Introduction
Diabetes, including type 1 and type 2 diabetes
mellitus, is one of the most serious health-threatening diseases
worldwide. Transplantation of pancreatic β-cells (pβCs), which are
responsible for producing insulin, is an effective strategy for the
treatment of type 1 and type 2 diabetes (1). However, the availability of pβCs is
limited and therefore, the production of more pβCs is urgently
required. The technology of differentiating stem cells into certain
cell types holds great promise. Previous studies have focused on
inducing embryonic stem cells (ESCs) to differentiate into
pancreatic β-like cells (pβLCs) (2).
However, due to several concerns, including ethics, immunological
rejection and tumorigenesis potential, the suitability and safety
of pβLCs derived from ESCs remain under debate (3).
Human umbilical cord mesenchymal stem cells (HuMSCs)
isolated from human umbilical cord express the surface markers of
stem cells, including CD29, CD44, CD59 and CD105 (4,5). Studies
from our and other groups have demonstrated that HuMSCs were able
to be induced into several types of cell, including osteocytes,
chondrocytes, lipocytes, neuron-like cells, cardiomyocyte-like
cells and spermatogonium-like cells (6–13).
Furthermore, a previous study by our group indicated that
vector-mediated overexpression of pancreatic and duodenal homeobox
1 (Pdx1), MAF bZIP transcription factor A (MafA) and neurogenin 3
(Ngn3) in HuMSC significantly promoted the expression of pancreatic
genes (13). Therefore, HuMSCs have
multi-lineage differentiation potential and may be used for
generating pβLCs.
The differentiation of stem cells is tightly
regulated by the temporal and spatial expression of transcription
factors; in other words, stem cell differentiation is able to be
determined by the transcription factors introduced. In addition to
stem cells, terminally differentiated cells may be reprogramed to
other cell types (14). Numerous
cell types have been trans-differentiated into pβLCs by forcing the
expression of three transcription factors, Pdx1, Ngn3 and MafA,
which have important roles in promoting the development of the
pancreas (15–17). Pax4 is also an indispensable
transcription factor for the generation, differentiation,
development and survival of pβCs, evidenced by the observation that
Pax4-knockout mice lack a pancreas and die 1–2 days after birth
(18). Recent studies highlight the
important role of the Pax4 gene in driving the formation of pβLCs
from other cell types, including pancreatic δ- and α-cells
(19,20). Based on these results, it may be
proposed that the Pax4 gene synergistically acts with the
transcription factors Pdx1, Ngn3 and MafA to promote the
development of pβCs. To this end, the present study aimed to
determine whether co-transfecting the above-mentioned 4
transcription factors in HuMSC increases the efficiency of pβLC
formation.
In the present study, HuMSCs were cultured and
co-transfected with Pdx1, Ngn3, MafA and Pax4, and the function of
the resulting pβLCs was assessed. It was demonstrated that Pax4
synergistically acts with Pdx1, Ngn3 and MafA to enhance the
differentiation of HuMSCs to pβLCs.
Patients and methods
Plasmids, human umbilical cord and
reagents
Adenovirus plasmid carrying Pdx1, Ngn3 and MafA
(Ad-3F) and Pax4 (Ad-Pax4) were kindly provided by Professor Chiju
Wei from the Multidisciplinary Research Center of Shantou
University (Shantou, China) and were originally obtained from the
Beta Cell Biology Consortium. Adenovirus plasmid carrying only EGFP
was used as a transfection control.
The umbilical cord was obtained from normal
full-term pregnant women with cesarean section at the Second
Affiliated Hospital of Shantou University (Shantou, China).
Maternal donors and their families provided written informed
consent and the present study was approved by the ethics committee
of Shanghai Children's Hospital (Shanghai, China; ethical approval
no. SHMC201707561) and the ethics committee of the Second
Affiliated Hospital of Shantou University Medical College (Shantou,
China).
H-Dulbecco's modified Eagle's medium (DMEM)/F12 and
fetal bovine serum (FBS) were purchased from Gibco (Thermo Fisher
Scientific, Inc.); human epidermal growth factor (EGF) and human
basic fibroblast growth factor (bFGF) were purchased from
Invitrogen (Thermo Fisher Scientific, Inc.); goat anti-human
insulin polyclonal antibody (cat. no. sc-7839) was obtained from
Santa Cruz Biotechnology; mouse anti-human glucagon monoclonal
antibody (cat. no. BM1621) was from Boster Biotechnology; rabbit
anti-human C-peptide polyclonal antibody (cat. no. ab82696) was
procured from Abcam; cyanine 3 (CY3)-labeled donkey anti-goat IgG
(cat. no. A0502) was purchased from Beyotime Institute of
Biotechnology; CY3-labeled goat anti-mouse IgG (cat. no. AP181C)
was obtained from Sigma-Aldrich (Merck KGaA); and horseradish
peroxidase-labeled goat anti-rabbit IgG (cat. no. G-21234) was from
Molecular Probes (Thermo Fisher Scientific, Inc.). Primers were
synthesized by Nanjing Kingsley Biotechnology Co., Ltd. The kit for
reverse transcription (RT; HiScript II Q Select RT SuperMix for
qPCR; cat. no. R233-01) and real-time PCR (AceQ qPCR SYBR Green
Master Mix; cat. no. Q141-02) was purchased from Vazyme
Biotechnology Co. Human insulin ELISA kit (cat. no. EZHI-14K) was
purchased from Sigma-Aldrich (Merck KGaA).
HuMSC culture
Umbilical cords of healthy fetuses from full-term
pregnancies were obtained and washed with PBS. Amniotic membrane
and blood vessels were removed and the remaining tissue was cut
into pieces (1.0 mm3). The pieces were cultured in
DMEM/F12 containing 10% FBS, EGF (5 ng/ml) and Bfgf (5 ng/ml), and
maintained at 37°C with 5% CO2 in a humidified
atmosphere. The culture medium was replaced every 2 days. When the
cells reached 80–90% confluence, the cells were digested with 0.25%
trypsin-EDTA (Sigma-Aldrich; Merck KGaA) and passaged.
