luciferase activity was assessed using
the dual-luciferase assay system (Promega Corporation, Madison, WI,
USA), according to the manufacturer's protocol. Firefly luciferase
activity was normalized to Renilla luciferase activity.
Cell transfection
An miR-150-5p inhibitor and its control (inhibitor
control) were synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). BGC-823 cells were seeded into 6-well plates
(1×106 cells per well) and cultured at 37°C for 24 h.
Cells were transfected with 100 nM miR-150-5p inhibitor
(5′-CACUGGUACAAGGGUUGGGAGA-3′), 100 nM inhibitor control
(5′-CAGUACUUUUGUGUAGUACAA-3′), 10 µM control-siRNA (cat no.
abx941273), 10 µM SRCIN1-siRNA (cat no. abx905269; both Abbexa,
Ltd.) or miR-150-5p inhibitor+SRCIN1-siRNA using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol.
Transfection efficiency was detected following 48-h transfection
via reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) or/and western blotting. Cells without any treatment were
used as the control.
Cell counting kit (CCK)-8 assay
The current study utilized the CCK8 method to detect
cell viability. Following 48-h transfection, BGC-823 cell
suspension was adjusted to 1×104 cells/ml and 100 µl
cell suspension was added to each well of a 96-well plate. Cells
were cultured at 37°C with 5% CO2 for 24 h.
Subsequently, 10 µl CCK-8 reagent (Sigma-Aldrich; Merck KGaA) was
added to each well. Absorbance at the wavelength of 450 nm was
measured using an automatic enzyme-linked immune detector following
2 h of incubation at 37°C. The experiment was repeated three
times.
Flow cytometry
Following 48-h transfection, BGC-823 cells were
digested with 0.2% trypsin, washed with PBS and fixed with 70%
ethanol overnight at 4°C. The apoptosis condition of cells was
detected using the annexin V-Fluorescein isothiocyanate
(FITC)/propidium iodide (PI) kit (cat no. 70-AP101-100; Hangzhou
MultiSciences (Lianke) Biotech Co., Ltd., Hangzhou, China)
according to the manufacturer's protocol. Cell apoptosis rate was
measured using a FACS Calibur flow cytometer with FlowJo software
(version 7.6.1; FlowJo LLC, Ashland, OR, USA). Cells without any
treatment were used as the control group. The assay was performed
in triplicate.
Western blotting
Cells (MGC-803, SGC-7901, BGC-823 and GES-1) were
washed with ice cold PBS. Total protein was extracted from cells
using radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Shanghai, China) at 4°C for 1 h. Protein samples
were collected via centrifugation at a speed of 12,000 × g for 5
min at 4°C. Total protein was quantified using a bicinchoninic acid
protein assay kit (Beyotime Institute of Biotechnology) and 30 µg
protein/lane was separated via SDS PAGE on a 10% gel. The separated
proteins were transferred onto polyvinylidene difluoride membranes
and blocked with 5% non-fat milk at room temperature for 2 h. The
membranes were incubated with primary antibodies against SRCIN1
(1:1,000; cat no. 3757), N-cadherin (1:1,000; cat no. 13116),
vimentin (1:1,000; cat no. 12,826), β-catenin (1:1,000; cat no.
25362), E-cadherin (1:1,000; cat no. 3195), ZO-1 (1:1,000; cat no.
13663) and β-actin (1:1,000; cat no. 4970; all Cell Signaling
Technology, Inc., Danvers, MA, USA) overnight at 4°C. Membranes
were washed four times with PBST. Following primary incubation,
membranes were incubated with horseradish peroxidase-conjugated
anti-rabbit Immunoglobulin G secondary antibody (cat no. 7074;
1:2,000; Cell Signaling Technology, Inc.) for 2 h at room
temperature. Membranes were washed four times with PBST. Protein
bands were visualized using an ECL reagent (EMD Millipore,
Billerica, MA, USA). Protein expression was quantified using
AlphaView 3.4.0 software (ProteinSimple, San Jose, CA, USA).
RNA isolation and RT-qPCR
Total RNA was extracted from tissue samples and
cells (MGC-803, SGC-7901, BGC-823 and GES-1) using the
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA concentration was detected using NanoDrop™ 2000
spectrophotometer (Thermo Fisher Scientific, Inc.) and samples were
stored at −80°C for further use. Total RNA was reverse transcribed
into cDNA using the miScript Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. qPCR was subsequently performed using
QuantiFast SYBR Green PCR kit (Takara Bio, Inc., Otsu, Japan) using
a CFX Connect Real-Time System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The primer sequences for qPCR were as follows:
miR-150-5p forward, 5′-TCGGCGTCTCCCAACCCTTGTAC-3′ and reverse,
5′-GTCGTATCCAGTGCAGGGTCCGAGGT-3′; SRCIN1 forward,
5′-AGCCCCGACAAAAGCAAAC-3′ and reverse,
5′-CCAAAGGAAGTCAATACAGGGATAG-3′; N-cadherin forward,
5′-TTTGATGGAGGTCTCCTAACACC-3′ and reverse,
5′-ACGTTTAACACGTTGGAAATGTG-3′; vimentin forward,
5′-GACGCCATCAACACCGAGTT-3′ and reverse,
5′-CTTTGTCGTTGGTTAGCTGGT-3′; β-catenin forward,
5′-AACAGGGTCTGGGACATTAGTC-3′ and reverse,
5′-CGAAAGCCAATCAAACACAAAC-3′; E-cadherin forward,
5′-CGAGAGCTACACGTTCACGG-3′ and reverse,
5′-GGGTGTCGAGGGAAAAATAGG-3′; ZO-1 forward,
5′-CCTCTGATCATTCCACACAGTC-3′ and reverse,
5′-TAGACATGCGCTCTTCCTCTCT-3′; GAPDH forward,
5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse,
5′-GTAGAGGCAGGGATGATGTTCT-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 10 min; 35 cycles of 95°C
for 15 sec and 55°C for 40 sec. Relative gene expression was
calculated using the 2−ΔΔCq method (28) and U6 and GAPDH served as internal
controls for miRNA and mRNA expression, respectively.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). Data
are presented as the mean ± standard deviation. One-way analysis of
variance followed by a Tukey's post-hoc test was used analyze
differences among multiple groups and an unpaired or paired
Student's t-test was used to analyze the statistical significance
between two groups. P<0.05 was considered to indicate a
statistically significant result.
