Introduction
Pterygium, a benign tumor of conjunctival tissues,
affects visual function and ultimately leads to blindness (1). Recurrent pterygium usually occurs
following primary pterygium excision, with recurrence rates
reported to range from 1–31.2% (2).
Pterygium is considered a degenerative condition (3) characterized by fibrovascular outgrowth
of the conjunctiva over the cornea and is associated with cytokine
and growth factor imbalance, immunological disturbances and genetic
mutations (3). A previous study
determined that human papillomavirus infection and genes associated
with DNA repair, cell proliferation, migration and angiogenesis
were critical in the development of pterygium (1).
The inadequate understanding of recurrent pterygium
pathogenesis means that prevention is far from effective. Various
surgical procedures and adjuvant treatments have been proposed to
reduce post-operative pterygium recurrence, including radiation
therapy, anti-metabolites, anti-angiogenetic factors and
conjunctival auto-grafting, which are considered effective
techniques to treat primary pterygium with low recurrence rates
(2,4). Recurrence not only depends on a
multitude of patient-related, clinical and surgical factors but is
also closely associated with cellular molecular changes (4). The development of drug targets for
pterygium treatment relies on a novel understanding of the specific
appropriate molecular pathways; however, studies on the molecular
mechanisms involved in recurrent pterygium are limited.
In the present study, expression profiling
microarray analysis was performed to investigate the differentially
expressed genes related to recurrent pterygium and their possible
contributions to find a novel treatment target for recurrent
pterygium.
Materials and methods
Sample collection
Conjunctival tissue samples were collected during
surgery from 18 patients with pterygium and ocular trauma who
underwent surgery between December 2014 and June 2017 in the Second
Hospital of Dalian Medical University (Dalian, China) and Yantai
Affiliated Hospital of Binzhou Medical University (Yantai, China).
Patients included 10 males and 8 females between the ages of 34 to
67 years. The inclusion criteria consisted of patients diagnosed
with primary pterygium (n=6), recurrent pterygium (n=6) and ocular
trauma (n=6). Those with other medical diseases were excluded.
Patients were divided into the primary group, recurrence group and
control group, respectively. Patients in the primary and recurrent
groups were subjected to pterygium excision, whilst those in the
control group received conjunctiva excision. Tissue samples were
collected from the patients and stored in a nitrogen canister
(−196°C). The study was approved by the Institutional Ethical
Committee of Yantai Affiliated Hospital of Binzhou Medical
University and written informed consent was obtained from all
patients.
Microarray analysis
Tissues from the primary, recurrent and control
groups were homogenized and TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to isolate RNA. Total RNA was
quantified using NanoDrop 2000 (Thermo Fisher Scientific, Inc.).
RNA samples (300 ng/sample) were used for the analysis of
differentially expressed genes using the Gene Chip™ Human Genome
U133 Plus 2.0 Array (Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. When compared with the
corresponding genes of two groups, those with absolute value of
log2 fold change >1 and false discovery rate <0.05
were considered differentially expressed genes. The primary samples
were assessed with the control and then the recurrent samples.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from primary pterygium,
recurrent pterygium and normal conjunctival tissues with
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). A High Capacity cDNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to synthesize
cDNA with the following protocol: 94°C for 3 min, then 94°C for 30
sec; 58°C for 30 sec and 72°C for 5 sec for a total of 35 cycles.
qPCR was performed with the Power SYBR Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 94°C for 3 min, 40 cycles
of 94°C for 20 sec and 60°C for 60 sec, and then 72°C for 10 min.
