Introduction
Gene therapy has become an increasingly important
strategy for treating various hepatic diseases (1). The liver is an important organ with
potential therapeutic targets, including cholesterol biosynthesis,
fibrosis, and hepatitis. For example, in hepatic gene therapy,
mutated genes that cause hepatic diseases can be replaced by the
transduction of mutation-corrected genes into hepatocytes. However,
a major requirement for hepatic gene therapy is the efficient
delivery of DNA into hepatocytes by systemic injection (2). Based on the type of vector used for
gene delivery, vectors can be divided into viral and non-viral
vectors (3,4). Non-viral vectors are less efficient but
safer than viral vectors; therefore, non-viral vectors are an
attractive alternative method for gene therapy. Cationic liposomes
are an example of a non-viral vector, and among them, cationic
liposomes composed of cationic lipids, such as
1,2-dioleoyl-3-trimethylammonium-propane (DOTAP), have often been
used for the in vivo transduction of plasmid DNA (pDNA)
(5,6). However, the positive charge of cationic
liposome/pDNA complexes (pDNA lipoplexes) leads to interactions
with albumin and other serum proteins (7), and the agglutinates contribute to high
entrapment of pDNA lipoplexes in the highly extended lung
capillaries (8), resulting in
expression mostly in the lung when injected intravenously.
Therefore, it is necessary to develop a delivery system to
efficiently target pDNA lipoplexes to the liver.
Receptor-mediated targeting is a promising approach
to deliver pDNA lipoplexes to hepatocytes. Hepatocytes express the
asialoglycoprotein receptor on their surface, which recognizes the
galactose residue of asialoglycoproteins. Therefore,
galactose-modified cationic liposomes have been utilized for
liver-targeting pDNA delivery (9).
Furthermore, hepatocytes play a key role in lipid and lipoprotein
metabolism, and some apolipoproteins, such as apolipoprotein B
(ApoB) and apolipoprotein E (ApoE), serve as ligands for the uptake
of lipoprotein by hepatocytes (10).
ApoE is a constituent of chylomicron, very low-density lipoprotein
(VLDL), low-density lipoprotein (LDL), and high-density lipoprotein
(11), and it is a high-affinity
ligand for several ApoE receptors such as LDL receptor, VLDL
receptor, and lipoprotein receptor-related protein 1 (LRP1), which
have been shown to be essential for hepatic clearance of VLDL and
remnant lipoprotein (12). It has
been reported that ApoE can bind to liposomes (13), and it mediated the uptake of
liposomes by hepatocytes in mice (14). Tamaru et al (15) found that recombinant human
ApoE3-modification increases the uptake of pDNA entrapped in
liposomes in neuroblastoma Neuro2a cells. However, recombinant ApoE
protein is too large (34 kDa) to use as a ligand for the
liver-targeting of pDNA lipoplexes. Previous studies showed that
dApoE peptide containing amino acid sequence 141–151 of human ApoE
in a tandem dimer (16,17) and ApoE fragment peptide (ApoE-F)
containing amino acid sequence 151–173 of human ApoE4 (18) bind to cells expressing the LDL
receptor. Therefore, we speculated that ApoE-derived
peptide-modified pDNA lipoplexes may improve pDNA delivery to the
liver after systemic injection.
In this study, we synthesized two types of
ApoE-derived peptide, dApoE-R9 and ApoE-F-R9, which included nine
arginine residues at the terminus of the peptides, for interaction
with pDNA, and evaluated transfection efficiency in vitro
and in vivo by ternary complexes with pDNA, cationic
liposomes, and ApoE-R9 peptide. To the best of our knowledge, there
are no reports on the application of ApoE-derived peptides for pDNA
delivery into hepatic cells. Here, we found that ternary complexes
increased the transfection efficiency in hepatic cells by inclusion
of the ApoE-R9 peptide, although the in vivo optimal
liposomal formulation in ternary complexes with the ApoE-R9 peptide
were different from the in vitro one.
Materials and methods
Materials
1,2-Dioleoyl-3-trimethylammonium-propane methyl
sulfate salt (DOTAP) was obtained from Avanti Polar Lipids Inc.
1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE,
COATSOME ME-8181) was obtained from NOF Co. Ltd. Cholesterol (Chol)
was purchased from Wako Pure Chemical Industries, Ltd. Quaser670
carboxylic acid was obtained from Biosearch Technologies, Inc. All
other chemicals were of the finest grade available.
Synthesis of ApoE-derived peptide
dApoE peptide contained amino acid sequence 141–151
of human ApoE in a tandem dimer comprising
WG-(LRKLRKRLLR)2-NH2 (16,17). The
ApoE fragment (ApoE-F) peptide contained the amino acid sequence of
the binding domain of human ApoE4 (amino acids 151–173) comprising
YLRVRLASHLRKLRKRLLRDADDLY (18). For
the detection of ApoE-derived peptides, dApoE and ApoE-F peptides
were labeled with Quaser670 via three glycine residues as a spacer
at the N-terminus of dApoE and C-terminus of ApoE-F
[Quaser670-labeled dApoE (Q-dApoE) and Quaser670-labeled ApoE-F
(Q-ApoE-F), GenScript Biotech Corp., Piscataway, NJ, USA] (Table I). Quasar670 is an indocarbocyanine
that exhibits fluorescence in the red region of the visible
spectrum. The purities of Q-dApoE and Q-ApoE-F were 79.3 and 71.7%,
respectively, by HPLC analysis, and their molecular weights were
3,578.56 and 3,920.47, respectively, by MALDI-TOF mass
spectrometry. For the interaction of ApoE-derive peptides with
pDNA, nine arginine residues were included at the N-terminus of
dApoE and C-terminus of ApoE-F via three glycine residues as a
spacer (dApoE-R9 and ApoE-F-R9; Medical & Biological
Laboratories Co., Ltd.) (Table I).
The purities of dApoE-R9 and ApoE-F-R9 were 96.9 and 99.7%,
respectively, by HPLC analysis, and their molecular weights were
4505.25 and 4718.55, respectively, by MALDI-TOF mass
spectrometry.
 | Table I.Apo E-derived peptides used in the
present study. |
Table I.
Apo E-derived peptides used in the
present study.
| Peptide | Sequence |
|---|
| Q-dApoE |
Quasar670-GGGWGLRKLRKRLLRLRKLRKRLLR-NH2 |
| dApoE-R9 |
RRRRRRRRRGGGWGLRKLRKRLLRLRKLRKRLLR-NH2 |
| Q-ApoE-F |
YLRVRLASHLRKLRKRLLRDADDLYGGG-Lys-Quasar670-CONH2 |
| ApoE-F-R9 |
YLRVRLASHLRKLRKRLLRDADDLYGGGRRRRRRRRR |
Cell culture
Human hepatoblastoma HepG2 cells were donated by
Prof. Kei-ichi Ozaki (Education and Research Center for
Pharmaceutical Sciences, Osaka University of Pharmaceutical
Sciences, Osaka, Japan). Human lung adenocarcinoma A549 cells were
kindly provided by OncoTherapy Science, Inc.
HepG2 cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) with 10% heat-inactivated fetal bovine serum
(FBS) and kanamycin (100 µg/ml) in a humidified atmosphere
containing 5% CO2 at 37°C. A549 cells were grown in
RPMI-1640 medium supplemented with 10% heat-inactivated FBS and
kanamycin (100 µg/ml) at 37°C in a 5% CO2 humidified
atmosphere.
Cellular uptake of ApoE-derived
peptides
HepG2 cells were plated into 35-mm culture dishes at
a density of 3×105 cells 24 h prior to each experiment.
Quaser670, Q-dApoE, and Q-ApoE-F were diluted in 1 ml of culture
medium to final concentrations of 1, 10, and 10 µg/ml (2, 2.8, and
2.6 µM), respectively, and they were incubated with cells for 3 h.
After the incubation, the cells were fixed with 10% formaldehyde.
The localization of Quaser670, Q-dApoE, and Q-ApoE-F was visualized
using an Eclipse TS100-F microscope (Nikon).
Biodistribution of ApoE-derived
peptides in mice
All animal experiments were conducted in accordance
with the ‘Guide for the Care and Use of Laboratory Animals’ adopted
by the Institutional Animal Care and Use Committee of Hoshi
University (Tokyo, Japan) (which is accredited by the Ministry of
Education, Culture, Sports, Science, and Technology, Japan).
Ethical approval for this study was obtained from the Institutional
Animal Care and Use Committee of Hoshi University (Permission no.
30-072). A total of six female BALB/c mice (18–20 g, 8 weeks of
age; Sankyo Labo Service Corp., Tokyo, Japan) were housed in a
temperature-(24°C) and humidity-(55%) controlled room with a 12 h
light/dark cycle (lights on at 8:00 a.m.) with ad libitum
access to food and water.