Cell transfection
HuMSCs were seeded into 12-well plates at a density
of 2×105 cells/well. After culture for 12 h, Ad-3F
and/or Ad-Pxa4 (2 µl; multiplicity of infection, 10) were added to
the respective wells and served as the 3F, Pax4 or 3F/Pax4 group.
After incubation for 12 h, the cells were washed three times with
PBS and cultured in H-DMEM containing 1% FBS for 4 days. The
culture medium was replaced every 2 days.
Immunofluorescence
HuMSCs of the 4th generation were digested and
seeded in a 96-well plate. After reaching 60–70% confluence, the
cells were transfected with various Ad plasmids as mentioned above.
The cells were fixed with 4% paraformaldehyde at room temperature
for 5 min and permeabilized with Triton X-100 at room temperature
for 30 min. Subsequently, cells were incubated with primary
antibodies, including goat anti-human insulin (dilution, 1:100),
mouse anti-human glucagon (dilution, 1:100) and rabbit anti-human
C-peptide (dilution, 1:100) at 37°C for 2 h. Cells were then
incubated with CY3-labeled donkey anti-goat IgG (dilution,
1:1,000), CY3-labeled goat anti-mouse IgG (dilution, 1:1,000) and
CY3-labeled goat anti-rabbit IgG (dilution, 1:1,000) at room
temperature for 1 h in the dark. Following staining with DAPI, the
fluorescence was examined with a fluorescence microscope (DMi8
Fluorescence Imaging; Leica Microsystems GmbH). The percentage of
insulin-, C-peptide- or glucagon-positive cells was calculated as
the ratio of positive cells among total cells. The percentage of
insulin- or C-peptide-positive cells was used to evaluate the
differentiation efficiency of HuMSCs into pβCs.
RT-quantitative (q)PCR
The total RNA was extracted with a TRIzol kit
(Invitrogen; Thermo Fisher Scientific, Inc.). The total RNA was
used as template to synthesize complementary (c)DNA by using the
commercial RT kit. The expression of target genes was detected by
qPCR. The thermocycling protocol was as follows: Denaturation at
95°C for 1 min; followed by 40 cycles of 95°C for 15 sec, 60°C for
15 sec and 72°C for 45 sec, and then a final elongation at 72°C for
10 min. RT-qPCR was performed in a 7300 Real-time PCR instrument
(Applied Biosystems; Thermo Fisher Scientific, Inc.) and
standardized by the value of β-actin in the same sample. Data were
quantified by using the 2−ΔΔCq method (21). The sequences of primers were as
follows: PDX1 (GenBank ID, NM_000209.4; product length, 193 bp;
forward (F)-5′-ATCTCCCCATACGAAGTGCC-3′, reverse
(R)-5′-CGTGAGCTTTGGTGGATTTCAT-3′; NGN3 (XM_017016280.1; product
length, 86 bp; F-5′-CTAAGAGCGAGTTGGCACTGA-3′,
R-5′-GAGGTTGTGCATTCGATTGCG-3′); MAFA (NM_201589.4; product length,
150 bp; F-5′-GTACAGGACGTGGACACCAG-3′,
R-5′-AATCACCGTTCTCCGCTCAA-3′); PAX4 (NM_001366111.1; product
length, 127 bp; F-5′-ATACCCGGCAGCAGATTGTG-3′,
R-5′-AAGACACCTGTGCGGTAGTAA-3′); glucagon (GCG; NM_002054.5; product
length, 108 bp; F-5′-AGCTGCCTTGTACCAGCATT-3′,
R-5′-TGCTCTCTCTTCACCTGCTC-3′); insulin (INS; NM_001291897.1;
product length, 130 bp; F-5′-CAATGCCACGCTTCTGC-3′,
R-5′-TTCTACACACCCAAGACCCG-3′); neuronal differentiation 1 (NEUROD1;
NM_002500.4; product length, 92 bp; F-5′-ATCAGCCCACTCTCGCTGTA-3′,
R-5′-GCCCCAGGGTTATGAGACTAT-3′); NK6 homeobox 1 (NKX6.1;
NM_006168.2; product length, 106 bp; F-5′-CGAGTCCTGCTTCTTCTTGG-3′,
R-5′-GGGGATGACAGAGAGTCAGG-3′); ISL LIM homeobox 1 (ISL1;
NM_002202.3; product length, 108 bp; F-5′-TCACGAAGTCGTTCTTGCTG-3′,
R-5′-CATGCTTTGTTAGGGATGGG-3′); PAX6 (NM_001310161.1; product
length, 101 bp; F-5′-TCCGTTGGAACTGATGGAGT-3′,
R-5′-GTTGGTATCCGGGGACTTC-3′); SLC2A2 (NM_001278658.1; product
length, 93 bp; F-5′-GACAGTGAAAACCAGGGTCC-3′,
R-5′-TGTGCCACACTCACACAAGA-3′); glucokinase (GCK; NM_000162.5;
product length, 97 bp; F-5′-CCTTCTTCAGGTCCTCCTCC-3′,
R-5′-ATGCTGGACGACAGAGCC-3′); PCSK1 (NM_000439.5; product length,
101 bp; F-5′-CGGGTCATACTCAGAGGTCC-3′, R-5′-CTCTGGCTGCTGGCATCT-3′);
proprotein convertase subtilisin/kexin type 2 (PCSK2; NM_002594.5;
product length, 99 bp; F-5′-TTTCGGTCAAATCCTTCCTG-3′,
R-5′-TGCAAAGGCCAAGAGAAGAC-3′); β-ACTIN (NM_001101.5; product
length, 93 bp; F-5′-GTTGTCGACGACGAGC-3′,
R-5′-GCACAGAGCCTCGCCTT-3′).
