Introduction
Hepatocellular carcinoma (HCC) refers to malignant
tumors of the liver, including primary and metastatic HCC (1). In recent years, the incidence of HCC
has increased on a global scale, ranking as the fifth most common
malignant tumors (2). The number of
cases of HCC in China accounted for ~55% of the world's total
occurrence and presented with a high-risk of mortality (3). At present, surgical treatments,
including liver resection and transplantation, are the primary
means of treating patients with HCC (4). However, existing treatment techniques
frequently fail to achieve favorable results due to a high
incidence of metastasis and recurrence (5). Therefore, the underlying biological
mechanisms need to be determined to develop improved therapeutic
options.
At present, targeted therapy has been widely
researched. In April 2018, antisense nucleic acid drug ‘CT102 for
injection’ entered clinical trials in China and provided a new
gene-targeted therapy for patients with HCC (6). Long noncoding RNAs (lncRNAs) are a
group of noncoding RNAs over 200 nucleotides in length with little
or no protein-coding ability (7).
Various lncRNAs have been demonstrated to participate in the
pathogenesis of HCC (7,8). In addition, lncRNA overexpression in
HCC was closely related with other types of cancer, including
colorectal cancer, gastric cancer, colorectal cancer and
osteosarcoma (9). Xu et al
(10) showed that lncSHRG promotes
HCC progression through the transcription cofactor HES-6 pathway.
Huang et al (11) found that
lncAKHE contributes to HCC migration and invasion by activating the
NOTCH signaling pathway. Upregulation of histone H2B ubiquitin
ligase complex expression resulted in an interaction with miR-372,
augmenting the cAMP-dependent protein kinase pathway and thus,
promoting hepatocellular carcinoma (8). Furthermore, lncRNA metallothionein 1D,
pseudogene was found to inhibit the expression of forkhead box
(Fox)A1 in HCC cells by negatively regulating transcriptional
coactivator YAP1 and runt-related transcription factor 2 (12). Various lncRNAs can regulate several
biological processes in tumor cells, including apoptosis,
proliferation, migration and invasion (12). Therefore, it is important to
investigate and understand the functions of the numerous
dysregulated lncRNAs in cancer.
LncRNA MAFG-antisense 1 (AS1) has been screened as a
novel target for treating patients with cancer (13). Zhang et al (13) confirmed that lncRNA MAFG-AS1
influenced the proliferation of osteosarcoma cells and also
regulated the expression level of various downstream proteins. Cui
et al (14) showed that
MAFG-AS1 promotes colorectal cancer development. Additionally, Jia
et al (15) determined that
MAFG-AS1 promotes metastasis of lung cancer (15). However, the molecular mechanism of
lncRNA MAFG-AS1 in other types of cancer is still unknown. In the
present study, the role of lncRNA MAFG-AS1 in HCC was explored, and
potential regulatory factors and associated mechanisms were
examined. LncRNA MAFG-AS1 may be a potentially novel target for
treating patients with HCC.
Materials and methods
The Cancer Genome Atlas (TCGA) dataset
analysis and cell culture
Two sets of HCC data were downloaded from the The
Cancer Genome Atlas (TCGA) database (tcga-data.nci.nih.gov/tcga) with one set containing 40
HCC samples and 25 controls, and the other containing 200 tumor
samples and 50 controls. The human HCC cells (Hep3B and Huh7) and a
human liver cell line (L02) were purchased from Shanghai Institute
of Biochemistry and Cell Biology, and cultured using DMEM medium
(Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS (Invitrogen; Thermo Fisher Scientific, Inc.) with 100 U/ml
penicillin (Invitrogen; Thermo Fisher Scientific, Inc.) and
streptomycin (100 µg/ml; Invitrogen; Thermo Fisher Scientific,
Inc.) in an incubator with 5% CO2 at 37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to detect the expression level and
distribution of MAFG-AS1. The cultured cells were digested and
centrifuged at 4°C for 2 min at 3,000 × g. Total RNA was extracted
using TRlzol® reagent according to the manufacturer's
protocol (Invitrogen; Thermo Fisher Scientific, Inc.) and
quantified using NanoDrop 2000 (Thermo Fisher Scientific, Inc.). A
total 1 mg RNA was used for cDNA synthesis using PrimeScript RT
Master mix system (Takara Biotechnology Co., Ltd.) according to the
manufacturer's protocol. The reaction was performed with incubation
at 42°C for 1 h, and the enzyme was subsequently inactivated by
incubation at 85°C for 5 min. The total reaction volume was 20 µl
and the mixture was prepared as previously described (16). For qPCR, the PCR solution contained 1
ml cDNA, 1 ml primers and 10 ml SYBR Green and 5 ml RT-qPCR Master
mix (all from Invitrogen; Thermo Fisher Scientific, Inc.). The
final volume was adjusted to 20 µl using RNase-free water. The
amplification was carried out in an ABI FAST 7500 system (Applied
Biosystems; Thermo Fisher Scientific Inc.). The relative expression
level of each gene was normalized to U6 and calculated using the
2−ΔΔCq method (17). The
thermocycling conditions were as follows: initial denaturation step
of 94°C for 2 min, followed by 30 cycles of 94°C for 30 sec, 59°C
for 30 sec and 72°C for 2 min with a final extension step at 72°C
for 10 min. Primers for PCR amplification were: MAFG-AS1 forward,
5′-ATGACGACCCCCAATAAAGGA-3′ and reverse, 5′-CACCGACATGGTTACCAGC-3′;
miR-6852 forward, 5′-AACGAGACGACGACAGAC-3′ and reverse,
5′-CCCTGGGGTTCTGAGGACATG-3′; U6 forward, 5′-AACGAGACGACGACAGAC-3′
and reverse, 5′-GCAAATTCGTGAAGCGTTCCATA-3′.