Q-dApoE [20 or 100 µg (5.6 or 27.9 nmol)] or
Q-ApoE-F [20 or 100 µg (5.1 or 25.5 nmol)] were administered
intravenously via the lateral tail vein into female BALB/c mice
(n=1 for 20 and 100 µg ApoE peptide, respectively). As a control,
Quaser670 [2 or 10 µg, (4.0 or 20.1 nmol)] was administered
intravenously via the lateral tail vein into mice (n=1 for 2 and 10
µg Quaser670, respectively). Ten or 60 min after the injection,
mice were sacrificed, and Quaser670 fluorescence imaging of the
tissues was performed using a NightOWL LB981 NC100 system (Berthold
Technologies). In Quaser670 fluorescence imaging, the excitation
and emission filters were set at 630/20 and 680/30 nm,
respectively. The exposure time for fluorescence was 1 sec. A
grayscale body-surface reference image was collected using a
NightOWL LB981 CCD camera. The images were analyzed using IndiGo2
software (version 2.0.1.0) provided with the in vivo imaging
system (Berthold Technologies). The tissues after fluorescence
imaging were frozen on dry ice and sliced into 16 µm sections. The
localization of Quaser670, Q-dApoE, and Q-ApoE-F was examined using
an Eclipse TS100-F microscope.
Preparation of plasmid DNA
The pCMV-luc plasmid encoding the firefly
luciferase gene under the control of the cytomegalovirus (CMV)
promoter was constructed as described previously (19). A protein-free preparation of pDNA was
purified after alkaline lysis using a QIAGEN Plasmid Maxi Kit
(Qiagen, Hilden, Germany).
Preparation of cationic liposomes,
pDNA lipoplexes, and ternary complexes
Cationic liposomes (LP-DOTAP/Chol and LP-DOTAP/DOPE)
were prepared from DOTAP:Chol or DOTAP:DOPE at a molar ratio of 1:1
using a dry-film method (20).
Briefly, all lipids were dissolved in chloroform, which was removed
by evaporation. The thin film was hydrated with water at 60°C by
vortex mixing and sonication. The particle size distributions and
ζ-potentials were determined by the dynamic light scattering method
(ELS-Z2; Otsuka Electronics, Osaka, Japan) at 25°C after diluting
the dispersion to an appropriate volume with water.
Cationic liposome/pDNA complexes (pDNA lipoplexes)
were prepared by mixing pDNA with cationic liposomes at a charge
ratio (−:+) of 1:4, as reported previously (21). Binary complexes of pDNA and ApoE-R9
peptide were prepared by mixing pDNA with dApoE-R9 or ApoE-F-R9 at
charge ratios (−:+) of 1:1, 1:2, and 1:3 (3.2, 6.5, and 9.7 µg
dApoE-R9 or 3.4, 6.8, and 10.2 µg ApoE-F-R9 for 2 µg pDNA,
respectively). Ternary complexes of pDNA, cationic liposomes, and
ApoE-R9 peptide were prepared by mixing ApoE-R9 peptide with
cationic liposomes, followed by mixing with pDNA at charge ratios
(−:+:+) of pDNA:cationic liposomes:ApoE-R9 peptide of 1:4:1, 1:4:2,
and 1:4:3. The complexes were shaken gently and stood for 15 min at
room temperature. The charge ratio (−:+) of pDNA:Cationic liposomes
was expressed as the molar ratio of pDNA phosphate to DOTAP. The
charge ratio (−:+) of pDNA:dApoE-R9 or pDNA:ApoE-F-R9 was expressed
as the molar ratio of pDNA phosphate to arginine residue at the
terminus of the ApoE-R9 peptides (9 arginine residues per
peptide).
Accessibility of pDNA in binary and
ternary complexes
pDNA association with ApoE-R9 peptide or cationic
liposomes was analyzed using an exclusion assay with
SYBR® Green I Nucleic Acid Gel Stain (Takara Bio Inc.).
Binary complexes of pDNA and ApoE-R9 peptide were formed at charge
ratios (−:+) of 1:1, 1:2, and 1:3. Ternary lipoplexes of pDNA,
cationic liposome, and ApoE-R9 peptide were formed at charge ratios
(−:+:+) of 1:4:1, 1:4:2, and 1:4:3. The binary or ternary complexes
of 0.5 µg of pDNA in a volume of 100 µl of Tris-HCl buffer (pH 8.0)
were mixed with 100 µl of 5,000-fold diluted SYBR® Green
I Nucleic Acid Gel Stain solution with Tris-HCl buffer, and then
incubated for 30 min. Fluorescence was measured at an emission
wavelength of 535 nm with an excitation wavelength of 485 nm using
a fluorescence plate reader (ARVO X2; Perkin Elmer). As a control,
the value of fluorescence obtained upon addition of free pDNA
solution was set as 100%. The amount of pDNA available to interact
with the SYBR® Green I was expressed as a percentage of
the control.
Luciferase activity in vitro
HepG2 and A549 cells were prepared by plating cells
in a 6-well plate 24 h prior to each experiment. pDNA lipoplexes of
pCMV-Luc (2 µg), binary complexes of pCMV-Luc (2 µg) and ApoE-R9
peptide or ternary complexes of pCMV-Luc (2 µg), cationic
liposomes, and ApoE-R9 peptide were transfected into cells.
Twenty-four hours after transfection, luciferase activity was
measured as counts per second (cps)/µg protein using the luciferase
assay system (PicaGene; Toyo Ink Manufacturing. Co., Ltd.) and
bicinchoninic acid (BCA) reagent (Pierce™ BCA Protein Assay kit;
Pierce; Thermo Fisher Scientific, Inc.) as reported previously
(22).
Cytotoxicity by ternary complexes
HepG2 cells were seeded in 96-well plates 24 h prior
to transfection. Ternary complexes of pDNA, LP-DOTAP/DOPE, and
ApoE-R9 peptide were formed at charge ratios (−:+:+) of 1:4:1,
1:4:2, and 1:4:3. Each ternary complex with 2 µg of pDNA was
diluted in 1 ml of medium supplemented with 10% FBS, and then the
mixture (100 µl) was added to the cells at 50% confluency in the
well. After a 24-h incubation period, cell numbers were determined
using a Cell Counting Kit-8 (Dojindo Laboratories). Cell viability
was expressed as relative to the absorbance at 450 nm of
untransfected cells.
Transfection activity in vivo
pDNA lipoplexes with 30 µg pCMV-Luc and cationic
liposomes were prepared at a charge ratios (−:+) of 1:4, and
ternary complexes with 30 µg pCMV-Luc, cationic liposomes, and
dApoE-R9 (145.5 µg for 30 µg pDNA) were prepared at a charge ratio
(−:+:+) of 1:4:3. The pDNA lipoplexes or ternary complexes with 30
µg of pCMV-Luc were administered intravenously via the lateral tail
vein into a total of 12 female BALB/c mice (8 weeks of age) (n=3
for each the complex). At 24-h post-injection, mice were sacrificed
by cervical dislocation, and tissues were removed for analysis.
Three microliters of ice-cold reporter lysis buffer (Promega
Corporation) per 1 mg of tissue was added, and then homogenized
immediately. The homogenate samples were centrifuged at 15,000 rpm
for 3 min at 4°C. Aliquots of 10 µl of the supernatants were mixed
with 50 µl of luciferase assay system (PicaGene), and counts per
second (cps) were measured using a chemoluminometer (ARVO X2). The
protein concentration of each supernatant was determined using a
BCA protein assay (Microplate BCA Protein Assay kit-Reducing Agent
Compatible; Pierce; Thermo Fisher Scientific, Inc.) with bovine
serum albumin as the standard, and luciferase activity was
calculated as cps/mg protein.
Statistical analysis
Data are presented as the mean + standard deviation
of three independent experiments. The statistical significance of
differences between mean values was determined by Student's t-test
using GraphPad Prism 4.0 (GraphPad Software Inc.). Multiple
measurement comparisons were performed by analysis of variance
followed by one-way analysis of variance on ranks with post hoc
Tukey-Kramer's test using GraphPad Prism 4.0. P<0.05 was
considered to indicate a statistically significant difference.