All experiments were independently replicated three times with
GAPDH as a control. Relative mRNA levels were quantified using the
2−ΔΔCq method (5). Primer
sequences were as follows: PP7080 forward,
5′-CCTGGTGATGTCCGACCTG-3′ and reverse, 5′-CCATGAGCGCATCGCAATC-3′;
small proline-rich protein 2A (SPRR2A) forward,
5′-AGTGCCAGCAGAAATATCCTCC-3′ and reverse,
5′-TGCTCTTGGGTGGATACTTTGA-3′; small proline-rich protein 2F
(SPRR2F) forward, 5′-AGTGCCAGCAGAAATATCCTCC-3′ and reverse,
5′-GAACGAGGTGAGCCAAATATCC-3′; defensin β4A (DEFB4A) forward,
5′-CAGGATTGAAGGGACCTGTT-3′ and reverse, 5′-CTTCACTTGGCCTGTGTGTC-3′;
serpin family A member 3 (SERPINA3) forward,
5′-TGCCAGCGCACTCTTCATC-3′ and reverse,
5′-TGTCGTTCAGGTTATAGTCCCTC-3′; S100 calcium-binding protein A7
(S100A7) forward, 5′-GATTGACAAGCCAAGCCTGC-3′ and reverse,
5′-CAAAGACGTCGGCGAGGTAA-3′; Fc fragment of IgG binding protein
(FCGBP) forward, 5′-CCTATGGAGCTGGTGGATACTC-3′ and reverse,
5′-GCATAGTCAGAATGGATCACCA-3′; BPI Fold Containing Family A Member 1
(BPIFA1) forward, 5′-CTTGGCCTTGTGCAGAGC-3′ and reverse,
5′-CAACAGACTTGCACCGACC-3′; H19 imprinted maternally expressed
transcript (H19) forward, 5′-TACAACCACTGCACTACCTG-3′ and reverse,
5′-TGGAATGCTTGAAGGCTGCT-3′; secretoglobin family 2A member 1
(SCGB2A1) forward, 5′-ACTCCTGGAGGACATGGTTGA-3′ and reverse,
5′-TCTGAGCCAAACGCCTTGGGT-3′; keratin 24 (KRT24) forward,
5′-GCTATGATGGGGGCCTTCTC-3′ and reverse, 5′-GCCACCTCCTCGTGGTTC-3′
and GAPDH forward, 5′-CACTGCCAACGTGTCAGTGGTG-3′ and reverse,
5′-GTAGCCCAGGATGCCCTTGAG-3′. All primers were designed by Primer
Premier 5.0 (Premier Biosoft International) and synthesized by
Sangon Biotech Co., Ltd.
Protein-protein interaction (PPI) and
gene ontology (GO) analysis
To identify the key genes that interact with S100A7,
a PPI network was constructed using the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) online database
(https://string-db.org/; version 10.5). All genes