Results
miR-150-5p is highly expressed in GC
tissues and cells and is associated with advanced GC and lymph node
metastasis
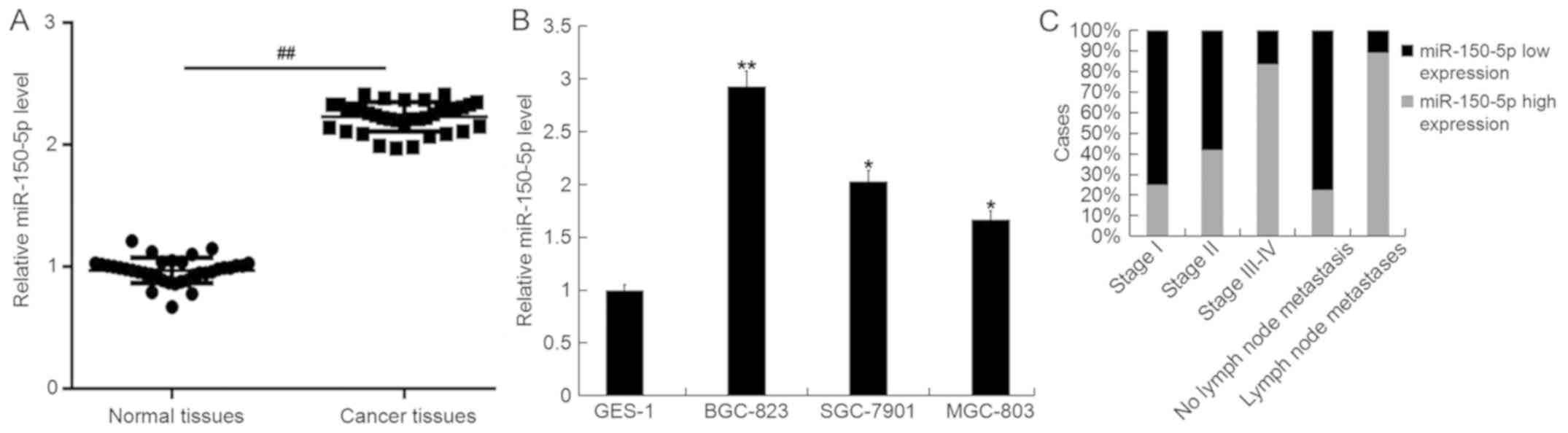
The level of miR-150-5p in GC tissues, paired normal
tissues, GC cell lines (MGC-803, SGC-7901 and BGC-823) and the
normal gastric epithelial cell line (GES-1) were detected using
RT-qPCR. The results demonstrated that compared with normal
tissues, miR-150-5p was upregulated in GC tissue (Fig. 1A). Additionally, compared with GES-1
cells, miR-150-5p was significantly upregulated in all GC cell
lines and was most abundantly expressed in BGC-823 cells (Fig. 1B). Furthermore, it was revealed that
higher levels of miR-150-5p were exhibited in patients with
advanced GC, with evidence of lymph node metastases (Fig. 1C). The expression of miR-150-5p
gradually increased from Stage I, Stage II, Stage III–IV, non-lymph
node metastasis and lymph node metastasis.