ELISA
Transfected cells were incubated with Krebs-Ringer
buffer (KRB) medium containing 2.8 or 20 mM glucose (Beijing
Maichen Science and Technology) at 37°C for 3 h. The KRB medium was
collected for detection of secreted insulin. The remaining cells
were scraped off in 35% ethanol solution containing 0.18 M
hydrochloric acid. The cells were then homogenized by ultrasound
(20 kHZ, ultrasound utilized for 10 sec with 60 sec intervals. The
procedure was repeated 6 times) and put under vortex in a shaker
(100 times/min) at 4°C overnight. The homogenate was centrifuged at
300 × g for 30 min at 4°C. The supernatant was collected to
determine the intracellular insulin content and insulin secretion
was normalized to it. The insulin assay was performed by using the
commercial ELISA kit.
Statistical analysis
Values are expressed as the mean ± standard
deviation. Student's t-test was used to determine the statistical
significance of differences between two groups, and for more than
two groups, one-way analysis of variance followed by a
least-significant difference post-hoc test was used. P<0.05 was
considered to indicate statistical significance. All data were
analyzed by SPSS 20.0 (IBM Corp.).
Results
Morphology of cultured HuMSCs and cell
transfection
After the pieces of umbilical cord tissues had been
cultured for 5–7 days, cells that migrated out from the surrounding
tissues were fibroblast-like (Fig.
1A). After culture for 10–14 days, the cells reached 80–90%
confluence and were passaged (Fig.
1B). After the second generation, the cells grew rapidly and
were subjected to subculture once every 3–5 days. In the 3rd-5th
generation, cells had a stable fibroblast-like morphology (Fig. 1C and D), and a previous study by our
group demonstrated that these cells highly expressed adult stem
cell markers, including CD29, CD44 and CD59 (2). However, after the 10th generation, the
cells tended to grow slowly and become wide. Therefore, cells in
the 3rd-5th generation were used in the present study. A diagram of
the Ad-3F and Ad-Pax4 construct is presented in Fig. 1E. The results of EGFP
immunofluorescence revealed that the adenovirus plasmid
transfection efficiency was >80% (Fig. 1F-I). Furthermore, the results from
RT-qPCR suggested that the transfected genes were efficiently
expressed in HuMSCs (Fig. 1J).
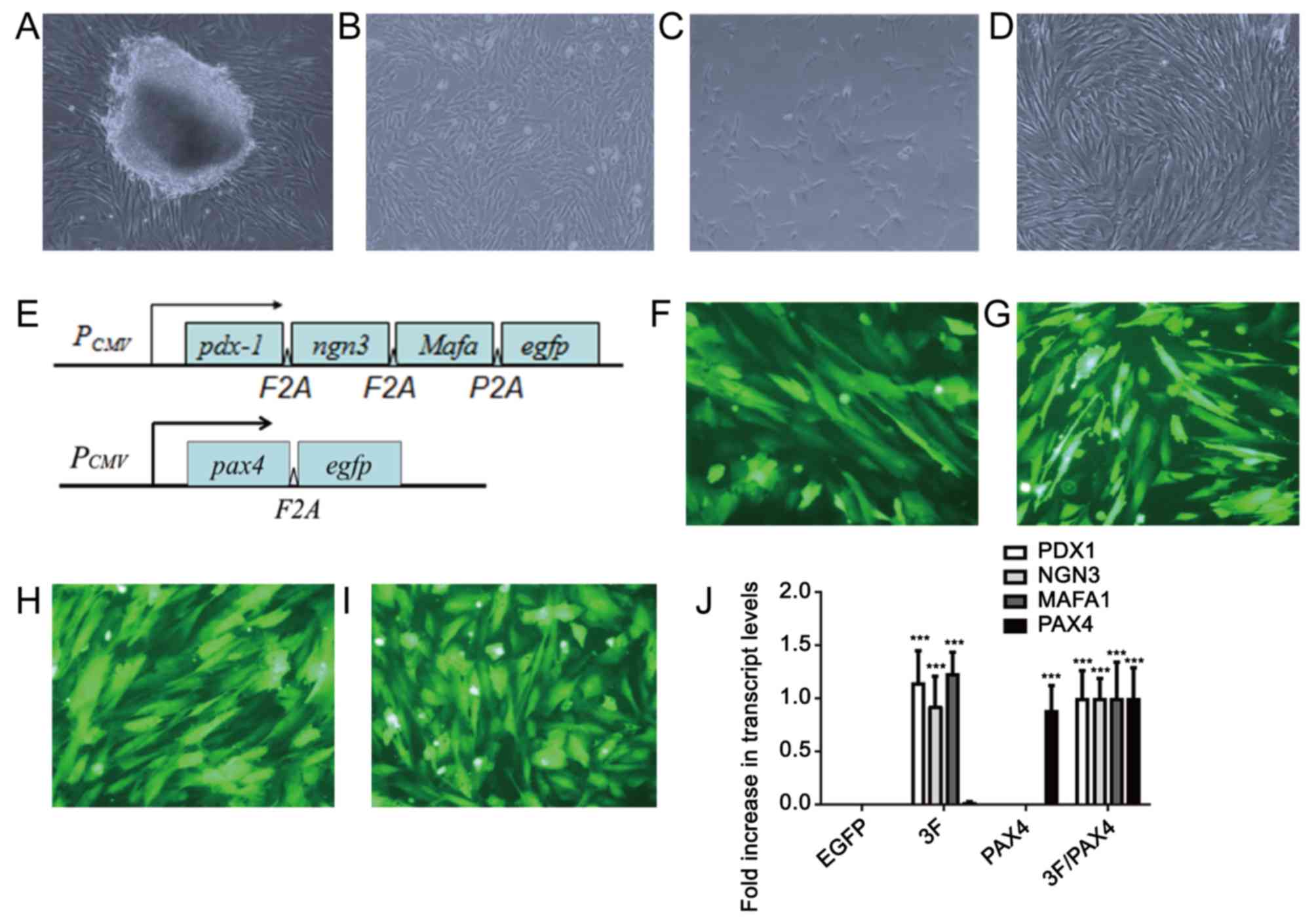 | Figure 1.Morphology of cultured HuMSCs and cell
transfection. (A) Primary culture of HuMSCs at day 7. Fibroblasts
of HuMSCs migrated out of the tissue block. (B-D) Fibroblast-like
HuMSCs of the (B) 1st, (C) 3rd and (D) 5th passage (magnification,
×100). (E) Diagram of the Ad-3F and Ad-Pax4 constructs. (F-I) EGFP
immunofluorescence of HuMSCs transfected with (F) Ad-EGFP, (G)
Ad-Pax4, (H) Ad-3F and (I) Ad-Pxa4/Ad-3F at 48 h (magnification,
×200). (J) Expression of genes, including PDX1, NGN3, MAFA and
PAX4, in HuMSCs transfected with Ad-EGFP, Ad-Pax4, Ad-3F and
Ad-Pax4/Ad-3F at 48 h. ***P<0.001 vs. EGFP. Pdx1, pancreatic and