Transient transfection
The siRNA (50 nM) targeting MAFG-AS1
(5′-GGGCAAUUCCAACCAAGAAAC-3′), negative control siRNA (50 nM;
5′-AAUUCUCCGAACGUGUCACGU-3′), miR-6852 mimics
(5′-CCCUGGGGUUCUGAGGACAUG-3′; 50 nM), miR-6852 inhibitors
(5′-CAUGUCCUCAGAACCCCAGGG-3′; 50 nM), inhibitor negative controls
(5′-GCGUAACUAAUACAUCGGAUUCGU-3′; 50 nM) and mimic negative controls
(5′-ACAUCUGCGUAAGAUUCGAGUCUA-3′; 50 nM) were obtained from
Guangzhou RiboBio Co., Ltd. For MAFG-AS1 overexpression, the
sequence of MAFG-AS1 was inserted into a pcDNA3.1 vector
(Invitrogen; Thermo Fisher Scientific, Inc.). The cultured Hep3B
and Huh7 cells were transfected using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Following 48 h of incubation, the
transfection efficiency was analyzed by RT-qPCR.
Cell Counting Kit-8 (CCK8) assay
CCK8 assay was used to analyze the proliferation of
HCC cell lines. The cells transfected for 48 h were plated at a
density of 2×103 cells per well in a 24-well plate, and
three replicate wells were used for each condition. The cells were
cultured at 37°C in a 5% CO2 incubator. Cell
proliferation assays were performed at 0, 24, 48 and 72 h. A total
of 10 ml CCK8 reagent was added to each well, incubated for 4-h at
room temperature and the absorbance was measured at 450 nm (A)
using a microplate reader.
Transwell migration and invasion
assays
As described previously (18), 2×104 cells per well were
cultured in the upper Transwell chamber of a 24-well plate
(18). Matrigel (BD Biosciences) was
used to coat the upper side of the membrane at 37°C for 30 min. A
total of 2×104 cells cultured in the upper chamber were
placed in 200 µl FBS-free DMEM (Invitrogen; Thermo Fisher
Scientific, Inc.) and 600 µl DMEM with 10% FBS was placed in the
lower chamber. After a 17-h incubation, the cells were fixed in 70%
methanol at room temperature for 30 min and subsequently stained
with 0.1% crystal violet at room temperature for 30 min. The cells
on the lower chamber were counted using light microscopy to
calculate the invasion at ×200 magnification. Five random fields of
views were counted.
The experimental procedures of the migration assay
were the same as invasion assay except that Matrigel was not
used.
Luciferase reporter assay
Bioinformatics analysis and luciferase reporter
assay were used to screen binding sites and confirm the target of
miR-6852, respectively. By using the miRDB tool (http://mirdb.org/miRDB/index.html), miR-6852 was
identified as the most likely candidate target of MAFG-AS1. The 3
end of MAFG-AS1 containing the miR-6852 binding site was cloned
into a luciferase reporter vector. A MAFG-AS1 3 UTR wild type (WT)
plasmid (MAFG-AS1 3 UTR-WT) was constructed based on the 3-end
primer sequence of MAFG-AS1. Using the MAFG-AS1 3 UTR-WT plasmid, a
binding site was mutated to construct a MAFG-AS1 3 UTR mutant (MUT)
plasmid (MAFG-AS12 3 UTR-MUT). The construction and sequencing of
the plasmid was performed by Sangon Bioengineering Co., Ltd. The
constructed luciferase reporter plasmids pmirGLO-MAFG-AS1-WT,
pmirGLO-MAFG-AS1-MUT and miR-6852 mimics, mimics negative control
were co-transfected into Hep3B and Huh7 cell lines using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, the luciferase activity was measured
using a dual luciferase activity assay kit (Promega Corporation)
and results were normalized to Renilla luciferase
activity.
Western blotting
HCC cells were lysed using radioimmunoprecipitation
assay buffer (Thermo Fisher Scientific, Inc.) and the total protein
lysates were obtained. Protein concentration was determined using a
bicinchoninic acid assay. Total protein (20 µg) was separated using
SDS-PAGE on a 4–20% gel and then transferred to polyvinylidene
difluoride membranes. Membranes were subsequently blocked in 5%
non-fat milk/Tris-buffered saline containing 0.1% Tween-20 at 25°C
for 1 h and then incubated at 4°C overnight with the primary
antibodies. The following primary antibodies were used: Anti-PI3K
(1:2,000; cat. no. 4292), anti-p-PI3K (1:2,000; cat. no. 4228),
anti-p-STAT3 (1:2,000; cat. no. 9145), anti-STAT3 (1:2,000; cat.
no. 12640), anti-MYC (1:2,000; cat. no. 5605) and GAPDH (1:2,000;
cat. no. 5174) all from Cell Signaling Technology, Inc.