SRCIN1 is a target of miR-150-5p,
which is lowly expressed in GC tissues and cells
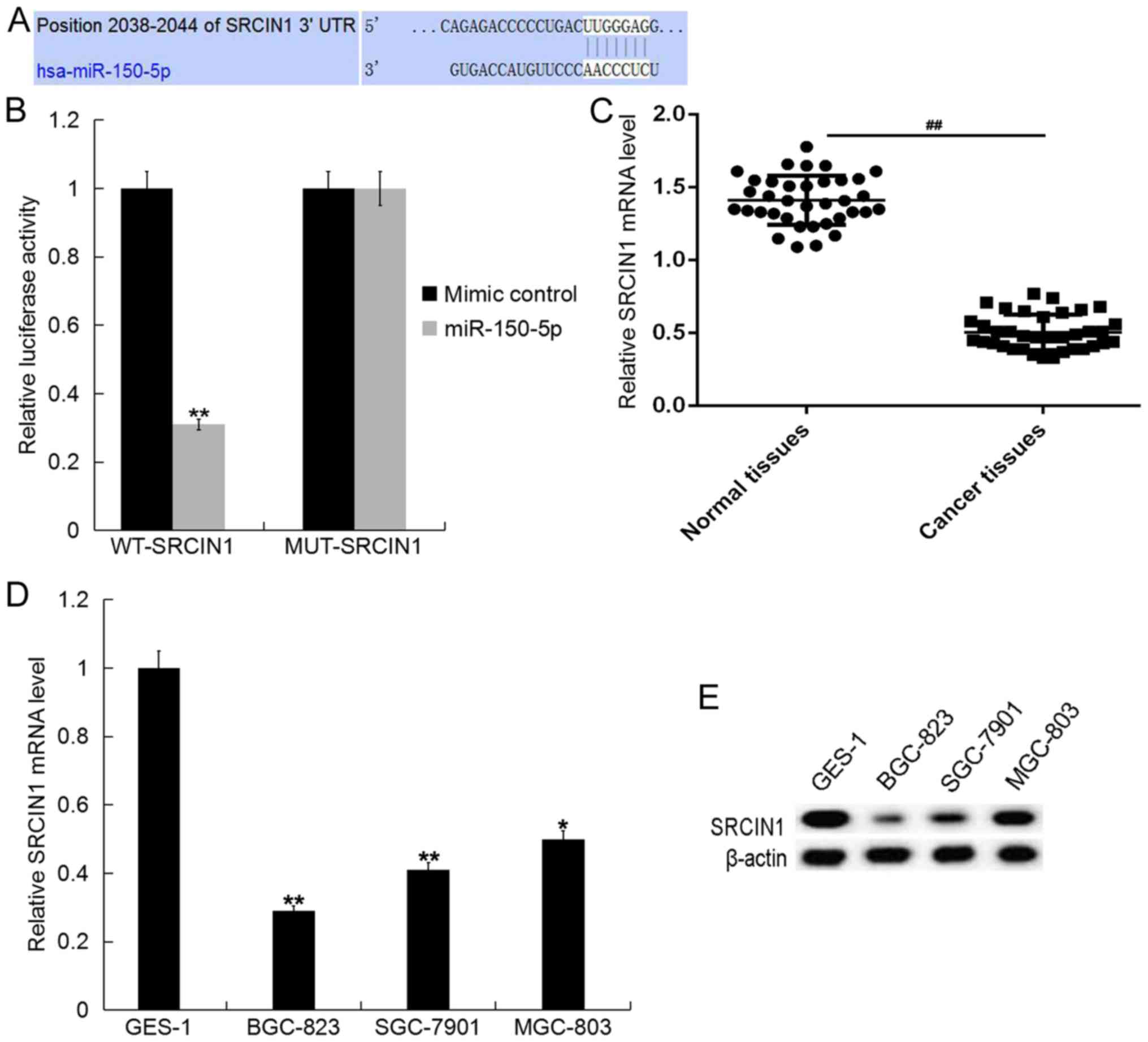
To assess the target genes of miR-150-5p, the
current study utilized TargetScan (www.targetscan.org/vert_71). The results demonstrated
the existence of binding sites between SRCIN1 and miR-150-5p
(Fig. 2A). Additionally, to reveal
whether miR-150-5p directly binds to SRCIN1, a dual luciferase
reporter assay was performed. The miR-150-5p-SRCIN1-WT or
miR-150-5p-SRCIN1-MUT reporter plasmids were co-transfected into
BGC-823 cells with miR-150-5p mimics or mimic controls. The results
revealed that luciferase activity significantly decreased in the
BGC-823 cells co-transfected with miR-150-5p mimic and
miR-150-5p-SRCIN1-WT, but not with miR-150-5p-SRCIN1-MUT (Fig. 2B). The data indicates that SRCIN1 is
a target gene of miR-150-5p.
It was also demonstrated that the mRNA level of
SRCIN1 in GC tissues was lower than that of normal tissues
(Fig. 2C). SRCIN1 was also
downregulated in all GC cell lines of the current study when
compared with GES-1 cells and the expression was lowest in BGC-823
cells (Fig. 2D and E).
miR-150-5p inhibitor suppresses GC
cell viability and induces apoptosis
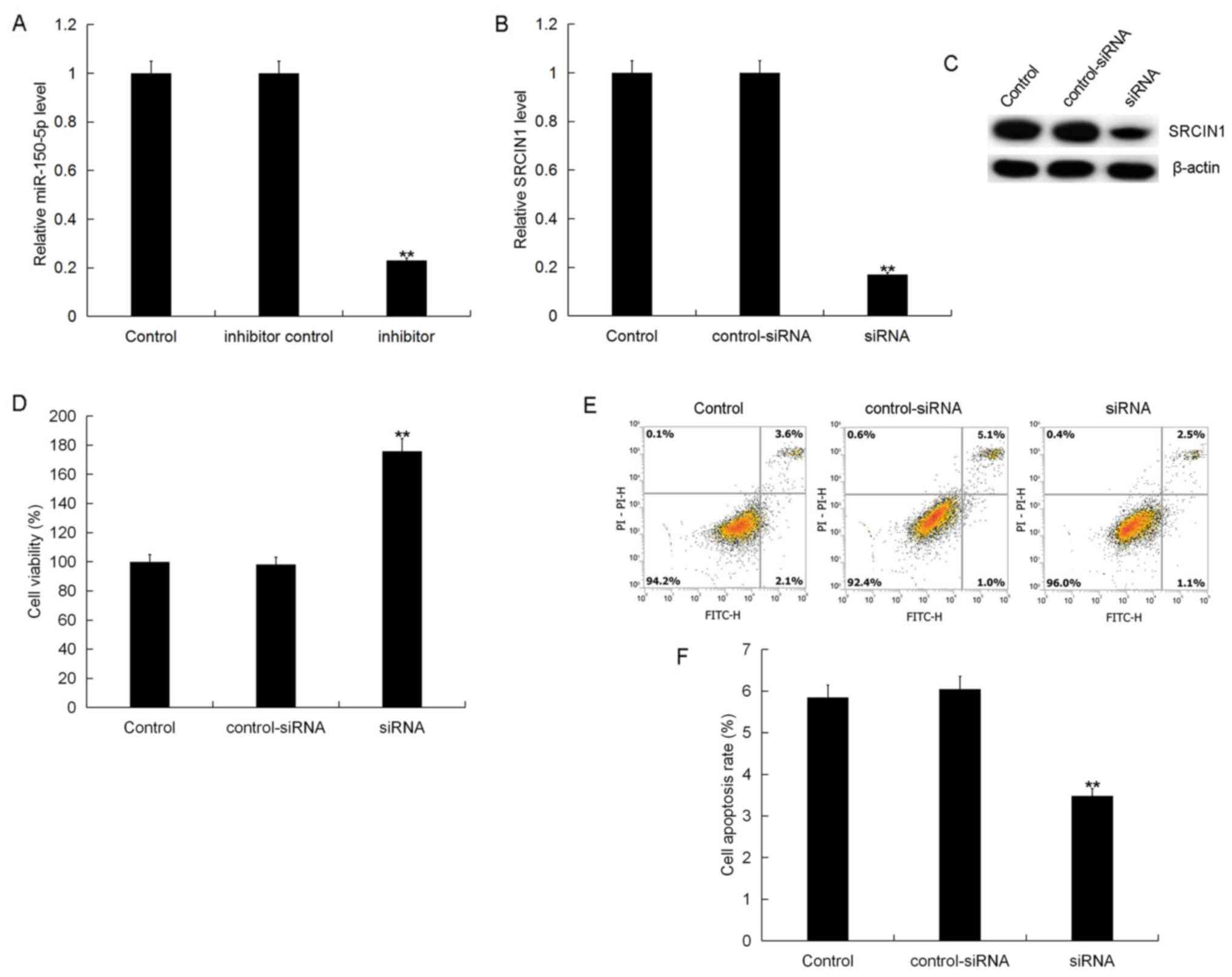