duodenal homeobox 1; MafA, MAF bZIP transcription factor A; Ngn3,
neurogenin 3; Pax4, paired box 4; Ad-Pax4, adenovirus expressing
Pax4; Ad-3F, Ad-Pdx1/MafA/Ngn3; HuMSCs, human umbilical cord
mesenchymal stem cells; PCMV, cytomegalovirus plasmid;
EGFP, enhanced green fluorescence plasmid. |
Effect of Ad-3F in combination with Ad-Pax4 on the
differentiation of HuMSCs to pβLCs by assessing crucial genes
associated with differentiation. To investigate whether Ad-Pax4
further promotes the differentiation of Ad-3F-transfected HuMSCs to
pβLCs, a panel of genes involved in converting the progenitors of
pβCs into pβCs were detected, including NKX6-1, SLC2A2, GCK, PCSK1,
NEUROD1, ISL1, PAX6 and PCSK2. The results demonstrated that
compared with that in the control group, transfection with Ad-Pax4
alone had little effect on the expression of the above-mentioned
genes, while transfection with Ad-3F or Ad-3F/Pax-4 increased the
expression of these genes. Of note, Ad-Pax4 further promoted the
expression of NKX6-1, SLC2A2, GCK and PCSK1 in Ad-3F-transfected
cells (Fig. 2).
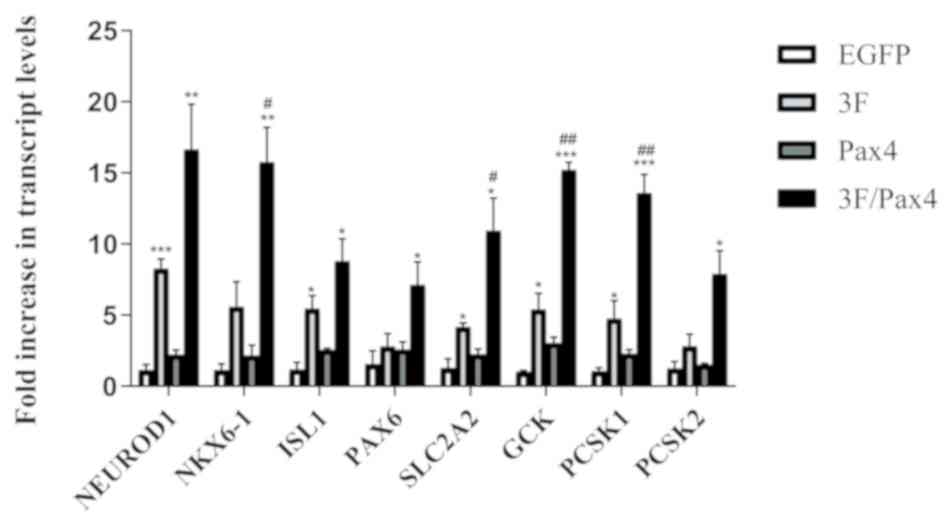 | Figure 2.Effect of 3F plasmid in combination
with Pax4 overexpression on the expression of genes associated with
the differentiation of human umbilical cord mesenchymal stem cells
to pancreatic β-like cells. The genes assessed included NKX6-1,
SLC2A2, GCK, PCSK1, NEUROD1, ISL1, PAX6 and PCSK2. Values are
expressed as the mean ± standard deviation (n=5). *P<0.05,
**P<0.01, ***P<0.001 vs. EGFP group; #P<0.05,
##P<0.01 vs. 3F group. Groups: EGFP, cells
transfected with EGFP plasmid; 3F, cells transfected with
Pdx1/MafA/Ngn3 plasmid; Pax4, cells transfected with Pax4 plasmid;
3F/Pax4, cells transfected with Pdx1/MafA/Ngn3 plasmid and Pax4
plasmid. Pdx1, pancreatic and duodenal homeobox 1; MafA, MAF bZIP
transcription factor A; Ngn3, neurogenin 3; Pax4, paired box 4;
EGFP, enhanced green fluorescence plasmid; GCK, glucokinase;
NKx6.1, NK6 homeobox 1; SLC2A2, glucokinase; PCSK1, proprotein
convertase subtilisin/kexin type 1; NEUROD1, neuronal
differentiation 1; ISL1, ISL LIM homeobox 1; PCSK2, proprotein
convertase subtilisin/kexin type 2. |
Effect of Ad-3F in combination with Ad-Pax4 on the
expression of C-peptide, insulin and glucagon. To investigate the
effect of Ad-3F in combination with Ad-Pax4 on the expression of
C-peptide, insulin and glucagon, RT-qPCR and immunofluorescence
analyses were performed. The RT-qPCR results indicated that
compared with that in the control group, the gene expression of INS
and GCG was significantly enhanced in the Ad-3F and Ad-3F/Pax-4
group (Fig. 3A and B). Compared with
that in the Ad-3F group, the gene expression of INS was
significantly increased, while that of GCG was significantly
reduced in the Ad-3F/Pax-4 group (Fig.
3A and B). The immunofluorescence results indicated that
compared with those in the control group, the glucagon-, insulin-
and C-peptide-positive cells were significantly increased in the
Ad-3F and Ad-3F/Pax4 groups (Fig.