Subsequently, the membranes were incubated with goat anti-rabbit
horseradish peroxidase-conjugated secondary antibody (1:2,000; cat.
no. 7074; Cell Signaling Technology, Inc.) for 2 h at 25°C. Signals
were visualized using the Pierce™ ECL Western Blotting Substrate
(Thermo Fisher Scientific, Inc.). Densitometry analysis was
performed using ImageJ version 1.41 (National Institutes of
Health).
Statistical analysis
All data were analyzed using SPSS 18.0 statistical
software (SPSS, Inc.). All data were expressed as the mean ±
standard deviation of at least three repeats. A Student's t-test
was performed for comparison between two groups. A one-way ANOVA
followed by Tukey's post-hoc test was performed for comparisons
between multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression levels of MAFG-AS1 in HCC
tissues and cell lines
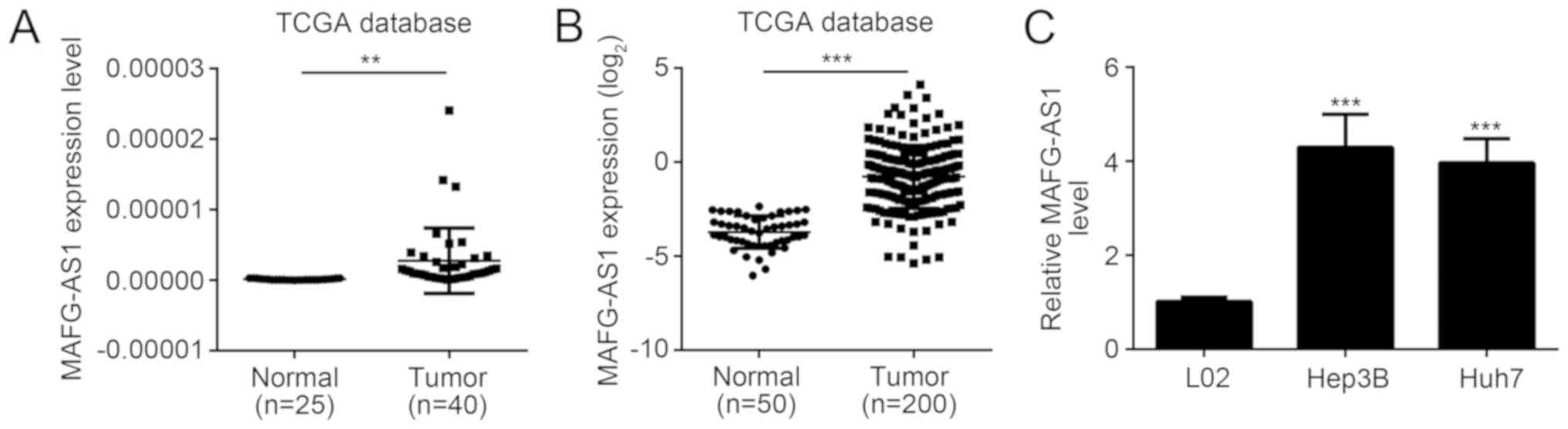
Based on two sets of data obtained from TCGA
database, the expression of MAFG-AS1 in HCC tissues was