Results and Discussion
Uptake of ApoE-derived peptide in
hepatic cells
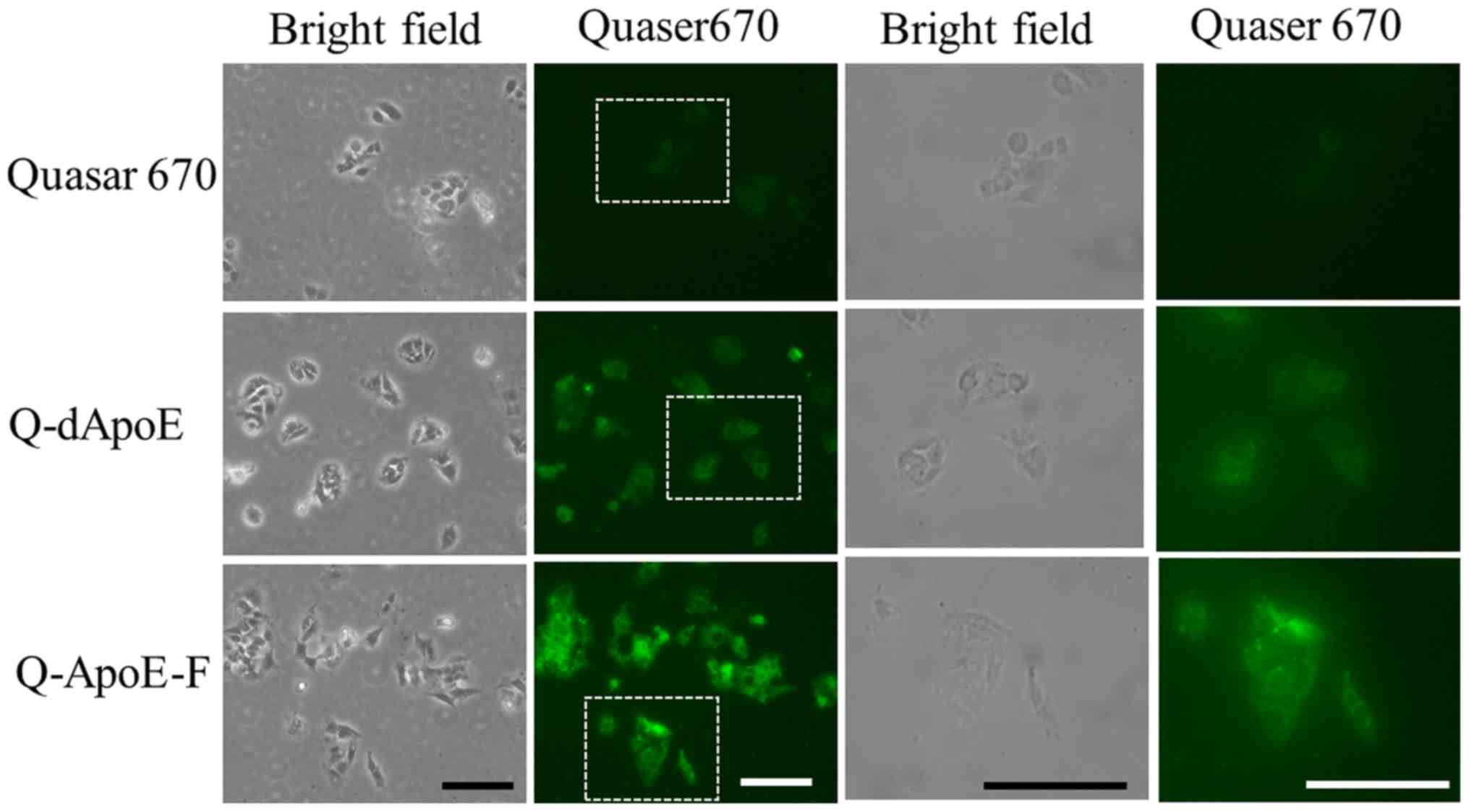
First, to confirm the uptake of the ApoE-derived
peptide in hepatic cells, we synthesized two types of
Quaser670-labeled ApoE-derived peptides, Q-dApoE and Q-ApoE-F
(Table I), and examined the
localization of their peptides in HepG2 cells after a 3-h
incubation. HepG2 cells are one of the most commonly used cell
lines as hepatic cells. Both peptides were detected throughout the
cytoplasm strongly and diffusively in most cells, although free
Quaser670 was not taken up by the cells (Fig. 1), indicating that ApoE-derived
peptides were effectively taken up by the cells.
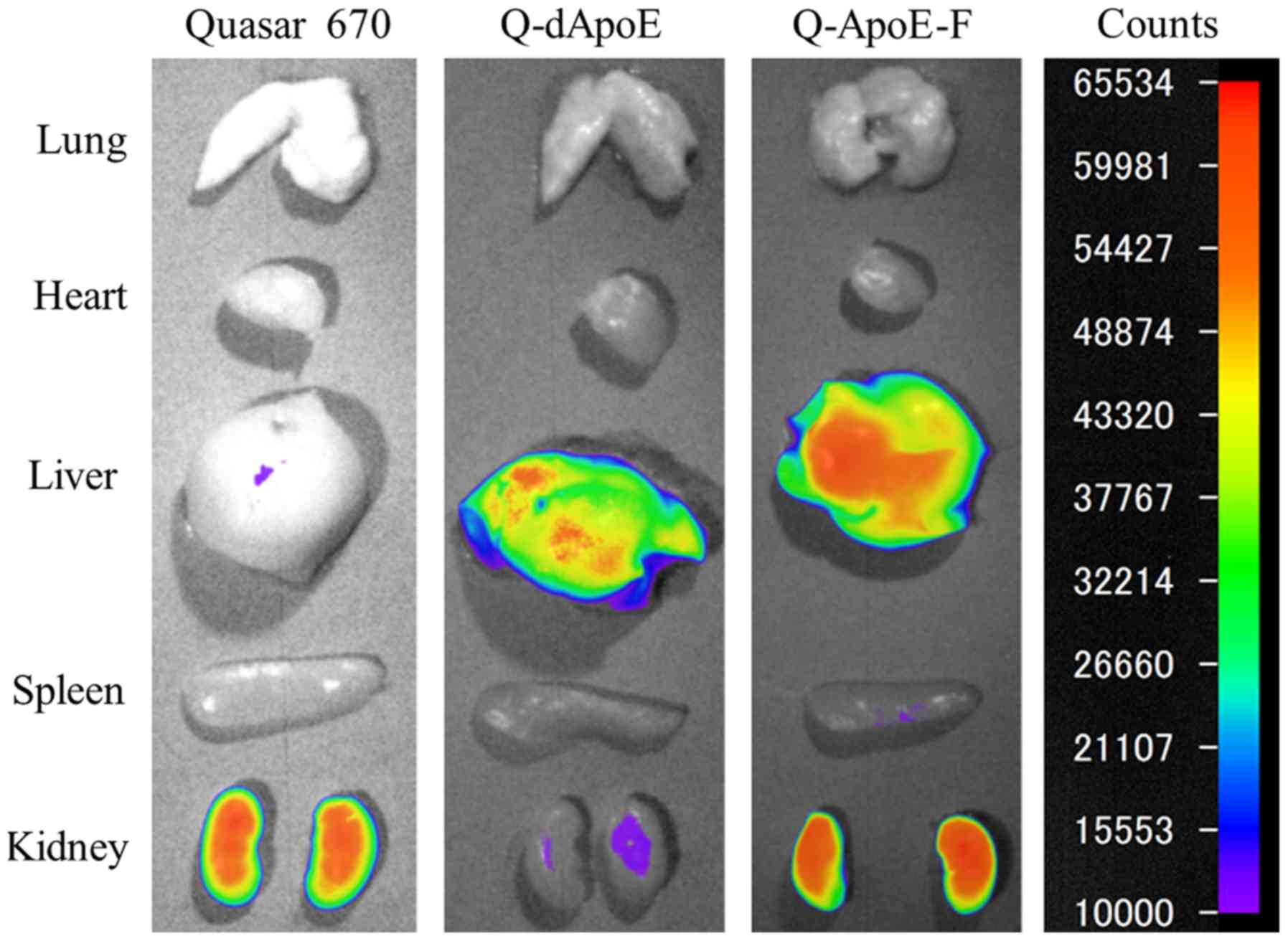
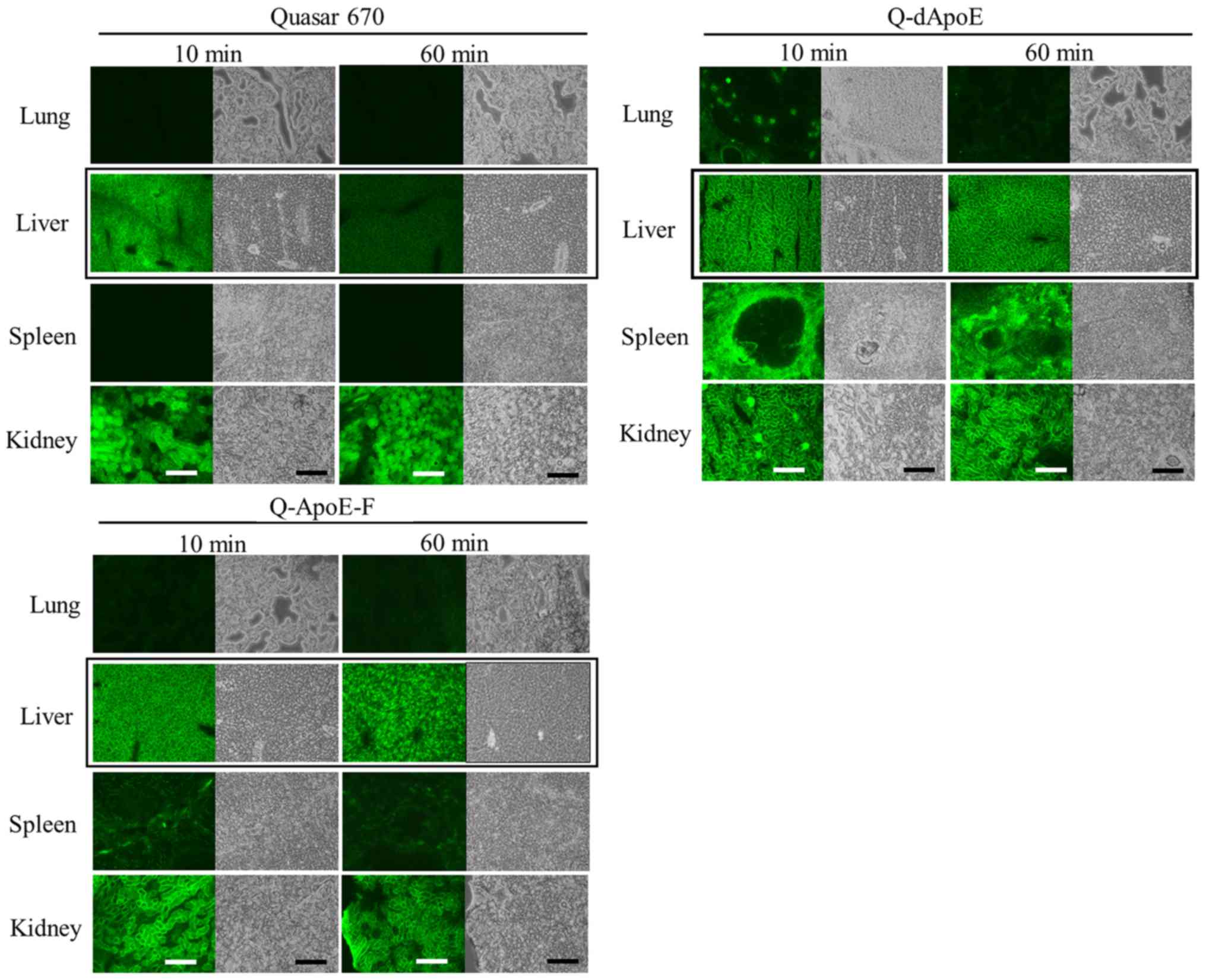
Next, to investigate whether ApoE-derived peptides
may be taken up into the liver, we injected Q-dApoE and Q-ApoE-F
intravenously into mice and observed their biodistributions at 10
and 60 min after injection (Figs. 2
and 3). Free Quaser670 accumulated
mainly in the liver and kidneys at 10 min after injection (Fig. 3); however, it was detected only in
kidneys at 60 min (Figs. 2 and
3), indicating that free Quaser670
(MW 497.69) was rapidly excreted from the kidneys after intravenous
injection. In contrast, Q-dApoE was detected mainly in the liver,
spleen, and kidneys, and Q-ApoE-F was found in the liver and
kidneys at 60 min after injection (Figs.