3B-H). Compared with those in the Ad-3F group, insulin- and
C-peptide-positive cells were significantly increased, while
glucagon-positive cells were significantly reduced in Ad-3F/Pax4
group (Fig. 3B-H). Furthermore, the
differentiation efficiency of HuMSCs co-transfected with Ad-3F and
Ad-PAX4 into pβLCs was 30–40%, as reflected by the insulin- or
C-peptide-positive cells (Fig. 3C, D, F
and G).
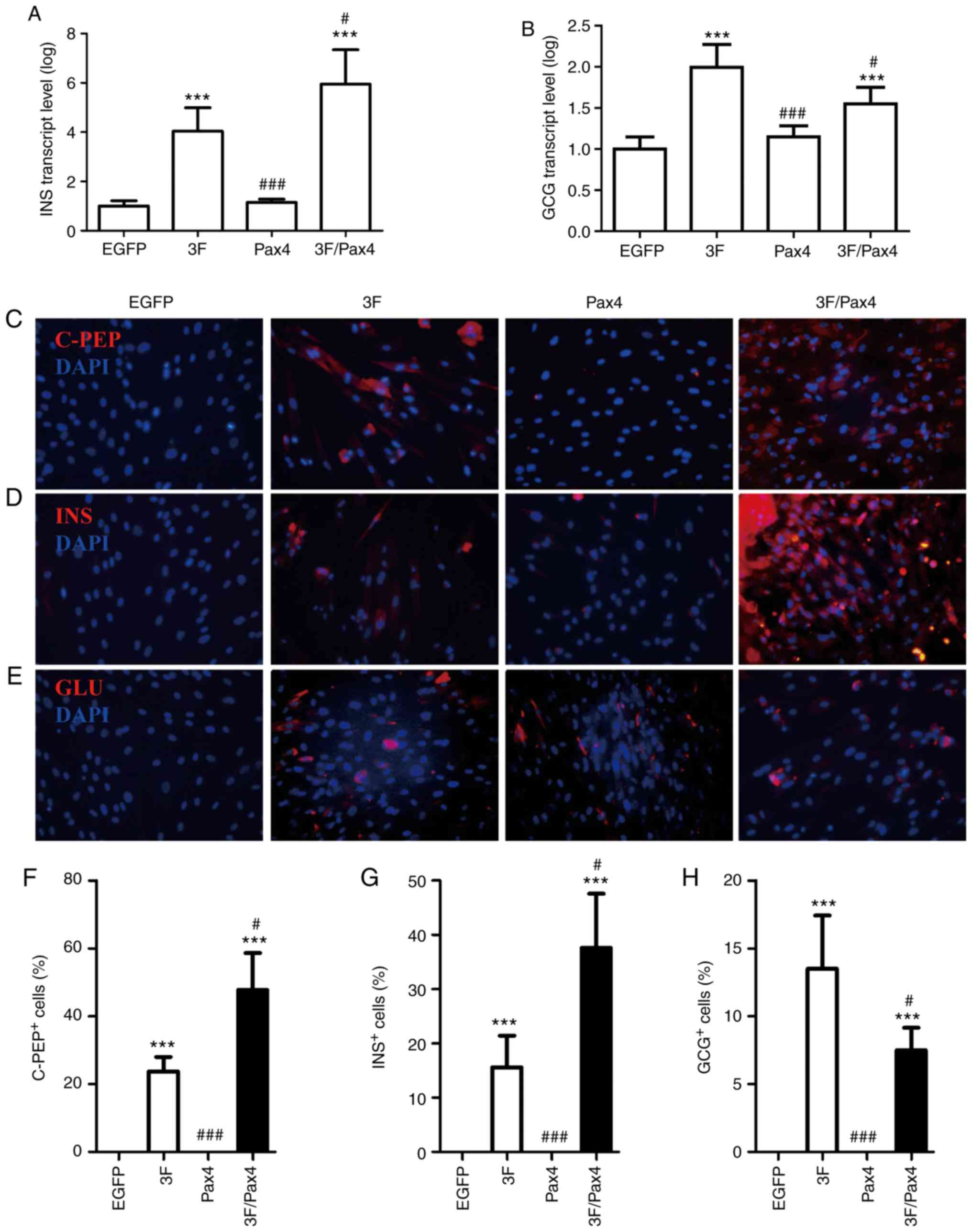 | Figure 3.Effect of 3F plasmid in combination
with Pax4 overexpression on the expression of C-PEP, INS and
glucagon. (A and B) Gene expression of (A) INS and (B) GCG in the
different groups compared with the EGFP group. (C-E)
Immunofluorescence detection of (C) C-PEP (red), (D) INS and (E)
GCG (red) (magnification, ×200). (F-H) Quantification of (F) C-PEP-
(G) INS- and (H) GCG-positive cells from C-E, respectively. Values
are expressed as the mean ± standard deviation (n=5). ***P<0.001
vs. EGFP group; #P<0.05, ###P<0.001 vs.
3F group. Groups: EGFP, cells transfected with EGFP plasmid; 3F,
cells transfected with Pdx1/MafA/Ngn3 plasmid; Pax4, cells
transfected with Pax4 plasmid; 3F/Pax4, cells transfected with
Pdx1/MafA/Ngn3 plasmid and Pax4 plasmid. Pdx1, pancreatic and
duodenal homeobox 1; MafA, MAF bZIP transcription factor A; Ngn3,
neurogenin 3; Pax4, paired box 4; EGFP, enhanced green fluorescence
plasmid; C-PEP, C-peptide; INS, insulin; GCG, glucagon. |
Effect of Ad-3F in combination with Ad-Pax4 on
glucose-stimulated insulin secretion (GSIS). To investigate the
effect of Ad-3F in combination with Ad-Pax4 on GSIS, the cells were
stimulated with low (2.8 mM) and high (20 mM) glucose. The results
suggested that compared with low glucose, the insulin secretion in
cells stimulated with high glucose was significantly increased in
the Ad-3F and Ad-3F/Pax4 groups (Fig.
4). Compared with that in the 3F group, the insulin secretion
was significantly increased in the Ad-3F/Pax-4 group (Fig. 4).