significantly increased compared with the normal controls
(P<0.01; Fig. 1A and B).
Similarly, the expression of MAFG-AS1 in Hep3B and Huh7 cell lines
were significantly higher compared with the L02 cell line
(P<0.01; Fig. 1C). MAFG-AS1
expression was increased in both HCC tissues and cell lines
compared with their respective controls.
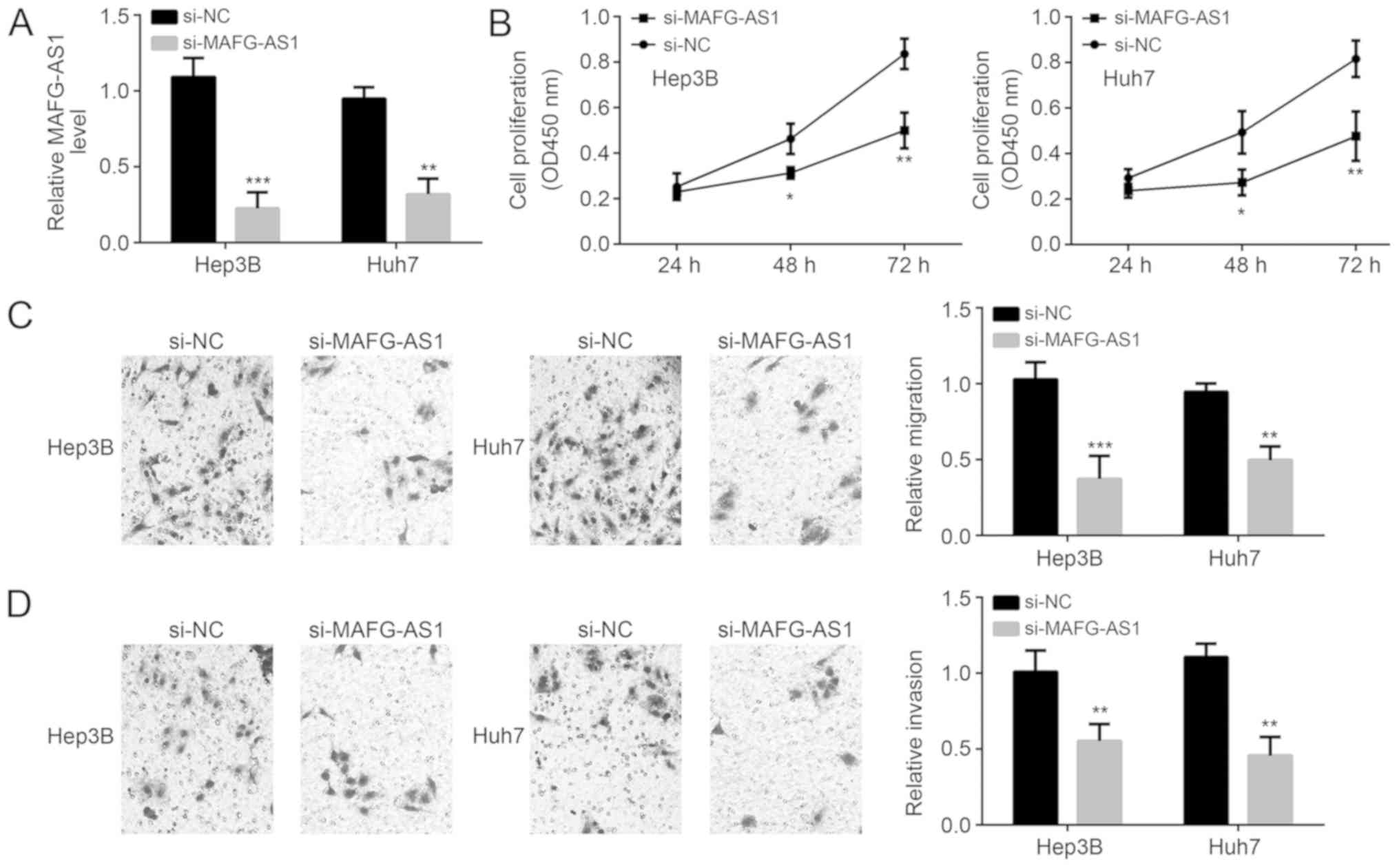
MAFG-AS1 knockdown inhibits
proliferation, invasion and migration of HCC cells
The relative MAFG-AS1 mRNA expression levels were
significantly decreased following MAFG-AS1 knockdown in both cell
lines confirming transfection was successful (Fig. 2A). Following MAFG-AS1 knockdown, the
proliferation, invasion and migration of both Hep3B and Huh7 cell
lines were significantly inhibited (Fig.
2B-D) suggesting that MAFG-AS1 was a critical gene in
pathogenesis of HCC.
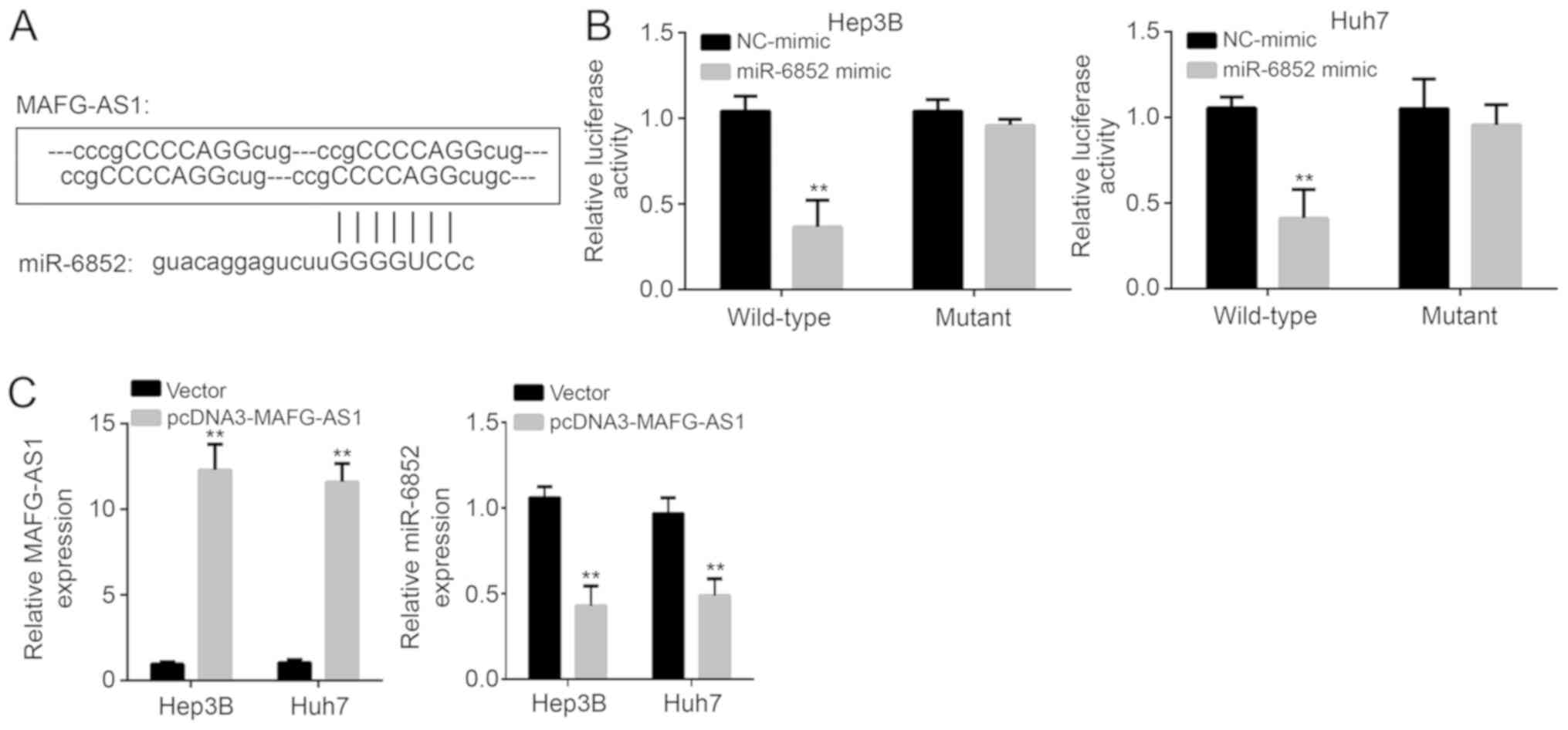
Cellular localization and target of
MAFG-AS1
A previous study demonstrated that MAFG-AS1 was