correlated with S100A7 were identified according to the strength of
the data support (thickness of the line connecting the two genes)
and a high-resolution bitmap was downloaded. For the interacting
genes in the network, GO analysis was performed using functional
enrichments function of the STRING database and biological
processes and molecular functions were exported.
Statistical analysis
SPSS 20.0 software (IBM Corp.) was used for
statistical analysis, and data are presented as the mean ± standard
error. The independent Student's t test was used to compare
relative mRNA levels in the microarray analysis. For multiple
comparisons between groups, one-way analysis of variance was
performed prior to running the Student-Newman-Keuls post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Identification of differentially
expressed genes associated with recurrent pterygium
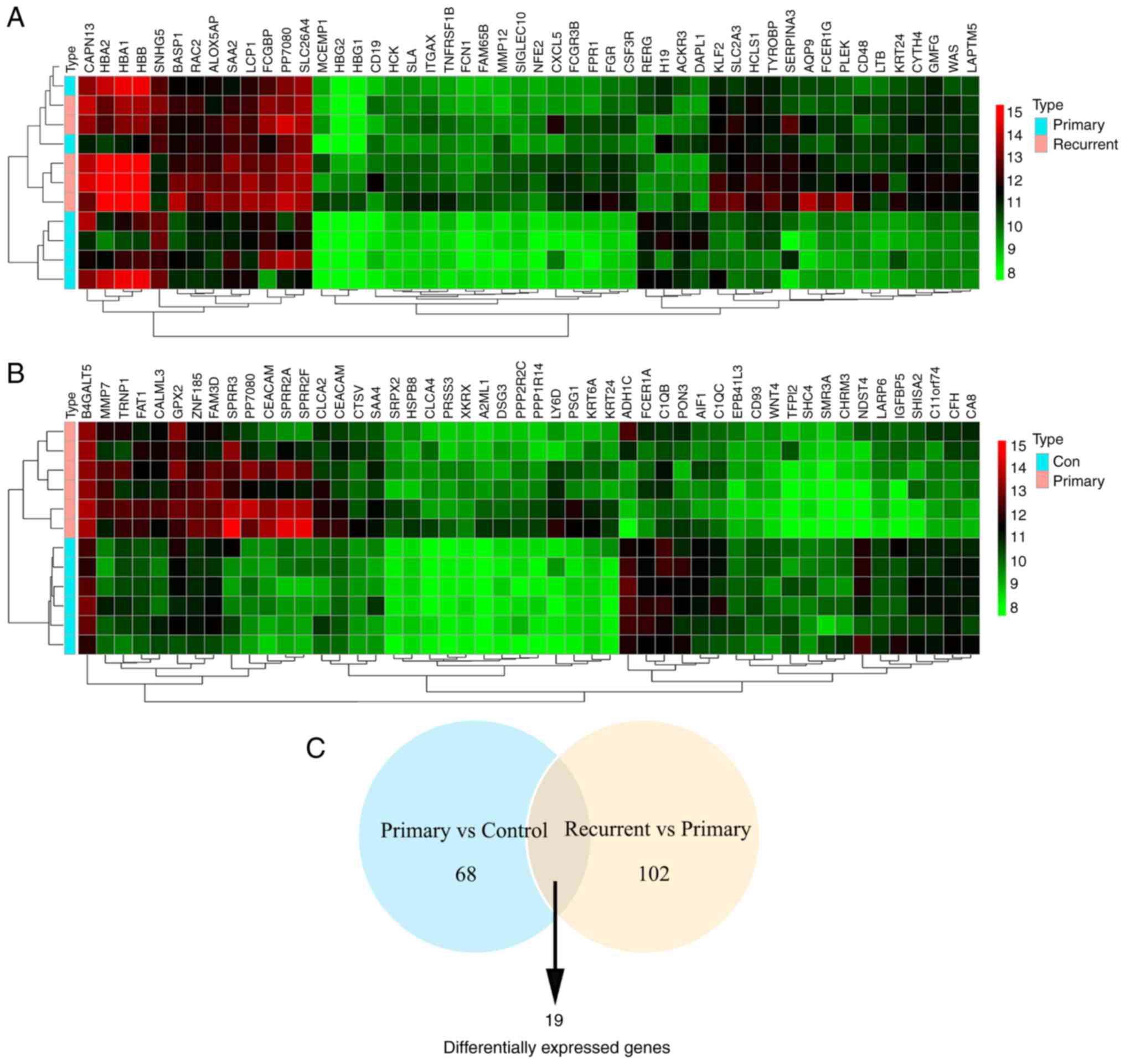
Compared with the control group, 78 genes were
upregulated and 45 genes were downregulated in primary pterygium
tissues determined using microarray analysis (P<0.05). The top
50 differential expressed genes are presented in Fig. 1A. There were 60 upregulated genes and
9 downregulated genes in recurrent pterygium tissues compared with
primary pterygium tissues (P<0.05; Fig. 1B). To identify consistently
differentially expressed genes associated with the recurrence of
pterygium, the common differentially expressed genes between the
three groups were selected (Table
I). Compared with the control and primary groups, PP7080,
SPRR2A, KRT24, SPRR2F, DEFB4A, SERPINA3, S100A7, FCGBP and BPIFA1
were consistently upregulated in recurrent pterygium whilst H19 and
SCGB2A1 were consistently downregulated (Fig. 1C).
 | Table I.Common differentially expressed genes
in control group, primary and recurrent groups. |
Table I.
Common differentially expressed genes
in control group, primary and recurrent groups.
|
| Primary vs.
Control | Recurrent vs.