2 and 3), indicating that
Q-dApoE and Q-ApoE-F were efficiently taken up via ApoE receptors
by hepatocytes in the mouse liver, although some injected peptides
were excreted from the kidneys due to their low molecular weight
(less than 4,000). However, we could not confirm whether Q-dApoE
and Q-ApoE-F were localized mainly in parenchymal or/and
non-parenchymal cells of the liver. These results suggested that
dApoE and ApoE-F may be useful as ligands for liver targeting.
Characterization of cationic liposomes
and ternary complexes
For pDNA delivery with ApoE-derived peptides, we
synthesized two further types of ApoE-derived peptides, dApoE-R9
and ApoE-F-R9 (Table I), which were
conjugated with nine arginine residues for interaction with pDNA,
and prepared ternary complexes of pDNA, cationic liposomes, and
ApoE-R9 peptides. The selected cationic liposomes, DOTAP/Chol
liposomes and DOTAP/DOPE liposomes, have often been used for pDNA
transfection in previous studies (23–25). In
addition, it has been shown that neutral helper lipids for cationic
liposomal formulation significantly affected the transfection
efficiency in vitro and in vivo (26,27).
Therefore, in this study, we used DOTAP as a cationic lipid, and
DOPE or Chol as a neutral helper lipid, and prepared two types of
cationic liposomes, LP-DOTAP/DOPE and LP-DOTAP/Chol for pDNA
delivery. LP-DOTAP/DOPE consisted of DOTAP and DOPE at a molar
ratio of 1:1, and LP-DOTAP/Chol consisted of DOTAP and Chol at a
molar ratio of 1:1.
Next, we measured the particle size and ζ-potential
of the cationic liposomes, pDNA lipoplexes, and ternary complexes.
The sizes of LP-DOTAP/DOPE and LP-DOTAP/Chol were approximately 100
nm, and the ζ-potentials were approximately 46–48 mV (Table II). When LP-DOTAP/DOPE and
LP-DOTAP/Chol were mixed with pDNA, the sizes were 152 and 184 nm,
respectively, and the ζ-potentials were approximately 37 and 34 mV,
respectively. For formation of the ternary complexes, LP-DOTAP/DOPE
or LP-DOTAP/Chol was mixed with dApoE-R9 or ApoE-F-R9, followed by
mixing with pDNA at charge ratios (−:+:+) of pDNA:cationic
liposome:ApoE-R9 peptide from 1:4:1 to 1:4:3 (Table II). Here, in order to make the
number of moles of ApoE peptides equal between the ternary
complexes with dApoE-R9 and ApoE-F-R9, we calculated the charge
ratio (−:+) of pDNA:dApoE-R9 or pDNA:ApoE-F-R9 as the molar ratio
of pDNA phosphate to arginine residue at a terminus of the ApoE-R9
peptides (9 arginine residues per peptide) although both dApoE-R9
and ApoE-F-R9 contained positively charged amino acids in the
sequence of ApoE (the net charges at pH 7 calculated by Innovagen's
peptide calculator were 22.0 and 14.1 in dApoE-R9 and ApoE-F-R9,
respectively). All the ternary complexes with dApoE-R9 or ApoE-F-R9
were approximately 130–150 nm in size with a monodisperse
distribution (polydispersity index: 0.16–0.19) and their
ζ-potentials were approximately 36–42 mV.
 | Table II.Particle size and ζ-potential of
ternary complexes with pDNA, cationic liposomes and ApoE-R9
peptides. |
Table II.
Particle size and ζ-potential of
ternary complexes with pDNA, cationic liposomes and ApoE-R9
peptides.
| Liposomes and
lipoplexes | Charge ratio
(−:+:+) | Sizea (nm) | PDI |
ζ-potentiala (mV) |
|---|
| LP-DOTAP/DOPE | – | 102.2±0.6 | 0.22±0.01 | 46.3±0.8 |
|
pDNA:LP-DOTAP/DOPE | 1:4:0 | 152.6±0.3 | 0.19±0.00 | 37.1±1.8 |
|
pDNA:LP-DOTAP/DOPE:dApoE-R9 | 1:4:1 | 142.5±1.7 | 0.19±0.01 | 41.9±0.4 |
|
pDNA:LP-DOTAP/DOPE:dApoE-R9 | 1:4:2 | 132.2±2.2 | 0.18±0.01 | 41.4±0.8 |
|
pDNA:LP-DOTAP/DOPE:dApoE-R9 | 1:4:3 | 134.4±0.6 | 0.18±0.01 | 38.0±2.5 |
|
pDNA:LP-DOTAP/DOPE:ApoE-F-R9 | 1:4:1 | 140.4±1.7 | 0.16±0.02 | 37.9±0.5 |
|
pDNA:LP-DOTAP/DOPE:ApoE-F-R9 | 1:4:2 | 132.0±0.8 | 0.18±0.01 | 40.7±0.8 |
|
pDNA:LP-DOTAP/DOPE:ApoE-F-R9 | 1:4:3 | 129.2±2.6 | 0.18±0.02 | 42.2±1.0 |
| LP-DOTAP/Chol | – | 104.4±0.8 | 0.21±0.01 | 47.7±0.6 |
|
pDNA:LP-DOTAP/Chol | 1:4:0 | 184.0±1.8 | 0.18±0.01 | 33.7±0.7 |
|
pDNA:LP-DOTAP/Chol:dApoE-R9 | 1:4:1 | 150.1±0.8 | 0.16±0.00 | 42.3±1.1 |
|