primarily localized in the cytoplasm and inhibits miRNAs in
colorectal cancer (14). Therefore,
it was hypothesized that MAFG-AS1 may utilize a similar mechanism
in HCC. Bioinformatics analysis was used to identify potential
binding targets. The analysis indicated that miR-6852 ranked top
among all the potential targets. MAFG-AS1 has four potential
binding sites with miR-6852 (Fig.
3A). A luciferase reporter assay revealed that upregulation of
miR-6852 decreased the luciferase activity of the wild-type
MAFG-AS1 reporter in both HCC cell lines (P<0.01; Fig. 3B). Upregulation of MAFG-AS1 decreased
the expression levels of miR-6852 (P<0.01; Fig. 3C). The above results demonstrate that
MAFG-AS1 targets and negatively regulates miR-6852.
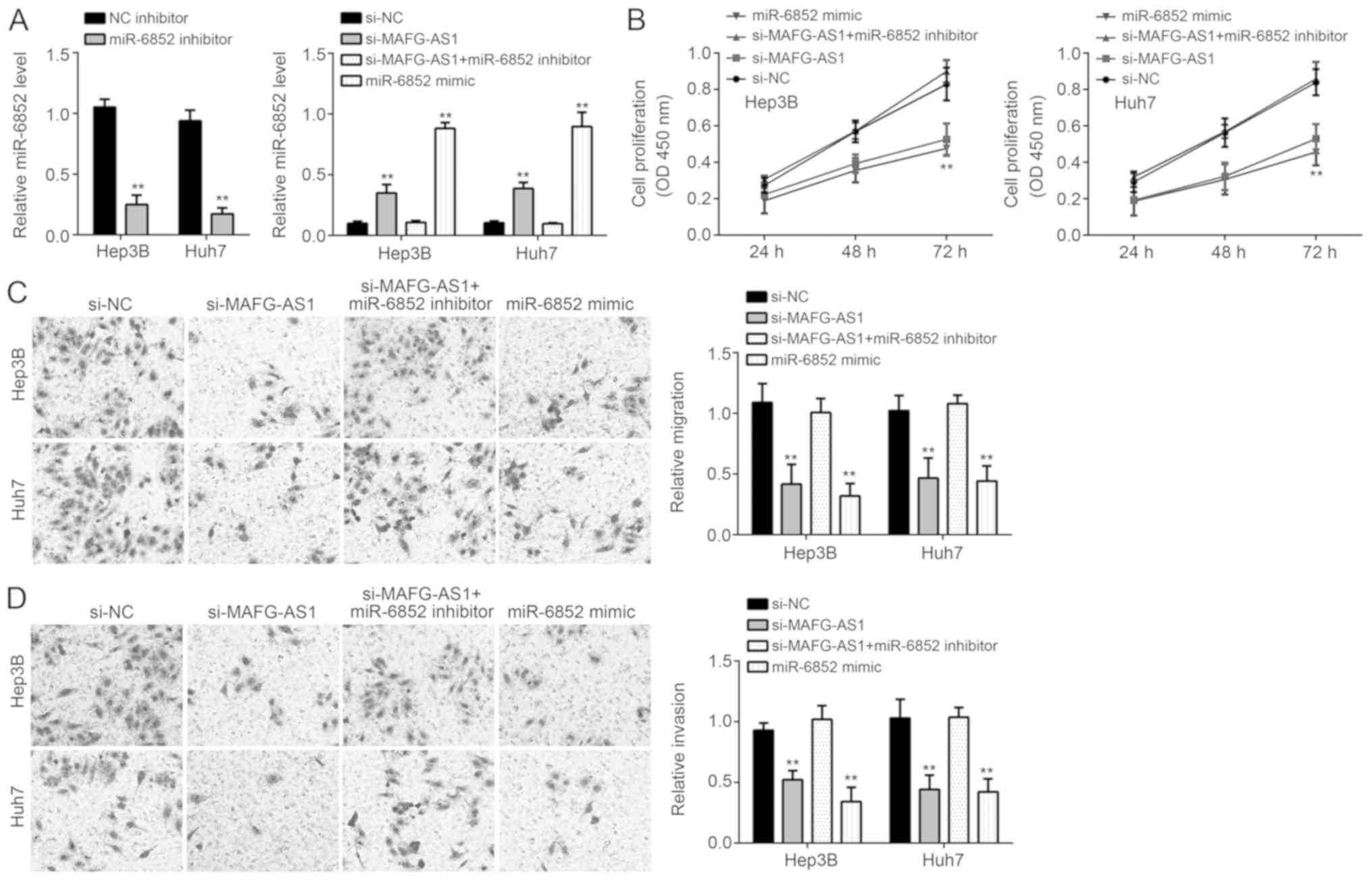
Inhibiting miR-6852 decreases
proliferation, migration and invasion of HCC cells
miR-6852 was knocked-down using miR-6852 inhibitors
in Hep3B and Huh7 cells (Fig. 4A).