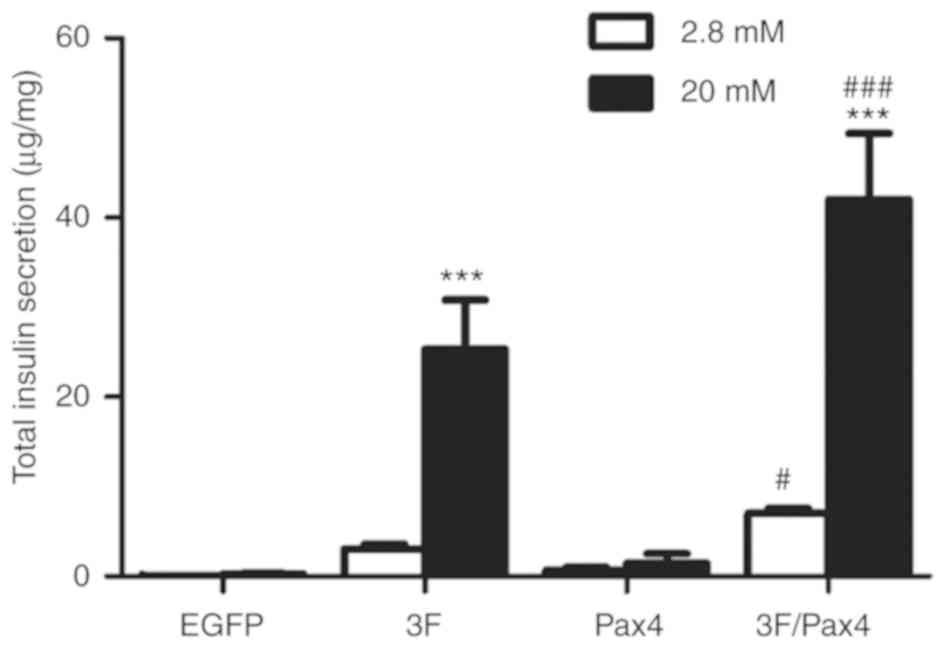 | Figure 4.Effect of 3F plasmid in combination
with Pax4 overexpression on insulin secretion in response to
glucose. Transfected cells were stimulated either with low-glucose
or high-glucose, and insulin secretion was determined by ELISA.
Experiments were performed 4 days after transduction. Values are
expressed as the mean ± standard deviation (n=5). ***P<0.001 vs.
low-glucose group; #P<0.05, ###P<0.001
vs. 3F group. Groups: EGFP, cells transfected with EGFP plasmid;
3F, cells transfected with Pdx1/MafA/Ngn3 plasmid; Pax4, cells
transfected with Pax4 plasmid; 3F/Pax4, cells transfected with
Pdx1/MafA/Ngn3 plasmid and Pax4 plasmid. Pdx1, pancreatic and
duodenal homeobox 1; MafA, MAF bZIP transcription factor A; Ngn3,
neurogenin 3; Pax4, paired box 4; EGFP, enhanced green fluorescence
plasmid. |
Discussion
In the present study, cultured HuMSCs were
successfully differentiated into functional pβLCs by introducing 4
transcription factors, Pdx1, Ngn3, MafA and Pax4. The results
demonstrated that, compared to Ad-3F-transfected cells, those
co-transfected with Ad-Pxa4 and Ad-3F expressed higher levels of
insulin, c-peptide and genes expressed in pancreatic β-precursor
cells, and secreted more insulin in response to glucose.
Furthermore, Ad-Pax4 significantly decreased the expression of
glucagon in Ad-3F-transfected HuMSC.
The embryonic pancreas development is regulated by
the interactions of numerous signaling pathways that contribute to
the proper initiation of the sequential expression of transcription
factors (22–25). Several transcription factors have
been demonstrated to be necessary for the development of a
functional pancreas, including Pdx1, Ngn3 and MafA. Pdx1, a member
of the ParaHox protein family, has an important role in driving the
formation of the pancreatic bud (26,27).
Ngn3, a member of the basic helix-loop-helix transcription factors,
is required for the formation of a common precursor for the four
pancreatic endocrine cell types. MafA, an eye-specific member of
the Maf family, has been demonstrated to have a vital role in
regulating insulin gene expression (28–30). It
has been previously reported that the 3 transcription factors
combined were able to reprogram other cell types into functional
pβLCs (17). Pax4 has been
demonstrated to have a critical role in converting the pancreatic
β-precursors to pancreatic endocrine cell types (31). In a previous study, exocrine tissue
was effectively converted to islet-like cells by using a cocktail
of transcription factors, Pdx1, Ngn3, MafA and Pax4 (32). Consistent with that, the present
study indicated that overexpression of Pax4 increased the
expression of INS, C-PEP and genes expressed in pancreatic
β-precursor cells, and enhanced glucose-stimulated insulin
secretion (GSIS) in Ad-3F transfected HuMSCs, indicating a
synergetic effect of Pax4 and Pdx1/Ngn3/MafA on converting HuMSCs
to functional pβLCs.
The role of the transcriptional factors Ngn3, Pdx1,
MafA and Pax4 in controlling the development of a functional
pancreas has been well established. Ngn3 facilitates the formation
of pancreatic endocrine cells. Pax4 directs the cell
differentiation towards pβCs. Pdx1 and MafA are necessary for
maintaining mature pβC function (31). The mechanism of Pax4 in promoting pβC
formation may rely on its role to suppress aristaless related
homeobox genes, and overexpression of Pax4 was sufficient to
convert pancreatic α-cells to β-cells (20,33). In
the present study, it was indicated that overexpression of Pax4
decreased glucagon expression in HuMSCs transfected with Ad-3F,
suggesting that Pax4 restricts the differentiation of HuMSCs into
paCs. However, glucagon-positive cells still existed in HuMSCs
co-transfected with Ad-3F and Ad-Pax4.
NKX6-1 has a key role in regulating insulin
secretion and β-cell proliferation (34). Overexpression of NKX6-1 in mature
β-cells enhanced proliferation and GSIS (35). It has been demonstrated that Glu2 was
involved in controlling insulin secretion (36,37).