To assess the role of miR-150-5p in GC, a miR-150-5p
inhibitor, SRCIN1-siRNA, or miR-150-5p inhibitor+SRCIN1-siRNA was
transfected into BGC-823 cells. Following 48-h transfection,
transfection efficiency was detected using RT-qPCR and/or western
blotting. The results demonstrated that, compared with the control
group, the miR-150-5p inhibitor significantly reduced miR-150-5p
expression (Fig. 3A) and
SRCIN1-siRNA significantly reduced the mRNA and protein level of
SRCIN1 (Fig. 3B and C). The results
also demonstrated that, compared with the control group,
SRCIN1-siRNA significantly enhanced BGC-823 cell viability
(Fig. 3D) and inhibited cell
apoptosis (Fig. 3E and F), while
control-siRNA had no effect on BGC-823 cell viability or apoptosis.
These results suggest that miR-150-5p may be involved in regulating
SRCIN1 expression in GC cells.
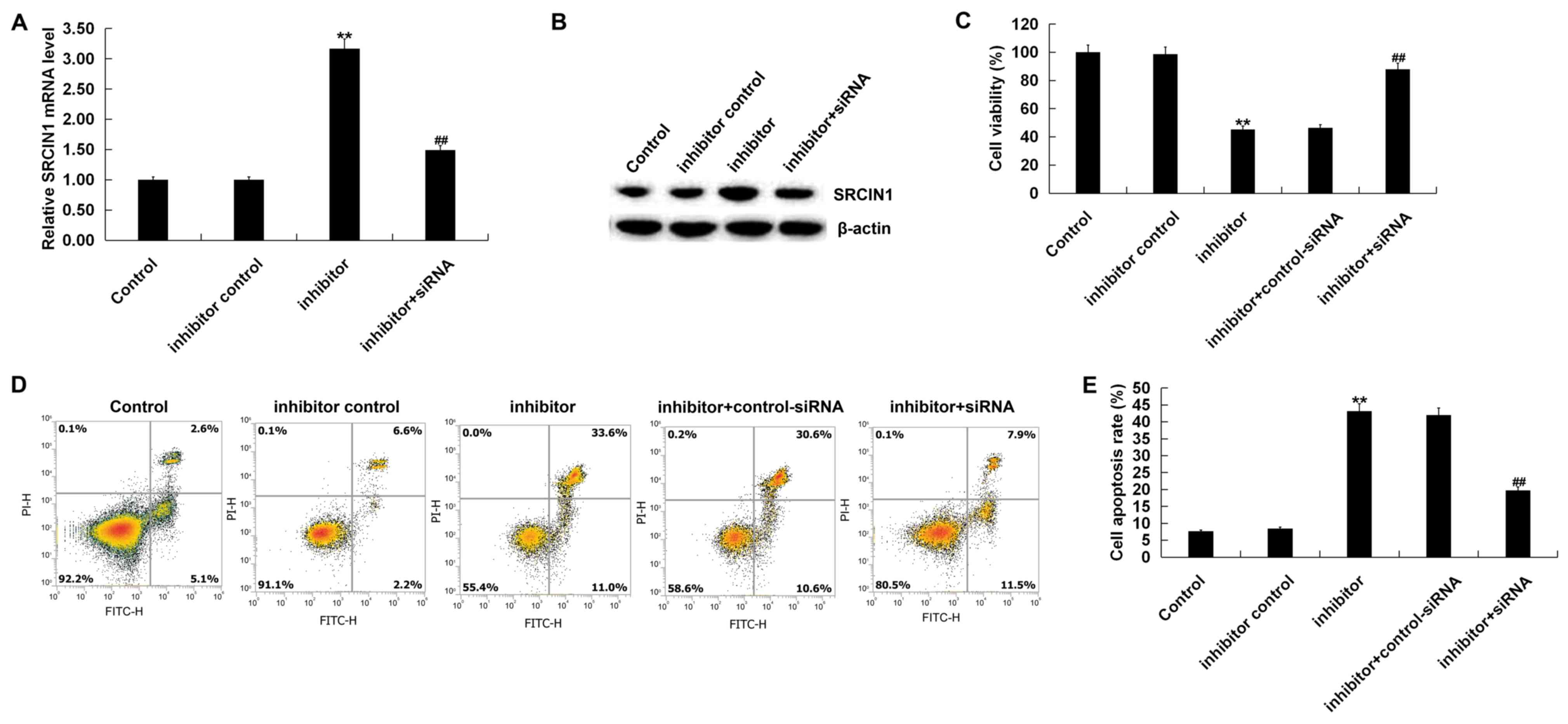
Additionally, compared with control group, the
miR-150-5p inhibitor significantly increased SRCIN1 mRNA and
protein expression, which was inhibited following SRCIN1-siRNA
treatment (Fig. 4A and B). The
results of the CCK-8 assay indicated that the miR-150-5p inhibitor
decreased BGC-823 cell viability, whilst SRCIN1-siRNA treatment
reversed this effect (Fig. 4C).