pDNA:LP-DOTAP/Chol:dApoE-R9 | 1:4:2 | 127.7±2.2 | 0.19±0.01 | 41.4±1.0 |
|
pDNA:LP-DOTAP/Chol:dApoE-R9 | 1:4:3 | 131.4±1.5 | 0.19±0.01 | 42.4±0.2 |
|
pDNA:LP-DOTAP/Chol:ApoE-F-R9 | 1:4:1 | 152.9±1.6 | 0.17±0.01 | 38.4±0.3 |
|
pDNA:LP-DOTAP/Chol:ApoE-F-R9 | 1:4:2 | 138.6±1.8 | 0.17±0.01 | 36.1±0.8 |
|
pDNA:LP-DOTAP/Chol:ApoE-F-R9 | 1:4:3 | 134.4±2.3 | 0.17±0.00 | 36.1±1.0 |
Association of pDNA with cationic
liposome and ApoE-R9 peptide
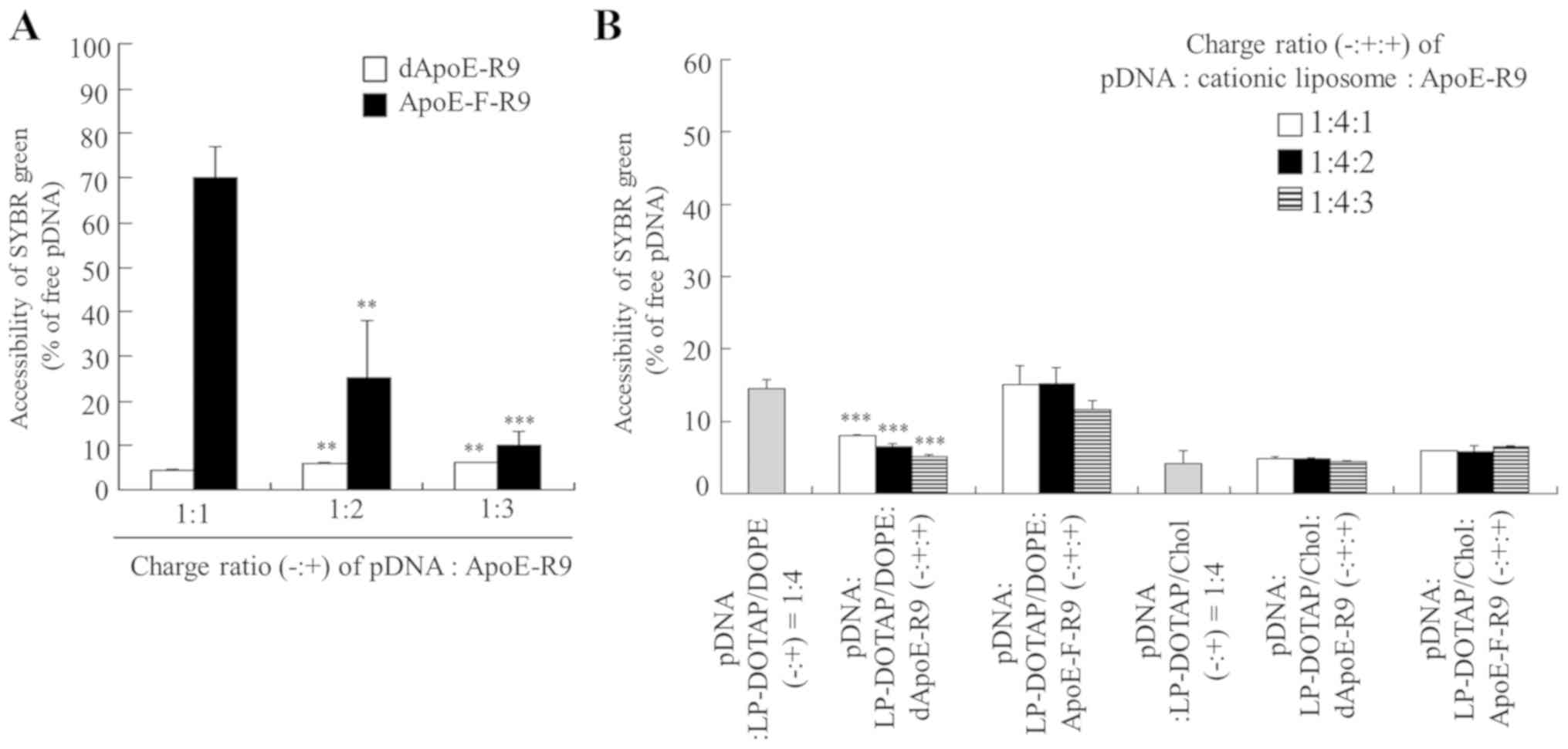
Next, we examined the effect of the charge ratio of
the ApoE-R9 peptide in ternary complexes on pDNA association.
SYBR® Green I is a DNA/RNA-intercalating agent, the
fluorescence of which is markedly enhanced upon binding to pDNA
that is not bound to cationic liposomes or ApoE-R9 peptide. In the
binary complex of pDNA and ApoE-F-R9, with an increase in the
charge ratio (−:+) of pDNA:ApoE-F-R9, ApoE-F-R9 interacted with
pDNA, and at a charge ratio (−:+) of 1:3, the fluorescence of
SYBR® Green I decreased markedly due to the formation of
complexes (Fig. 4A). In contrast, in
the binary complex of pDNA and dApoE-R9, already at a charge ratio
(−:+) of 1:1, fluorescence decreased markedly. These results
indicated that dApoE-R9 interacts more efficiently with pDNA than
ApoE-F-R9. Furthermore, in LP-DOTAP/DOPE and LP-DOTAP/Chol
lipoplexes, a decrease in fluorescence was observed at a charge
ratio (−:+) of 1:4 (Fig. 4B), and in
the ternary complexes, fluorescence decreased markedly above a
charge ratio (−:+:+) of pDNA:cationic liposome:ApoE-R9 peptide of
1:4:1 regardless of the liposomal formulation, indicating that pDNA
lipoplexes and ternary complexes completely bound to pDNA at the
charge ratios used in this study. These findings suggested that the
interaction between pDNA and cationic liposomes was not affected by
the inclusion of ApoE-R9 peptides.
Effect of ApoE-R9 peptide in ternary
complexes on in vitro gene transfection
To examine the effect of the ApoE-R9 peptide in
ternary complexes on gene expression in hepatic cells, the ternary
complexes were added into HepG2 cells. The inclusion of dApoE-R9 or
ApoE-F-R9 into LP-DOTAP/DOPE lipoplexes significantly increased
transfection activity in HepG2 cells with increasing amounts of
ApoE-R9 peptide. Ternary complexes with dApoE-R9 and ApoE-F-R9
exhibited 7.5- and 3.8-fold higher expression in the cells,
respectively, than pDNA lipoplexes without ApoE-R9 peptide, when
the ternary complexes were prepared at a charge ratio (−:+:+) of
1:4:3 (Fig. 5). However, the
inclusion of ApoE-R9 peptide into LP-DOTAP/Chol lipoplexes did not
increase transfection activity. Furthermore, to examine the effect
of the ApoE-R9 peptide in binary complexes on gene expression in
HepG2 cells, binary complexes were added into HepG2 cells. However,
the binary complexes of pDNA with dApoE-R9 or ApoE-F-R9 did not
induce high gene expression in the cells (Fig. 6A) compared with their ternary
complexes (Fig. 5). This indicated
that formation of ternary complex was necessary for efficient pDNA
transfer into hepatic cells by ApoE-R9 peptide. Moreover, to
investigate the effect of the ApoE-R9 peptide in ternary complexes
on gene expression in non-hepatic cells, the ternary complexes were
added to A549 cells, which express the LDL receptor at lower levels
than HepG2 cells (data not shown). As the result, the ternary
complexes did not increase the transfection activity in A549 cells
by inclusion of dApoE-R9 or ApoE-F-R9 (Fig. 6B), suggesting that the ternary
complexes with ApoE-R9 peptide improve gene expression in hepatic
cells. In particular, dApoE-R9 has repeated binding domains for the
LDL receptor; therefore, it may exhibit better transfection
efficiency in hepatic cells in the ternary complexes than
ApoE-F-R9.
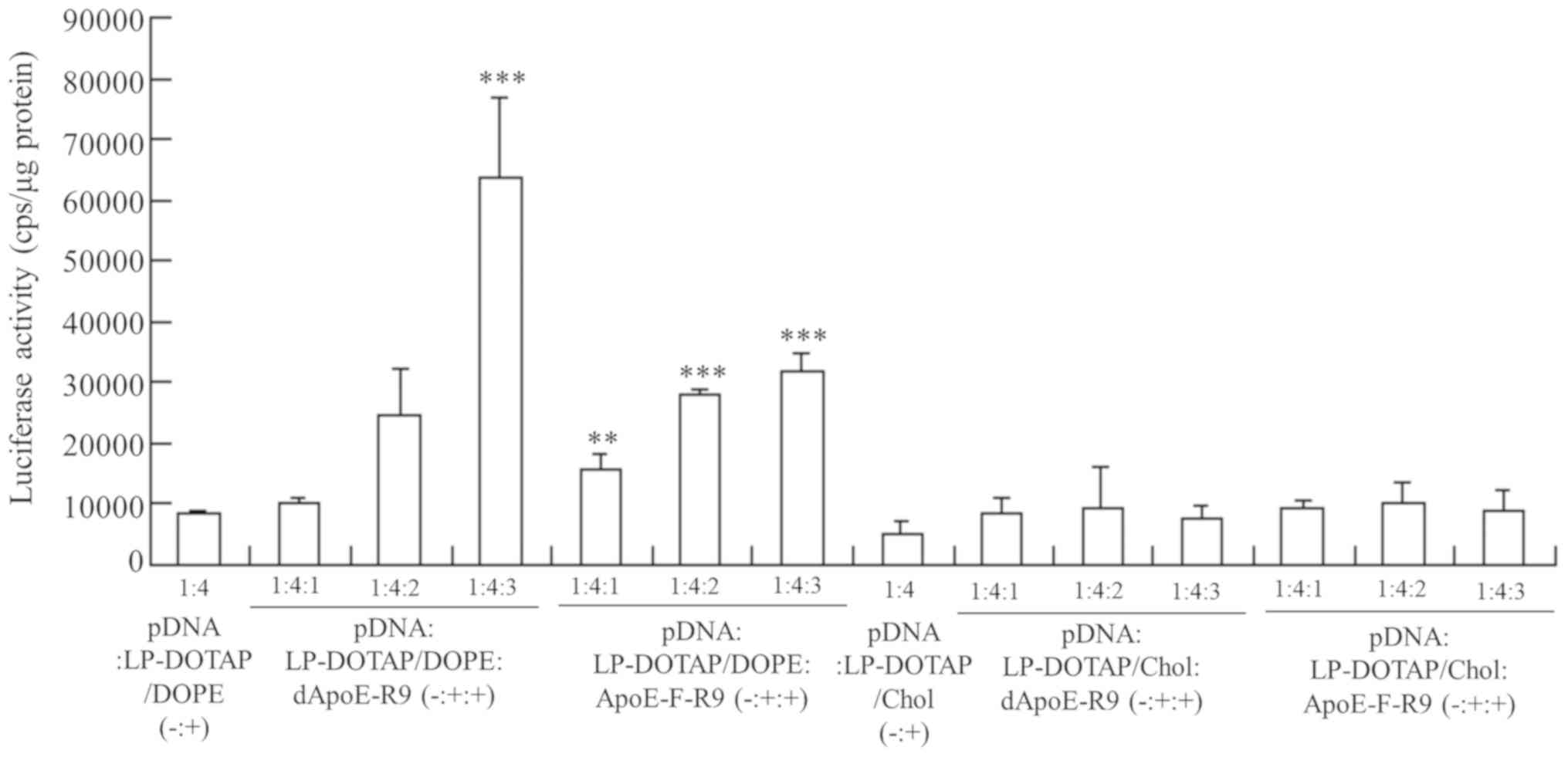 | Figure 5.Effect of charge ratio (−:+:+) of
pDNA, cationic liposome and ApoE-R9 peptide on the luciferase
activity of HepG2 cells at 24 h after transfection with the ternary
complex. pDNA lipoplexes were prepared by mixing pCMV-Luc with
LP-DOTAP/Chol or LP-DOTAP/DOPE at a charge ratio (−:+) of 1:4.