GCK, a key regulator of glucose metabolism, is crucial for GSIS and
overexpression of GCK in β-cells restored GSIS in a mouse model of
high-fat diet-induced diabetes (38). PCSK1 is necessary for processing
pro-insulin (39). In the present
study, it was indicated that Pax4 promoted the expression of all
these genes in the Ad-3F-transfected cells. In line with this,
overexpression of Pax4 increased GSIS of HuMSCs transfected with
Ad-3F. These results indicate that Pax4 synergistically acts with
Pdx1, Ngn3 and MafA to promote the expression of these genes,
thereby improving GSID. However, the underlying mechanisms remain
to be elucidated.
In conclusion, the present study demonstrated that
the Pax4 gene was able to synergistically act with the
transcription factors Pdx1, Ngn3 and MafA to convert HuMSCs to
functional pβCs. HuMSCs may be potential seed cells for generating
functional pβCs for the therapy of diabetes.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81671525 and
81070478), the Science and Technology Project from the Science
Technology and Innovation Committee of Shenzhen Municipality (grant
no. JCYJ20170817170110940) and the Sanming Project of Medicine in
Shenzhen (grant no. SZSM201512033).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
TZ, JS and LM designed the study and performed the
experiments. TZ, HW, TW and JY collected the data. CW, HJ and SJ
analyzed the data. TZ and HW prepared the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the ethics committees of
Shanghai Children's Hospital (Shanghai, China) and of the Second
Affiliated Hospital of Shantou University Medical College (Shantou,
China). Written informed consent was obtained from the maternal
donors and/or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Stanekzai J, Isenovic ER and Mousa SA:
Treatment options for diabetes: Potential role of stem cells.
Diabetes Res Clin Pract. 98:361–368. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bose B, Shenoy SP, Konda S and Wangikar P:
Human embryonic stem cell differentiation into insulin secreting
β-cells for diabetes. Cell Biol Int. 36:1013–1020. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fujikawa T, Oh SH, Pi L, Hatch HM, Shupe T
and Petersen BE: Teratoma formation leads to failure of treatment
for type I diabetes using embryonic stem cell-derived
insulin-producing cells. Am J Pathol. 166:1781–1791. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang H, Yang Y, Ho G, Lin X, Wu W, Li W,
Lin L, Feng X, Huo X, Jiang J, et al: Programming of human
umbilical cord mesenchymal stem cells in vitro to promote
pancreatic gene expression. Mol Med Rep. 8:769–774. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friedman R, Betancur M, Boissel L, Tuncer
H, Cetrulo C and Klingemann H: Umbilical cord mesenchymal stem
cells: Adjuvants for human cell transplantation. Biol Blood Marrow
Transplant. 13:1477–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sakamoto T, Ono H and Saito Y: Electron
microscopic histochemical studies on the localization of hyaluronic
acid in Wharton's jelly of the human umbilical cord. Nihon Sanka
Fujinka Gakkai Zasshi. 48:501–507. 1996.(In Japanese). PubMed/NCBI
|
|
7
|
Tsagias N, Koliakos I, Karagiannis V,
Eleftheriadou M and Koliakos GG: Isolation of mesenchymal stem
cells using the total length of umbilical cord for transplantation
purposes. Transfus Med. 21:253–261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Romanov YA, Svintsitskaya VA and Smirnov
VN: Searching for alternative sources of postnatal human
mesenchymal stem cells: Candidate MSC-like cells from umbilical
cord. Stem Cells. 21:105–110. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kadivar M, Khatami S, Mortazavi Y,
Shokrgozar MA, Taghikhani M and Soleimani M: In vitro
cardiomyogenic potential of human umbilical vein-derived
mesenchymal stem cells. Biochem Biophys Res Commun. 340:639–647.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma L, Feng XY, Cui BL, Law F, Jiang XW,
Yang LY, Xie QD and Huang TH: Human umbilical cord Wharton's
Jelly-derived mesenchymal stem cells differentiation into
nerve-like cells. Chin Med J (Engl). 118:1987–1993. 2005.PubMed/NCBI
|
|
11
|
Bao CS, Li XL, Liu L, Wang B, Yang FB and
Chen LG: Transplantation of Human umbilical cord mesenchymal stem
cells promotes functional recovery after spinal cord injury by
blocking the expression of IL-7. Eur Rev Med Pharmacol Sci.
22:6436–6447. 2018.PubMed/NCBI
|
|
12
|
Zhao Z, Chen Z, Zhao X, Pan F, Cai M, Wang
T, Zhang H, Lu JR and Lei M: Sphingosine-1-phosphate promotes the
differentiation of human umbilical cord mesenchymal stem cells into
cardiomyocytes under the designated culturing conditions. J Biomed
Sci. 18:372011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang P, Lin LM, Wu XY, Tang QL, Feng XY,
Lin GY, Lin X, Wang HW, Huang TH and Ma L: Differentiation of human
umbilical cord Wharton's jelly-derived mesenchymal stem cells into
germ-like cells in vitro. J Cell Biochem. 109:747–754.
2010.PubMed/NCBI
|
|
14
|
Zhou Q and Melton DA: Extreme makeover:
Converting one cell into another. Cell Stem Cell. 3:382–388. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cavelti-Weder C, Zumsteg A, Li W and Zhou
Q: Reprogramming of pancreatic Acinar cells to functional beta
cells by in vivo transduction of a polycistronic construct
containing Pdx1, Ngn3, MafA in mice. Curr Protoc Stem Cell Biol.
40:A.10.1–4A.10.12. 2017.
|
|
16
|
Yamada T, Cavelti-Weder C, Caballero F,
Lysy PA, Guo L, Sharma A, Li W, Zhou Q, Bonner-Weir S and Weir GC:
Reprogramming mouse cells with a pancreatic duct phenotype to
insulin-producing β-like cells. Endocrinology. 156:2029–2038. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Akinci E, Banga A, Tungatt K, Segal J,
Eberhard D, Dutton JR and Slack JM: Reprogramming of various cell
types to a beta-like state by Pdx1, Ngn3 and MafA. PLoS One.