Furthermore, it was revealed that the miR-150-5p inhibitor
significantly induced BGC-823 cell apoptosis, which was reversed
following SRCIN1-siRNA treatment (Fig.
4D and E).
miR-150-5p inhibitor suppresses EMT in
BGC-823 cells
Finally, the current study assessed whether
miR-150-5p affected GC cell EMT, interstitial cell markers
(vimentin, N-cadherin and β-catenin) and epithelial cell markers
(ZO-1 and E-cadherin). The results indicated that compared with the
control group, the miR-150-5p inhibitor reduced the expression of
interstitial cell markers (vimentin, N-cadherin and β-catenin) and
increased the expression of epithelial cell markers (ZO-1 and
E-cadherin), thus inhibiting gastric cancer cell EMT. All these
effects were reversed by SRCIN1-siRNA treatment (Fig. 5).
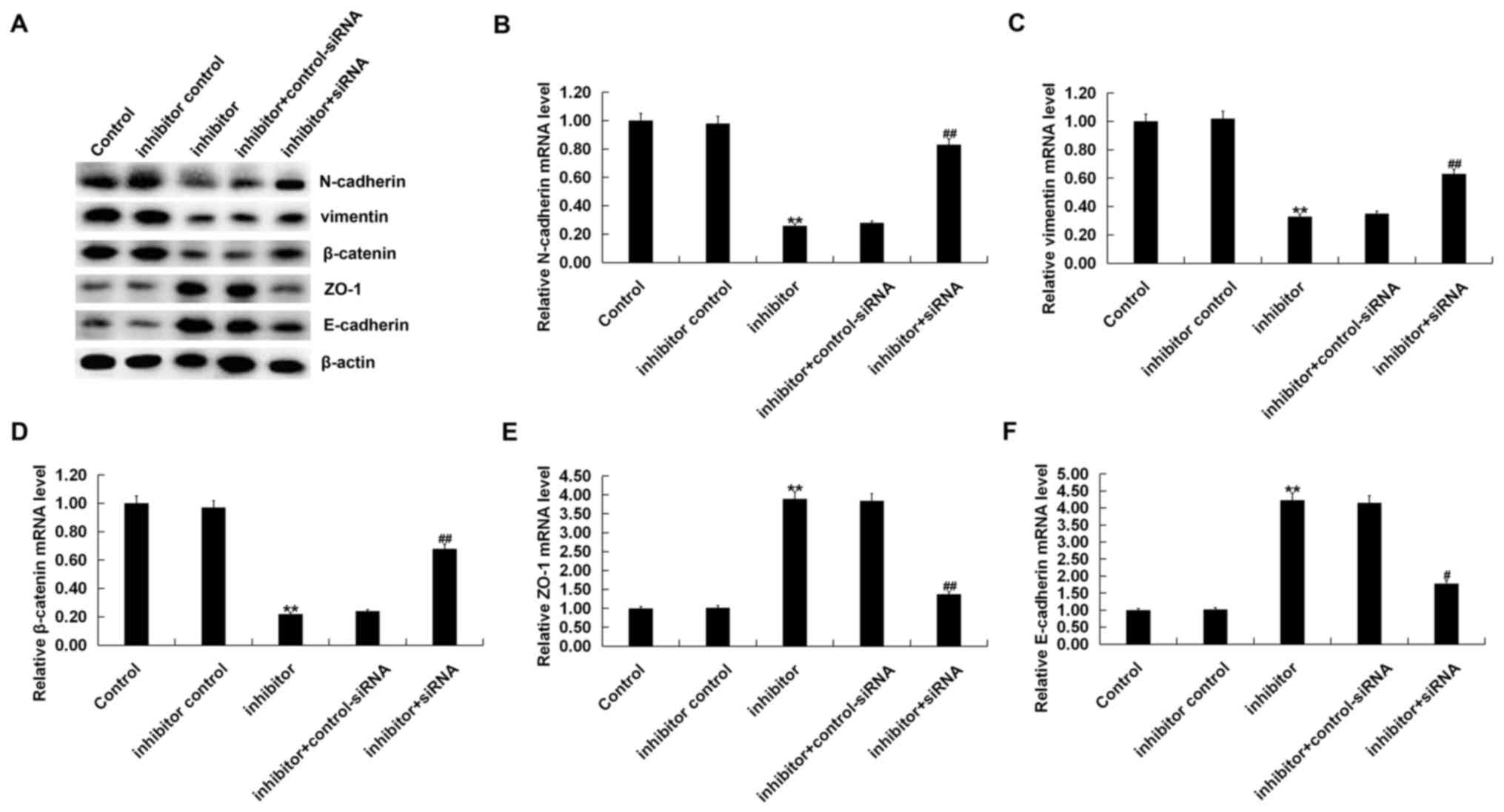 | Figure 5.miR-150-5p inhibitor inhibits BGC-823
cell EMT. BGC-823 cells were transfected with an inhibitor control,
a miR-150-5p inhibitor, and a miR-150-5p inhibitor+SRCIN1-siRNA for
48 h. The (A) protein and mRNA expression of EMT factors including
(B) N-cadherin, (C) vimentin and (D) β-catenin, and epithelial cell
markers (E) ZO-1 and (F) E-cadherin were detected via western
blotting and reverse transcription-quantitative polymerase chain
reaction. Data were presented as the mean ± standard deviation.