Ternary complexes of pCMV-Luc, cationic liposome and ApoE-R9
peptide were prepared at charge ratios (−:+:+) from 1:4:1-1:4:3.
Data are presented as the mean + standard deviation (n=3).
**P<0.01 and ***P<0.001 vs. LP-DOTAP/DOPE lipoplexes. pDNA,
plasmid DNA; ApoE, apolipoprotein E; LP, liposome; DOTAP,
1,2-dioleoyl-3-trimethylammonium-propane methyl sulfate salt; Chol,
cholesterol; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine. |
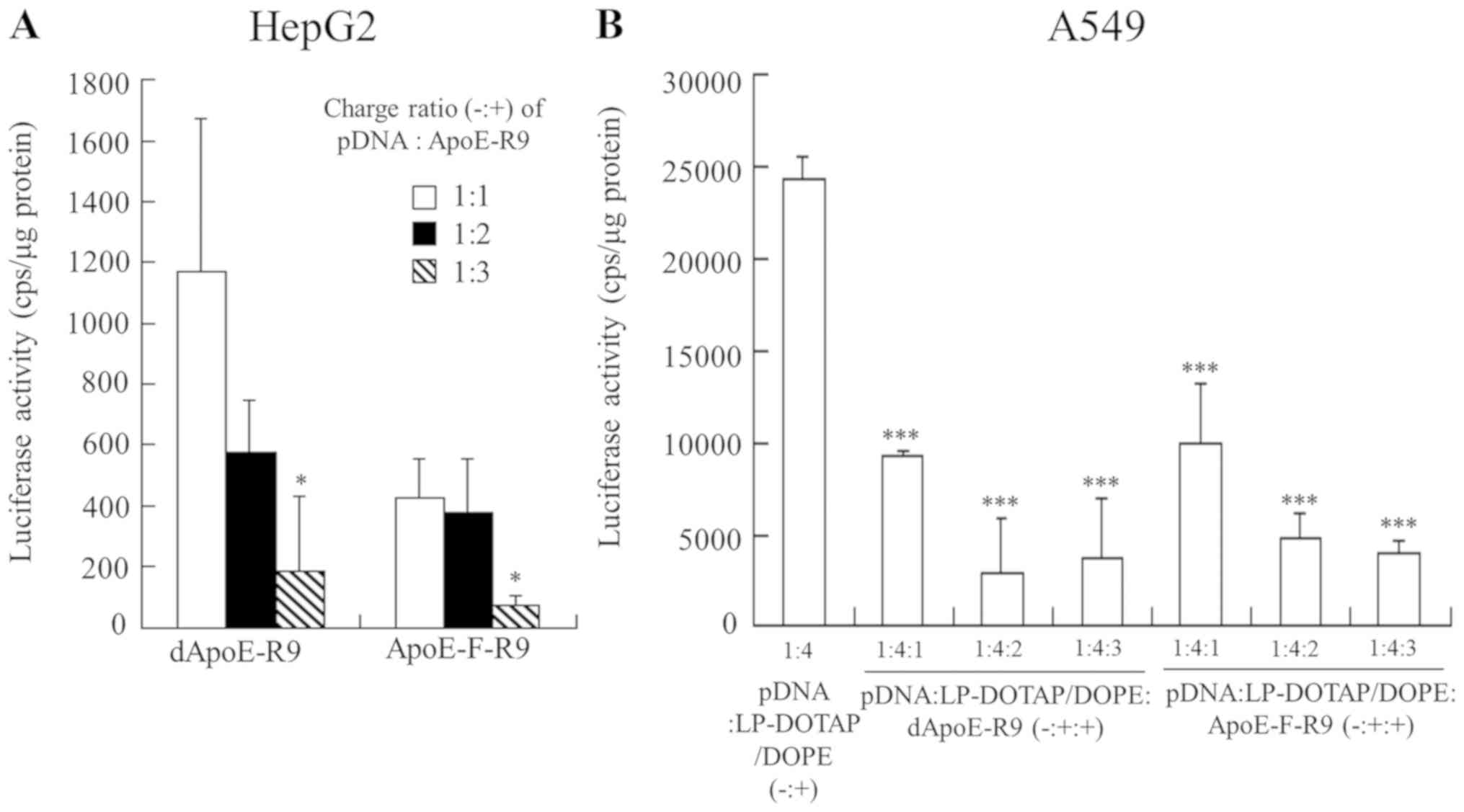 | Figure 6.Luciferase activity in HepG2 cells 24
h after transfection with binary complexes of pDNA and ApoE-R9
peptide, and in A549 cells 24 h after transfection with ternary
complexes. (A) binary complexes were prepared by mixing pCMV-Luc
with dApoE-R9 or ApoE-F-R9 at charge ratios (−:+) from 1:1-1:3.
*P<0.05 vs. binary complexes of dApoE-R9 or ApoE-F-R9 at a
charge ratio (−:+) of 1:1. (B) pDNA lipoplexes were prepared by
mixing pCMV-Luc with cationic liposomes (LP-DOTAP/DOPE or
LP-DOTAP/Chol) at a charge ratio (−:+) of 1:4. Ternary complexes of
pCMV-Luc, cationic liposome, and ApoE-R9 peptide were prepared at
charge ratios (−:+:+) from 1:4:1-1:4:3. ***P<0.001 vs.
LP-DOTAP/DOPE lipoplexes. Data are presented as the mean + standard
deviation (n=3). pDNA, plasmid DNA; ApoE, apolipoprotein E; LP,
liposome; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane methyl
sulfate salt; DOPE,
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine; Chol,
cholesterol. |
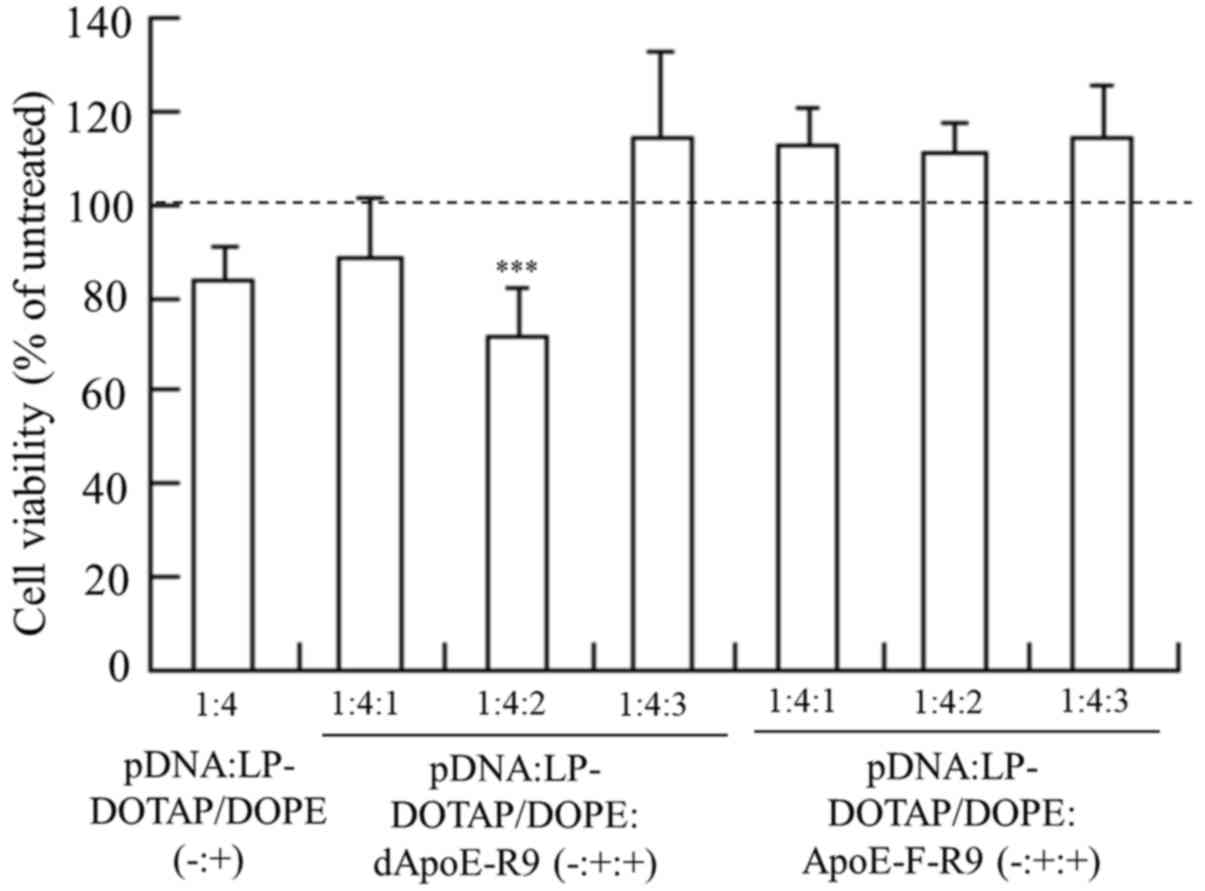
Cytotoxicity by ternary complex
To examine the effect of the ApoE-R9 peptide in
ternary complexes on cytotoxicity, we investigated cell viabilities
at 24 h after transfection into HepG2 cells with LP-DOTAP/DOPE
lipoplexes or ternary complexes. LP-DOTAP/DOPE lipoplexes exhibited
only very limited cytotoxicity (Fig.