In the si-MAFG-AS1 and miR-6852 mimics groups, proliferation,
migration and invasion of HCC cell lines were significantly
decreased (P<0.01; Fig. 4B).
However, inhibiting miR-6852 restored proliferation, migration and
invasion of the HCC cell lines (P<0.01; Fig. 4C and D). Taken together, these
results demonstrated that MAFG-AS1 may promote HCC progression
through inhibition of miR-6852. The effect of MAFG-AS1 on
expression of members of the STAT3, Wnt/β-catenin and PI3K/AKT
signaling pathways were determined, which are related to
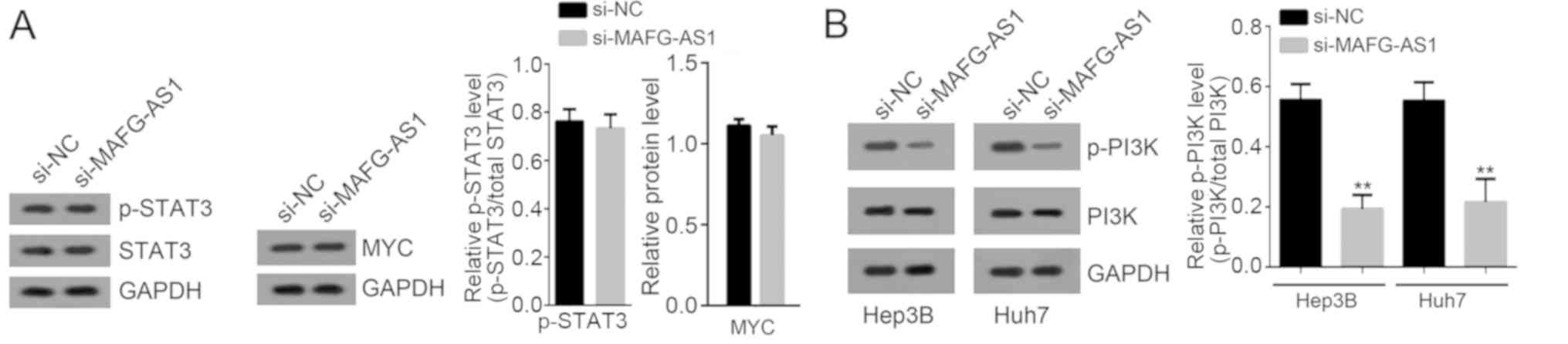
proliferation, migration and invasion of HCC (19). Western blotting showed that MAFG-AS1
silencing only suppressed the activation of the PI3K/AKT signaling
pathway (Fig. 5).
Discussion
Numerous lncRNAs are aberrantly expressed in HCC,
regulating various miRNAs and genes, and modulating a variety of
biological processes (20–22). Therefore, it is possible that one or
more of these lncRNAs may serve as a potential therapeutic target
for treating patients with HCC. In the present study, the
functional effects and potential underlying mechanism of lncRNA
MAFG-AS1 in HCC was examined. Based on the TCGA database and
RT-qPCR results, lncRNA MAFG-AS1 was highly expressed in HCC
tissues and cell lines. After knockdown of lncRNA MAFG-AS1, the
proliferation, migration and invasion of HCC cell lines were
significantly decreased. Interestingly, lncRNA MAFG-AS1 and
miR-6852 were demonstrated to exhibit a reciprocally negative
regulatory association with each other. By inhibiting the
expression of miR-6852, MAFG-AS1 promoted proliferation, migration
and invasion of HCC cells.
Zhang et al (23) processed a regulatory network analysis
of lncRNAs in colorectal cancer and showed that lncRNA MAFG-AS1 was
upregulated in this disease (23).
In addition, high-throughput data analysis and in vitro
experiments confirmed that lncRNA MAFG-AS1 was highly expressed and
affects the proliferation of osteosarcoma cells (13). Similarly, lncRNA MAFG-AS1 expression
was also upregulated in HCC tissues and primarily distributed in
the cytoplasm of HCC cells in the present study. After knockdown of
lncRNA MAFG-AS1, the proliferation, migration and invasion of HCC
cell lines were significantly inhibited. Therefore, lncRNA MAFG-AS1
may serve a critical role in pathogenesis of HCC.