**P<0.01 vs. control; #P<0.05 and
##P<0.01 vs. inhibitor. miR, microRNA, EMT,
epithelial mesenchymal transition; SRCIN1, SRC kinase signaling
inhibitor 1; siRNA, small interfering RNA; ZO-1, zonula
occluden-1. |
Discussion
GC is a malignant tumor that originates from the
epithelium of gastric mucosa (1).
The annual incidence of GC in China is ~400,000, which accounts for
42% of the total number of cases worldwide (4,29). GC
accounts for 10% of all diagnosed cancer types each year and
accounts for 12% of all cancer associated mortalities worldwide
(2,4). It is also one of the most common
malignancies (13,14). The current study assessed the role
and mechanism of miR-150-SRCIN1 in the pathogenesis of GC
development.
Many studies have determined the role of miRNAs in
the development of various diseases (7,30–32) and
further research has revealed that miRNAs are involved in the
development of various types of cancer (33–36).
Many studies have also identified the aberrant expression of
certain miRNAs in tumor cells and thus are considered novel
references for tumor diagnosis (37,38).
miRNAs are novel therapeutic targets and may serve as prognostic
indicators (34). In recent years, a
large number of studies have revealed that miRNAs serve a
significant role in the development of GC (39,40).
However, to the best of our knowledge, the role of miR-150-5p in
the pathological development of GC remains largely unclear.
Therefore, the current study was performed.
The present study utilized RT-qPCR to detect the
expression of miR-150-5p in different GC cell lines (including
well-differentiated gastric adenocarcinoma MGC-803, moderately
differentiated gastric adenocarcinoma SGC-7901 and poorly
differentiated gastric adenocarcinoma BGC-823) and in the normal
stomach mucosal epithelial cell line GES-1. The results revealed
that miR-150-5p was significantly upregulated in all GC cell lines
and the highest expression was exhibited in BGC-823 cells.
Additionally, 36 GC tissue samples and adjacent normal tissues
(including 12 cases of stage I GC, 12 cases of Stage II and 12
cases of Stage III–IV, with lymph node metastasis exhibited in 18
cases and no lymph metastasis in 18 cases) were utilized to assess
the association between miR-150-5p expression and GC staging. The
results revealed that the highest expression level of miR-150-5p
was observed in GC patients at Stage III–IV, and the expression
level of miR-150-5p was increased in GC patients with lymph node
metastasis. This indicated that an increased expression of
miR-150-5p was associated with advanced GC and lymph node
metastasis.
The current study predicted that SRCIN1 was a target
gene of miR-150-5p and this was confirmed by utilizing the dual
luciferase reporter gene system. Lower expression levels of SRCIN1
were also demonstrated in GC tissues and cells, with the lowest
expression exhibited in BGC-823 cells.
Furthermore, experiments were performed to assess
the effects of miR-150-5p downregulation on BGC-823 cells, which
exhibit the highest level of miR-150-5p among the GC cell lines
examined. The results indicated that the miR-150-5p inhibitor
decreased BGC-823 cell viability and induced cell apoptosis.
EMT is an important process of cancer cell migration
and metastasis that can be induced by various transcription factors
and signal transduction factors (21,22,24).
miRNAs, as small RNAs that regulate protein translation at the
post-transcriptional level, are also involved in the EMT process.
The present study assessed the effects of miR-150-5p treatment on
BGC-823 cell EMT. The results revealed that the expression of
vimentin, N-cadherin and β-catenin were significantly decreased,
while the expression of E-cadherin and ZO-1 were increased in
BGC-823 cells transfected with the miR-150-5p inhibitor, indicating
an inhibitory effect on GC cell EMT. In addition, the current study
determined that all the effects of the miR-150-5p inhibitor on
BGC-823 cells were reversed following SRCIN1-siRNA treatment.
Furthermore, the results indicated that control-siRNA exhibited no
significant effect on SRCIN1 expression at the mRNA or protein
level. Therefore, an inhibitor + control-siRNA group was not
included in the present study.
In conclusion, the results of the current study
revealed that the miR-150-5p inhibitor inhibited GC cell viability,
induced cell apoptosis and inhibited GC cell EMT by targeting
SRCIN1. Therefore, the miR-150-5p/SRCIN1 axis may be considered as
a potential clinical indicator and therapeutic target for the
treatment of GC. However, the current study only performed a
preliminary assessment of the role of miR-150-5p in GC. To validate
these results, further experimental research is required. For
example, the protein level of SRCIN1 in GC tissues should be
determined and the correlation between SRCIN1 and miR-150-5p should
be examined. In addition, the association between the expression
levels of SRCIN1 or miR-150-5p with the clinicopathological
features of patients with GC should be assessed. The current study
aims to determine these in the future.