7), and the inclusion of dApoE-R9 or ApoE-F-R9 into
LP-DOTAP/DOPE lipoplexes did not increase cytotoxicity.
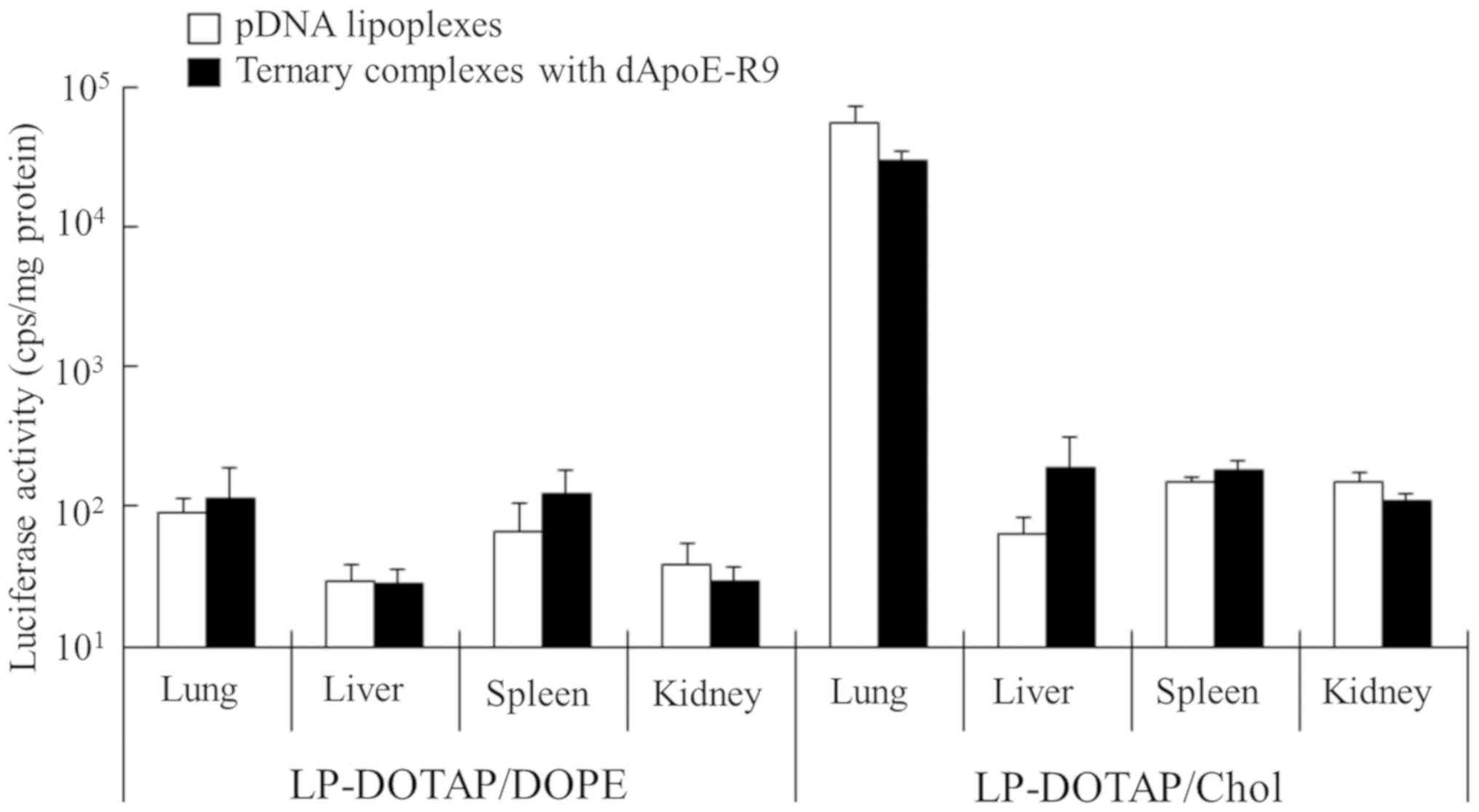
Gene expression in the liver after
injection of ternary complexes into mice
To investigate the effect of ApoE-R9 peptide in
ternary complexes on gene expression in the liver, we injected
ternary complexes with ApoE-R9 peptide intravenously into mice.
Here, we decided to use dApoE-R9, because ternary complexes with
dApoE-R9 exhibited higher gene expression in HepG2 cells than those
with ApoE-F-R9 (Fig. 5). Ternary
complexes were prepared at a charge ratio (−:+:+) of pDNA:cationic
liposomes:dApoE-R9 of 1:4:3. Injection of LP-DOTAP/Chol lipoplexes
induced gene expression mainly in the lungs (Fig. 8). In contrast, injection of the
ternary complexes with LP-DOTAP/Chol and dApoE-R9 showed 2-fold
lower expression in the lungs and 3-fold higher expression in the
liver, although the changes were not significant. These results
indicated that the inclusion of dApoE-R9 into the complexes
improved the uptake of pDNA by hepatocytes after intravenous
injection. However, LP-DOTAP/DOPE lipoplexes did not induce high
gene expression in any organs, and the inclusion of dApoE-R9 into
LP-DOTAP/DOPE lipoplexes did not increase gene expression in the
liver (Fig. 8). These results
indicate that the optimal in vivo liposomal formulation of
ternary complexes with ApoE-R9 peptide for pDNA transfection
differs from the in vitro one. Here, we confirmed that
ApoE-derived peptides could increase transfection activity of
ternary complexes in HepG2 cells (tumor cells) (Fig. 5) and mouse liver (Fig. 8). However, it will be important to
investigate the differences between normal and tumor cells in the
ability to bind ternary complexes. Therefore, the transfection
ability of ternary complexes to primary hepatic cells should be
examined in the future study.
In this study, ternary complexes with LP-DOTAP/Chol
induced higher gene expression in mice than those with
LP-DOTAP/DOPE, indicating that neutral helper lipid in the
liposomal formulation strongly affected in vivo transfection
activity. Generally, in in vitro transfection with pDNA
lipoplexes, DOPE is often used as a neutral helper lipid in
liposomal formulation, because DOPE is thought to improve
transfection efficiency by destabilizing the endosomal membrane
(28), thereby facilitating the
release of pDNA into the cytoplasm. In DOTAP-based cationic
liposomes, it has been reported that the in vitro
transfection efficiency was mainly influenced by liposomal
formulation, and the DOTAP/DOPE ratio determined transfection
efficiency (29), suggested that
DOPE is an important component of cationic liposomes in in
vitro transfection. Therefore, in in vitro transfection
of the ternary complexes, with an increase in cellular uptake with
the inclusion of dApoE-R9 into the ternary complexes, ternary
complexes with LP-DOTAP/DOPE may increase gene expression due to
the function of DOPE, compared with those with LP-DOTAP/Chol.
Furthermore, Hong et al (27)
found that cationic liposomes composed of dimethyl dioctadecyl
ammonium bromide (DDAB)/Chol exhibited higher gene expression in
the lung after intravenous injection of pDNA lipoplexes than those
of DDAB/DOPE, although they did not induce high gene expression
in vitro. In addition, Sakurai et al (26) reported that cationic liposomes
composed of
N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium
chloride (DOTMA)/Chol showed higher gene expression in the lung
after intravenous injection of pDNA lipoplexes than DOTMA/DOPE.
They speculated that the inclusion of DOPE in the liposomal
formulation caused fusion and aggregation of the lipoplexes with
erythrocytes in the blood circulation, resulting in a decrease in
in vivo transfection activity. Therefore, in in vivo
transfection of ternary complexes, the ternary complexes with
LP-DOTAP/DOPE may not exhibit strong transfection activities in any
organs, compared with LP-DOTAP/Chol. In the in vivo
transfection of ternary complexes with ApoE-R9 peptide,
LP-DOTAP/Chol may be more suitable as a liposomal carrier than
LP-DOTAP/DOPE, although they exhibited high gene expression in the
lung. Further studies should be performed to examine liposomal
formulations to decrease gene expression in the lungs by decreasing
the positive charge in ternary complexes and increase expression in
the liver using ApoE-R9 peptide after intravenous injection. In
addition, for the clinical application of the ternary complexes
with ApoE-R9 peptide, toxicity is an important factor. In a future
study, toxicity after intravenous injection of the ternary
complexes with ApoE-R9 peptide should be examined for the
development of a safe pDNA delivery system to the liver.