8:e824242013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sosa-Pineda B, Chowdhury K, Torres M,
Oliver G and Gruss P: The Pax4 gene is essential for
differentiation of insulin-producing beta cells in the mammalian
pancreas. Nature. 386:399–402. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Druelle N, Vieira A, Shabro A, Courtney M,
Mondin M, Rekima S, Napolitano T, Silvano S, Navarro-Sanz S, Hadzic
B, et al: Ectopic expression of Pax4 in pancreatic delta cells
results in beta-like cell neogenesis. J Cell Biol. 216:4299–4311.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Fava GE, Wang H, Mauvais-Jarvis
F, Fonseca VA and Wu H: PAX4 gene transfer induces α-to-β cell
phenotypic conversion and confers therapeutic benefits for diabetes
treatment. Mol Ther. 24:251–260. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hald J, Sprinkel AE, Ray M, Serup P,
Wright C and Madsen OD: Generation and characterization of Ptf1a
antiserum and localization of Ptf1a in relation to Nkx6.1 and Pdx1
during the earliest stages of mouse pancreas development. J
Histochem Cytochem. 56:587–595. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ding L, Han L, Li Y, Zhao J, He P and
Zhang W: Neurogenin 3-directed cre deletion of Tsc1 gene causes
pancreatic acinar carcinoma. Neoplasia. 16:909–917. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Babu DA, Chakrabarti SK, Garmey JC and
Mirmira RG: Pdx1 and BETA2/NeuroD1 participate in a transcriptional
complex that mediates short-range DNA looping at the insulin gene.
J Biol Chem. 283:8164–8172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang J, Elghazi L, Parker SE, Kizilocak H,
Asano M, Sussel L and Sosa-Pineda B: The concerted activities of
Pax4 and Nkx2.2 are essential to initiate pancreatic beta-cell
differentiation. Dev Biol. 266:178–189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ahlgren U, Jonsson J and Edlund H: The
morphogenesis of the pancreatic mesenchyme is uncoupled from that
of the pancreatic epithelium in IPF1/PDX1-deficient mice.
Development. 122:1409–1416. 1996.PubMed/NCBI
|
|
27
|
Guz Y, Montminy MR, Stein R, Leonard J,
Gamer LW, Wright CV and Teitelman G: Expression of murine STF-1, a
putative insulin gene transcription factor, in beta cells of
pancreas, duodenal epithelium and pancreatic exocrine and endocrine
progenitors during ontogeny. Development. 121:11–18.
1995.PubMed/NCBI
|
|
28
|
Gradwohl G, Dierich A, LeMeur M and
Guillemot F: neurogenin3 is required for the development of the
four endocrine cell lineages of the pancreas. Proc Natl Acad Sci
USA. 97:1607–1611. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kataoka K, Han SI, Shioda S, Hirai M,
Nishizawa M and Handa H: MafA is a glucose-regulated and pancreatic
beta-cell-specific transcriptional activator for the insulin gene.
J Biol Chem. 277:49903–49910. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sheets TP, Park KE, Park CH, Swift SM,
Powell A, Donovan DM and Telugu BP: Targeted mutation of NGN3 gene
disrupts pancreatic endocrine cell development in pigs. Sci Rep.
8:35822018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Napolitano T, Avolio F, Courtney M, Vieira
A, Druelle N, Ben-Othman N, Hadzic B, Navarro S and Collombat P:
Pax4 acts as a key player in pancreas development and plasticity.
Semin Cell Dev Biol. 44:107–114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lima MJ, Muir KR, Docherty HM, McGowan NW,
Forbes S, Heremans Y, Heimberg H, Casey J and Docherty K:
Generation of functional beta-like cells from human exocrine
pancreas. PLoS One. 11:e1562042016. View Article : Google Scholar
|
|
33
|
Collombat P, Hecksher-Sorensen J, Broccoli
V, Krull J, Ponte I, Mundiger T, Smith J, Gruss P, Serup P and
Mansouri A: The simultaneous loss of Arx and Pax4 genes promotes a
somatostatin-producing cell fate specification at the expense of
the alpha- and beta-cell lineages in the mouse endocrine pancreas.
Development. 132:2969–2980. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ray JD, Kener KB, Bitner BF, Wright BJ,
Ballard MS, Barrett EJ, Hill JT, Moss LG and Tessem JS:
Nkx6.1-mediated insulin secretion and β-cell proliferation is
dependent on upregulation of c-Fos. FEBS Lett. 590:1791–1803. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fueger PT, Schisler JC, Lu D, Babu DA,
Mirmira RG, Newgard CB and Hohmeier HE: Trefoil factor 3 stimulates
human and rodent pancreatic islet beta-cell replication with
retention of function. Mol Endocrinol. 22:1251–1259. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guillam MT, Dupraz P and Thorens B:
Glucose uptake, utilization and signaling in GLUT2-null islets.
Diabetes. 49:1485–1491. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Johnson JH, Ogawa A, Chen L, Orci L,
Newgard CB, Alam T and Unger RH: Underexpression of beta cell high
Km glucose transporters in noninsulin-dependent diabetes. Science.
250:546–549. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lu B, Kurmi K, Munoz-Gomez M, Jacobus
Ambuludi EJ, Tonne JM, Rakshit K, Hitosugi T, Kudva YC, Matveyenko
AV and Ikeda Y: Impaired β-cell glucokinase as an underlying
mechanism in diet-induced diabetes. Dis Model Mech. 11(pii):
dmm0333162018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vivoli M, Caulfield TR, Martinez-Mayorga
K, Johnson AT, Jiao GS and Lindberg I: Inhibition of prohormone
convertases PC1/3 and PC2 by 2,5-dideoxystreptamine derivatives.
Mol Pharmacol. 81:440–454. 2012. View Article : Google Scholar : PubMed/NCBI
|


