Acknowledgements
The authors would like to thank Dr Anlie Cai and Dr
Huaide Peng, Zhuzhou Central Hospital for their support for
experimental funding. The authors would like to thank Professor
Jiansi Zhu, Cancer Research Institute of Nanhua University for his
guidance on the experimental methods used in the current study, and
Professor Zhijiao Zhou, Department of Pathology, Third Xiangya
Hospital, for the help he provided during the preparation of the
manuscript.
Funding
No funding received.
Availability of data and materials
The analyzed datasets used and/or generated during
the present study are available from the corresponding author on
reasonable request.
Authors' contributions
XQ contributed to data collection, statistical
analysis, data interpretation and manuscript preparation. DC
contributed to the study design and literature search. ML
contributed to the study design, statistical analysis and data
interpretation; XC contributed to statistical analysis and
literature search; MH contributed to data collection and data
interpretation. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of the Zhuzhou Central Hospital and written informed
consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yu ZY, Wang Z, Lee KY, Yuan P and Ding J:
Effect of silencing colon cancer-associated transcript 2 on the
proliferation, apoptosis and autophagy of gastric cancer BGC-823
cells. Oncol Lett. 15:3127–3132. 2018.PubMed/NCBI
|
|
2
|
Salem M, Xiu J, Eldeiry W, Reddy S, Philip
P, Gatalica Z, Khan S, Denlinger C, Mikhail S, Smaglo B, et al:
Comparative molecular analyses of esophageal adenocarcinoma,
esophageal squamous cell carcinoma and gastric adenocarcinoma, and
impact of molecular profile on outcome. Ann Oncol. 27:ii119. 2016.
View Article : Google Scholar
|
|
3
|
Fu L, Yin F, Li XR, Han BK, Zhang C, Wang
JW, Wang YQ, Bi YF and Liu HM: Generation and characterization of a
paclitaxel-resistant human gastric carcinoma cell line. Anticancer
Drugs. 1:491–502. 2018. View Article : Google Scholar
|
|
4
|
Yang BF, Cai W and Chen B: LncRNA SNHG12
regulated the proliferation of gastric carcinoma cell BGC-823 by
targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci.
22:1297–1306. 2018.PubMed/NCBI
|
|
5
|
Liu L, Si N, Ma Y, Ge D, Yu X, Fan A, Wang
X, Hu J, Wei P, Ma L, et al: Hydroxysafflor-yellow A induces human
gastric carcinoma BGC-823 cell apoptosis by activating peroxisome
proliferator-activated receptor gamma (PPARγ). Med Sci Monit.
24:803–811. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rothmiller S, Wolf M, Worek F, Steinritz
D, Thiermann H and Schmidt A: Alteration of miRNA expression in a
sulfur mustard resistant cell line. Toxicol Lett. 293:38–44. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ultimo S, Martelli AM, Zauli G, Vitale M,
Calin GA and Neri LM: Roles and clinical implications of MicroRNAs
in acute lymphoblastic leukemia. J Cell Physiol. 233:5642–5654.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sheng N, Zhang L and Yang S: MicroRNA-429
decreases the invasion ability of gastric cancer cell line BGC-823
by downregulating the expression of heparanase. Exp Ther Med.
15:1927–1933. 2018.PubMed/NCBI
|
|
9
|
Meng Y, Tian H, Hu Q, Liang H, Zeng L and
Xiao H: MicroRNA repertoire and comparative analysis of Andrias
davidianus infected with ranavirus using deep sequencing. Dev Comp
Immunol. 85:108–114. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan J, Li X, Peng L, Shen X, Dang Y and
Zhang G: MicroRNA-150 as a potential biomarker in diagnosis of
cancer: A meta-analysis. Clin Lab. 63:1187–1197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li C, Du X, Xia S and Chen L: MicroRNA-150
inhibits the proliferation and metastasis potential of colorectal
cancer cells by targeting iASPP. Oncol Rep. 40:252–260.
2018.PubMed/NCBI
|
|
12
|
Tang W, Xu P, Wang H, Niu Z, Zhu D, Lin Q,
Tang L and Ren L: MicroRNA-150 suppresses triple-negative breast
cancer metastasis through targeting HMGA2. Onco Targets Ther.
11:2319–2332. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun X, Zhang C, Cao Y and Liu E: miR-150
suppresses tumor growth in melanoma through downregulation of MYB.
Oncol Res. 27:317–323. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Z, Wang J, Li J, Wang X and Song W:
MicroRNA-150 promotes cell proliferation, migration and invasion of
cervical cancer through targeting PDCD4. Biomed Pharmacother.