LncRNA MAFG-AS1 was found to negatively regulate the
expression of miR-6852 through four potential binding sites, and
further influence the proliferation, migration and invasion of HCC
cell lines. A luciferase reporter assay demonstrated that
overexpression of miR-6852 inhibited the activity of the wild-type
MAFG-AS1 reporter. As shown in previous studies, abnormal
expression of miR-6852 could regulate the expression of FoxM1, and
thus influence processes of cell cycle arrest and necrosis in
cervical cancer cells (24). Kopanja
et al (25) found that
upregulation of FoxM1 was associated with a poor prognosis in
patients with HCC possibly by participating in the Ras signaling
pathway. In colorectal cancer, miR-6852 was confirmed to target
lncRNA transcription factor 7 (TCF7), and suppress tumor metastasis
and growth (26). Furthermore,
miR-6852 modulated cell invasion and proliferation by regulating
the expression of FoxJ1 (27).
LncRNA TCF7 could activate the Wnt signaling pathway and promote
self-renewal of HCC cells (28). In
the present study, miR-6852 was inhibited by MAFG-AS1, and was a
critical factor for the proliferation, migration and invasion of
HCC cell lines. Notably, MAFG-AS1 was also reported to prevent
binding of miR-339-5p from MMP15 in non-small cell lung cancer
(15). Whether MAFG-AS1 similarly
suppressed miR-339-5p to upregulate MMP15 in HCC cells remains to
be determined.
In the present study, the TCGA database was used to
show the expression of lncRNA MAFG-AS1 in HCC tissues. TCGA was
jointly developed by he National Cancer Institute and the National
Human Genome Research Institute in 2006, and a total 36 types of
cancer were examined (29).
Large-scale sequencing-based genomic analysis technology was used
to aid in understanding the molecular mechanisms of cancer
(30). TCGA combined with other
bioinformatics platforms may improve our understanding of the
molecular basis of cancer and improve diagnosis, treatment and
prevention (31). In the present
study, results from the TCGA database analysis confirmed that
lncRNA MAFG-AS1 was highly expressed in HCC tissues, it was also
consistent with the results of the in vitro experiments.
In conclusion, lncRNA MAFG-AS1 was overexpressed in
patients with HCC. Furthermore, overexpression of lncRNA MAFG-AS1
downregulated the expression of miR-6852 and thus increased the
proliferation, migration and invasion of HCC cells. These results
may improve our understanding of the molecular mechanisms
underlying development and prognosis of HCC, and lncRNA MAFG-AS1
may be a valuable therapeutic target for treating patients with
HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the author on reasonable request.
Authors' contributions
HOY contributed to the conception and design of the
present study. In addition, HOY analyzed and interpreted the
results and wrote the manuscript. LZ, ZX and SM performed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Akyuz M, Yazici P, Yigitbas H, Dural C,
Okoh A, Aliyev S, Aucejo F, Quintini C, Fung J and Berber E:
Oncologic results of laparoscopic liver resection for malignant
liver tumors. J Surg Oncol. 113:127–129. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bai L, Liu Z, Fang Q, Yan Q, Shi O, Bao P,
Mu L, Chen X and Zhang T: The trends and projections in the
incidence and mortality of liver cancer in urban Shanghai: A
population-based study from 1973 to 2020. Clin Epidemiol.
10:277–288. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang SW, Zheng RS, Li N, Zeng HM, Dai Z,
Zou XN and Chen WQ: Analysis and prediction of liver cancer
incidence in China. Zhonghua Yu Fang Yi Xue Za Zhi. 46:5872012.(In
Chinese). PubMed/NCBI
|
|
4
|
Fuchs J, Cavdar S, Blumenstock G, Ebinger
M, Schäfer JF, Sipos B and Warmann SW: POST-TEXT III and IV
hepatoblastoma: Extended hepatic resection avoids liver
transplantation in selected cases. Ann Surg. 266:318–323. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Min BS, Kim NK, Jeong HC and Chung HC:
High levels of serum VEGF and TIMP-1 are correlated with colon
cancer liver metastasis and intrahepatic recurrence after liver
resection. Oncol Lett. 4:123–130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sadelain M, Brentjens R, Davila M, Riviere
I, Wang X, Bartido S, Park J, Bouhassira D, Curran K, Chung S, et
al: Abstract CT102: Efficacy and toxicity management of 19–28z CAR
T cell therapy in B cell acute lymphoblastic leukemia. Cancer Res.
74:CT1022014.
|
|
7
|
Li H, He Z, Gu Y, Fang L and Lv X:
Prioritization of non-coding disease-causing variants and long
non-coding RNAs in liver cancer. Oncol Lett. 12:39872016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma Z, Huang H, Xu Y, He X, Wang J, Hui B,
Ji H, Zhou J and Wang K: Current advances of long non-coding RNA
highly upregulated in liver cancer in human tumors. Onco Targets
Ther. 10:4711–4717. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu YC, Liang CJ, Zhang DX, Li GQ, Gao X,
Fu JZ, Xia F, Ji JJ, Zhang LJ, Li GM and Wu JX: LncSHRG promotes
hepatocellular carcinoma progression by activating HES6.