In conclusion, we synthesized two types of
ApoE-derived peptides, dApoE-R9 and ApoE-F-R9, and prepared ternary
complexes of pDNA, cationic liposomes, and ApoE-R9 peptides for
effective transfection into hepatic cells. In the in vitro
transfection assays, ternary complexes with dApoE-R9 and DOTAP/DOPE
liposomes increased gene expression in hepatic cells. In contrast,
in in vivo transfection analyses, ternary complexes with
dApoE-R9 and DOTAP/Chol liposomes increased gene expression in the
liver after intravenous injection. The optimal in vivo
liposomal formulation of ternary complexes with ApoE-R9 peptide for
pDNA transfection differed from the in vitro one. From these
findings, dApoE-R9 may have potential use as a ligand for liver
targeting with ternary complexes. This study provides valuable
information about liver targeting by ternary complexes for
efficient pDNA delivery into the liver.
Acknowledgements
The authors would like to thank Mr Yuki Yoshiike
(Department of Drug Delivery Research, Hoshi University, Tokyo,
Japan) for his experimental assistance (intracellular localization
and ex vivo imaging of ApoE-derived peptide).
Funding
This study was funded only by the resources of our
department.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YH conceived and designed the present study. The
experiments were primarily performed by YN. YH and HO analyzed and
interpreted the data. YH wrote the manuscript. HO reviewed and
edited the manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
All animal experiments were conducted in accordance
with the ‘Guide for the Care and Use of Laboratory Animals’ adopted
by the Institutional Animal Care and Use Committee of Hoshi
University (Tokyo, Japan; accredited by the Ministry of Education,
Culture, Sports, Science and Technology of Japan). Ethical approval
for the present study was obtained from the Institutional Animal
Care and Use Committee of Hoshi University (Permission no.
30-072).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Aravalli RN, Belcher JD and Steer CJ:
Liver-targeted gene therapy: Approaches and challenges. Liver
Transpl. 21:718–737. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu J, Nantz MH and Zern MA: Targeting
hepatocytes for drug and gene delivery: Emerging novel approaches
and applications. Front Biosci. 7:d717–d725. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nayerossadat N, Maedeh T and Ali PA: Viral
and nonviral delivery systems for gene delivery. Adv Biomed Res.
1:272012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Palaschak B, Herzog RW and Markusic DM:
AAV-mediated gene delivery to the liver: Overview of current
technologies and methods. Methods Mol Biol 1950. 333–360. 2019.
View Article : Google Scholar
|
|
5
|
Zhang Y, Bradshaw-Pierce EL, Delille A,
Gustafson DL and Anchordoquy TJ: In vivo comparative study of
lipid/DNA complexes with different in vitro serum stability:
Effects on biodistribution and tumor accumulation. J Pharm Sci.
97:237–250. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu C, Stewart DJ, Lee JJ, Ji L, Ramesh R,
Jayachandran G, Nunez MI, Wistuba II, Erasmus JJ, Hicks ME, et al:
Phase I clinical trial of systemically administered
TUSC2(FUS1)-nanoparticles mediating functional gene transfer in
humans. PLoS One. 7:e348332012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Eliyahu H, Servel N, Domb AJ and Barenholz
Y: Lipoplex-induced hemagglutination: Potential involvement in
intravenous gene delivery. Gene Ther. 9:850–858. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Simberg D, Weisman S, Talmon Y, Faerman A,
Shoshani T and Barenholz Y: The role of organ vascularization and
lipoplex-serum initial contact in intravenous murine lipofection. J
Biol Chem. 278:39858–39865. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pathak A, Vyas SP and Gupta KC:
Nano-vectors for efficient liver specific gene transfer. Int J
Nanomedicine. 3:31–49. 2008.PubMed/NCBI
|
|
10
|
Getz GS and Reardon CA: Apoprotein E and
reverse cholesterol transport. Int J Mol Sci. 19:34792018.
View Article : Google Scholar
|
|
11
|
Fazio S, Linton MF and Swift LL: The cell
biology and physiologic relevance of ApoE recycling. Trends
Cardiovasc Med. 10:23–30. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mortimer BC, Beveridge DJ, Martins IJ and
Redgrave TG: Intracellular localization and metabolism of
chylomicron remnants in the livers of low density lipoprotein
receptor-deficient mice and apoE-deficient mice. Evidence for slow
metabolism via an alternative apoE-dependent pathway. J Biol Chem.
270:28767–28776. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rensen PC, Schiffelers RM, Versluis AJ,
Bijsterbosch MK, Van Kuijk-Meuwissen ME and Van Berkel TJ: Human
recombinant apolipoprotein E-enriched liposomes can mimic
low-density lipoproteins as carriers for the site-specific delivery
of antitumor agents. Mol Pharmacol. 52:445–455. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yan X, Kuipers F, Havekes LM, Havinga R,
Dontje B, Poelstra K, Scherphof GL and Kamps JA: The role of
apolipoprotein E in the elimination of liposomes from blood by
hepatocytes in the mouse. Biochem Biophys Res Commun. 328:57–62.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tamaru M, Akita H, Nakatani T, Kajimoto K,
Sato Y, Hatakeyama H and Harashima H: Application of apolipoprotein
E-modified liposomal nanoparticles as a carrier for delivering DNA
and nucleic acid in the brain. Int J Nanomedicine. 9:4267–4276.
2014.PubMed/NCBI
|
|
16
|
Re F, Cambianica I, Zona C, Sesana S,
Gregori M, Rigolio R, La Ferla B, Nicotra F, Forloni G, Cagnotto A,
et al: Functionalization of liposomes with ApoE-derived peptides at
different density affects cellular uptake and drug transport across
a blood-brain barrier model. Nanomedicine. 7:551–559. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sauer I, Dunay IR, Weisgraber K, Bienert M
and Dathe M: An apolipoprotein E-derived peptide mediates uptake of
sterically stabilized liposomes into brain capillary endothelial
cells. Biochemistry. 44:2021–2029. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hülsermann U, Hoffmann MM, Massing U and
Fricker G: Uptake of apolipoprotein E fragment coupled liposomes by
cultured brain microvessel endothelial cells and intact brain
capillaries. J Drug Target. 17:610–618. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Igarashi S, Hattori Y and Maitani Y:
Biosurfactant MEL-A enhances cellular association and gene
transfection by cationic liposome. J Control Release. 112:362–368.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kato M, Hattori Y, Kubo M and Maitani Y:
Collagenase-1 injection improved tumor distribution and gene
expression of cationic lipoplex. Int J Pharm. 423:428–434. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hattori Y, Yamasaku H and Maitani Y:
Anionic polymer-coated lipoplex for safe gene delivery into tumor
by systemic injection. J Drug Target. 21:639–647. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hattori Y and Maitani Y: Folate-linked
nanoparticle-mediated suicide gene therapy in human prostate cancer
and nasopharyngeal cancer with herpes simplex virus thymidine
kinase. Cancer Gene Ther. 12:796–809. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li S, Tseng WC, Stolz DB, Wu SP, Watkins
SC and Huang L: Dynamic changes in the characteristics of cationic
lipidic vectors after exposure to mouse serum: Implications for
intravenous lipofection. Gene Ther. 6:585–594. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Martin B, Sainlos M, Aissaoui A, Oudrhiri
N, Hauchecorne M, Vigneron JP, Lehn JM and Lehn P: The design of
cationic lipids for gene delivery. Curr Pharm Des. 11:375–394.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yeeprae W, Kawakami S, Suzuki S, Yamashita
F and Hashida M: Physicochemical and pharmacokinetic
characteristics of cationic liposomes. Pharmazie. 61:102–105.
2006.PubMed/NCBI
|
|
26
|
Sakurai F, Nishioka T, Saito H, Baba T,
Okuda A, Matsumoto O, Taga T, Yamashita F, Takakura Y and Hashida
M: Interaction between DNA-cationic liposome complexes and
erythrocytes is an important factor in systemic gene transfer via
the intravenous route in mice: The role of the neutral helper
lipid. Gene Ther. 8:677–686. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hong K, Zheng W, Baker A and
Papahadjopoulos D: Stabilization of cationic liposome-plasmid DNA
complexes by polyamines and poly(ethylene glycol)-phospholipid
conjugates for efficient in vivo gene delivery. FEBS Lett.
400:233–237. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du Z, Munye MM, Tagalakis AD, Manunta MD
and Hart SL: The role of the helper lipid on the DNA transfection
efficiency of lipopolyplex formulations. Sci Rep. 4:71072014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim BK, Hwang GB, Seu YB, Choi JS, Jin KS
and Doh KO: DOTAP/DOPE ratio and cell type determine transfection
efficiency with DOTAP-liposomes. Biochim Biophys Acta 1848.
1996–2001. 2015.
|