97:511–517. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koshizuka K, Hanazawa T, Kikkawa N, Katada
K, Okato A, Arai T, Idichi T, Osako Y, Okamoto Y and Seki N:
Antitumor miR-150-5p and miR-150-3p, inhibit cancer cell
aggressiveness by targeting SPOCK1, in head and neck squamous cell
carcinoma. Auris Nasus Larynx. 45:854–865. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan R, Yang T, Zhai H, Zhou Z, Gao L and
Li Y: MicroRNA-150-5p affects cell proliferation, apoptosis and EMT
by regulation of the BRAFV600E mutation in papillary thyroid cancer
cells. J Cell Biochem. 119:8763–8772. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ergun S, Güney S, Temiz E, Petrovic N and
Gunes S: Significance of Mir-15a-5p and Cnksr3 as novel prognostic
biomarkers in non-small cell lung cancer. Anticancer Agents Med
Chem. 18:1695–1701. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu J, Feng Y, Wang Y and An R: Aryl
hydrocarbon receptor enhances the expression of miR-150-5p to
suppress in prostate cancer progression by regulating MAP3K12. Arch
Biochem Biophys. 654:47–54. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suetsugu T, Koshizuka K, Seki N, Mizuno K,
Okato A, Arai T, Misono S, Uchida A, Kumamoto T and Inoue H:
Downregulation of matrix metalloproteinase 14 by the antitumor
miRNA, miR-150-5p, inhibits the aggressiveness of lung squamous
cell carcinoma cells. Int J Oncol. 52:913–924. 2018.PubMed/NCBI
|
|
20
|
Wang S, Yan Y, Cheng Z, Hu Y and Liu T:
Sotetsuflavone suppresses invasion and metastasis in non-small-cell
lung cancer A549 cells by reversing EMT via the TNF-α/NF-κB and
PI3K/AKT signaling pathway. Cell Death Discov. 4:262018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zuo L, Zhao H, Yang R, Wang L, Ma H, Xu X,
Zhou P and Kong L: Lamin A/C might be involved in the EMT
signalling pathway. Gene. 663:51–64. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang Y, Gao M, Lin Z, Chen L, Jin Y, Zhu
G, Wang Y and Jin T: DEK promoted EMT and angiogenesis through
regulating PI3K/AKT/mTOR pathway in triple-negative breast cancer.
Oncotarget. 8:98708–98722. 2017.PubMed/NCBI
|
|
23
|
Pal M, Bhattacharya S, Kalyan G and Hazra
S: Cadherin profiling for therapeutic interventions in Epithelial
Mesenchymal Transition (EMT) and tumorigenesis. Exp Cell Res.
368:137–146. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma Z, Xin Z, Hu W, Jiang S, Yang Z, Yan X,
Li X, Yang Y and Chen F: Forkhead box O proteins: Crucial
regulators of cancer EMT. Semin Cancer Biol. 50:21–31. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu CY, Lin HH, Tang MJ and Wang YK:
Vimentin contributes to epithelial-mesenchymal transition cancer
cell mechanics by mediating cytoskeletal organization and focal
adhesion maturation. Oncotarget. 6:15966–15983. 2015.PubMed/NCBI
|
|
26
|
Shang S, Hua F and Hu ZW: The regulation
of β-catenin activity and function in cancer: Therapeutic
opportunities. Oncotarget. 8:33972–33989. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zihni C, Mills C, Matter K and Balda MS:
Tight junctions: From simple barriers to multifunctional molecular
gates. Nat Rev Mol Cell Biol. 17:564–580. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mellis D and Caporali A: MicroRNA-based
therapeutics in cardiovascular disease: Screening and delivery to
the target. Biochem Soc Trans. 46:11–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao DD, Li L and Chan WY: MicroRNAs: Key
regulators in the central nervous system and their implication in
neurological diseases. Int J Mol Sci. 17(pii): E8422016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mannucci C, Casciaro M, Minciullo PL,
Calapai G, Navarra M and Gangemi S: Involvement of microRNAs in
skin disorders: A literature review. Allergy Asthma Proc. 38:9–15.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu L, Ai J, Long H, Liu W, Wang X, Zuo Y,
Li Y, Wu Q and Deng Y: Intergrative microRNA and gene profiling
data analysis reveals novel biomarkers and mechanisms for lung
cancer. Oncotarget. 7:8441–8454. 2016.PubMed/NCBI
|
|
34
|
Tsai MM, Wang CS, Tsai CY, Huang HW, Chi
HC, Lin YH, Lu PH and Lin KH: Potential diagnostic, prognostic and
therapeutic targets of microRNAs in human gastric cancer. Int J Mol
Sci. 17(pii): E9452016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao H, Li M, Li L, Yang X, Lan G and
Zhang Y: MiR-133b is down-regulated in human osteosarcoma and
inhibits osteosarcoma cells proliferation, migration and invasion
and promotes apoptosis. PLoS One. 8:e835712013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kaukoniemi KM, Rauhala HE, Scaravilli M,
Latonen L, Annala M, Vessella RL, Nykter M, Tammela TL and
Visakorpi T: Epigenetically altered miR-193b targets cyclin D1 in
prostate cancer. Cancer Med. 4:1417–1425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schmidt A, Steinritz D, Thiermann H,
Meineke V and Abend M: Alteration of miRNA expression in early
endothelial cells after exposure with sub-lethal sulfur mustard
concentrations. Toxicol Lett. 244:88–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gharbi S, Khateri S, Soroush MR, Shamsara
M, Naeli P, Najafi A, Korsching E and Mowla SJ: MicroRNA expression
in serum samples of sulfur mustard veterans as a diagnostic gateway
to improve care. PLoS One. 13:e01945302018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma M, Zhao J, Wu Q, Xiao K, Li S, Zhu H,
Liu C, Xie H and Zuo C: MiRNA-545 negatively regulates the
oncogenic activity of EMS1 in gastric cancer. Cancer Med.
7:2452–2462. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liao Y, Liao Y, Li J, Xiao K, Li S, Zhu H,
Liu C, Xie H and Zuo C: Genetic variants in miRNA machinery genes
associated with clinicopathological characteristics and outcomes of
gastric cancer patients. Int J Biol Markers. 33:301–307. 2018.
View Article : Google Scholar : PubMed/NCBI
|