Oncotarget. 8:70630–70641. 2017.PubMed/NCBI
|
|
11
|
Huang G, Jiang H, Lin Y, Wu Y, Cai W, Shi
B, Luo Y, Jian Z and Zhou X: lncAKHE enhances cell growth and
migration in hepatocellular carcinoma via activation of NOTCH2
signaling. Cell Death Dis. 9:4872018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Wang H, Zhang Y, Zhen N, Zhang L,
Qiao Y, Weng W, Liu X, Ma L, Xiao W, et al: Mutual inhibition
between YAP and SRSF1 maintains long non-coding RNA, Malat1-induced
tumourigenesis in liver cancer. Cell Signal. 26:1048–1059. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang H, Ma Q, Yang T, Zhang T and Zhou Y:
The expression and function of lncRNA MAFG-AS1 in osteosarcoma. J
Modern Oncol. 3536–3540. 2018.
|
|
14
|
Cui S, Yang X, Zhang L, Zhao Y and Yan W:
LncRNA MAFG-AS1 promotes the progression of colorectal cancer by
sponging miR-147b and activation of NDUFA4. Biochem Biophys Res
Commun. 506:251–258. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jia YC, Wang JY, Liu YY, Li B, Guo H and
Zang AM: LncRNA MAFG-AS1 facilitates the migration and invasion of
NSCLC cell via sponging miR-339-5p from MMP15. Cell Biol Int.
43:384–393. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ye B, Hu B, Zheng Z, Zheng R and Shi Y:
The long non-coding RNA AK023948 enhances tumor progression in
hepatocellular carcinoma. Exp Ther Med. 14:3658–3664. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shin I, Kim S, Song H, Kim HR and Moon A:
H-Ras-specific activation of Rac-MKK3/6-p38 pathway: Its critical
role in invasion and migration of breast epithelial cells. J Biol
Chem. 280:14675–14683. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu P, Liu Z, Zhou J and Chen Y: Tanshinol
inhibits the growth, migration and invasion of hepatocellular
carcinoma cells via regulating the PI3K-AKT signaling pathway. Onco
Targets Ther. 12:87–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Z, Yu C, Zhan L, Pan Y, Chen L and
Sun C: LncRNA CRNDE promotes hepatic carcinoma cell proliferation,
migration and invasion by suppressing miR-384. Am J Cancer Res.
6:2299–2309. 2016.PubMed/NCBI
|
|
21
|
She K, Huang J, Zhou H, Huang T, Chen G
and He J: lncRNA-SNHG7 promotes the proliferation, migration and
invasion and inhibits apoptosis of lung cancer cells by enhancing
the FAIM2 expression. Oncol Rep. 36:26732016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han F, Wang C, Wang Y and Zhang L: Long
noncoding RNA ATB promotes osteosarcoma cell proliferation,
migration and invasion by suppressing miR-200s. Am J Cancer Res.
7:770–783. 2017.PubMed/NCBI
|
|
23
|
Zhang Y, Tao Y, Li Y, Zhao J, Zhang L,
Zhang X, Dong C, Xie Y, Dai X, Zhang X and Liao Q: The regulatory
network analysis of long noncoding RNAs in human colorectal cancer.
Funct Integr Genomics. 18:261–275. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Poudyal D, Herman A, Adelsberger JW, Yang
J, Hu X, Chen Q, Bosche M, Sherman BT and Imamichi T: A novel
microRNA, hsa-miR-6852 differentially regulated by Interleukin-27
induces necrosis in cervical cancer cells by downregulating the
FoxM1 expression. Sci Rep. 8:9002018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kopanja D, Pandey A, Kiefer M, Wang Z,
Chandan N, Carr JR, Franks R, Yu DY, Guzman G, Maker A and
Raychaudhuri P: Essential roles of FoxM1 in Ras-induced liver
cancer progression and in cancer cells with stem cell features. J
Hepatol. 63:429–436. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cui BH and Hong X: miR-6852 serves as a
prognostic biomarker in colorectal cancer and inhibits tumor growth
and metastasis by targeting TCF7. Exp Ther Med. 16:879–885.
2018.PubMed/NCBI
|
|
27
|
Yu H, Zhang J, Wen Q, Dai Y, Zhang W, Li F
and Li J: MicroRNA-6852 suppresses cell proliferation and invasion
via targeting forkhead box J1 in gastric cancer. Exp Ther Med.
16:3249–3255. 2018.PubMed/NCBI
|
|
28
|
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J,
Yan X, Ye B, Li C, Xia P, et al: The long noncoding RNA lncTCF7
promotes self-renewal of human liver cancer stem cells through
activation of Wnt signaling. Cell Stem Cell. 16:413–425. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Collins A: The Cancer Genome Atlas (TCGA)
pilot project. Cancer Res. 67:2007.
|
|
30
|
Huang Z, Duan H and Li H: TCGA4U: A
web-based genomic analysis platform to explore and mine TCGA
genomic data for translational research. Stud Health Technol
Inform. 216:658–662. 2015.PubMed/NCBI
|
|
31
|
Levine D and Mardis E; T. Cancer Genome
Atlas Research Network, : Late-breaking abstract 4: The Cancer
Genome Atlas (TCGA) project on endometrial carcinoma: Initial data
and preliminary genomic analysis. Gynecol Oncol. 125:7722012.
View Article : Google Scholar
|