Primary |
|---|
|
|
|
|
|---|
| Gene | Log2 fold
change | t | P-value | Log2 fold
change | t | P-value |
|---|
| PP7080 | 2.61 | 5.69 | 0.00 | 1.64 | 3.57 | 0.00 |
| SPRR2A | 2.87 | 4.65 | 0.00 | 1.35 | 2.19 | 0.04 |
| KRT24 | 1.42 | 4.25 | 0.00 | 1.04 | 3.12 | 0.01 |
| SPRR2F | 2.71 | 4.23 | 0.00 | 1.36 | 2.13 | 0.05 |
| TYROBP | −1.20 | −3.76 | 0.00 | 1.11 | 3.48 | 0.00 |
| DEFB4A | 2.85 | 3.69 | 0.00 | 1.82 | 2.36 | 0.03 |
| SERPINA3 | 1.68 | 3.53 | 0.00 | 1.91 | 4.03 | 0.00 |
| S100A7 | 3.36 | 3.42 | 0.00 | 2.12 | 2.17 | 0.04 |
| ALOX5AP | −1.07 | −3.21 | 0.00 | 1.01 | 3.03 | 0.01 |
| FCER1G | −1.02 | −3.10 | 0.01 | 1.21 | 3.66 | 0.00 |
| FCGBP | 1.40 | 2.91 | 0.01 | 1.47 | 3.05 | 0.01 |
| PLA2G2A | −1.27 | −2.82 | 0.01 | 1.17 | 2.58 | 0.02 |
| H19 | −1.19 | −2.79 | 0.01 | −1.67 | −2.46 | 0.00 |
| BPIFA1 | 1.53 | 2.57 | 0.02 | 1.29 | 2.18 | 0.04 |
| CAPN13 | −1.08 | −2.52 | 0.02 | 1.43 | 3.34 | 0.00 |
| KRT12 | 1.34 | 2.47 | 0.02 | −1.29 | −2.37 | 0.03 |
| SCGB2A1 | −1.04 | −2.46 | 0.02 | −1.02 | −2.41 | 0.03 |
| CYP1B1 | −1.72 | −2.29 | 0.03 | 1.91 | 2.54 | 0.02 |
| FPR1 | −1.02 | −2.17 | 0.04 | 1.47 | 3.12 | 0.01 |
S100A7 is a marker associated with
recurrence of pterygium
To further validate the aforementioned
differentially expressed genes, RT-qPCR was performed on normal
conjunctival, primary and recurrent pterygium tissues. Results
revealed that S100A7 was upregulated in primary (P<0.05) and
recurrent pterygium tissues (P<0.05); however, there were no
significant differences identified for any other genes (Fig. 2). These results suggested that S100A7
was closely associated with the recurrence of pterygium.
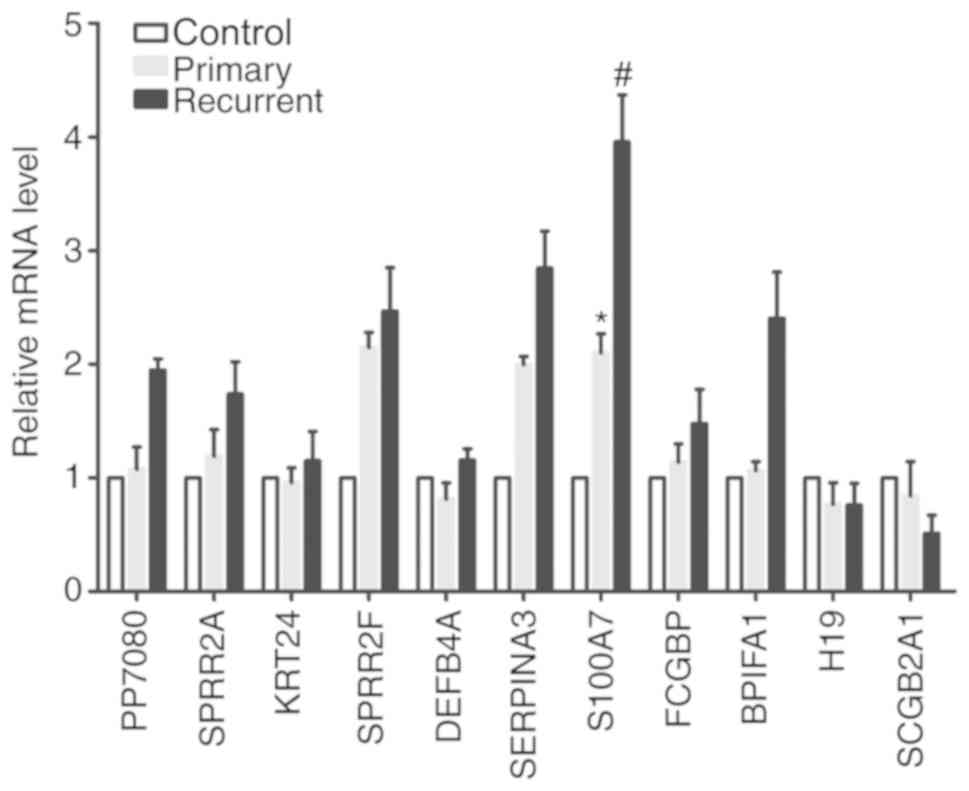 | Figure 2.Differentially expressed genes were
verified by reverse transcription-quantitative PCR analysis.
*P<0.05 vs. Control and #P<0.05 vs. Primary.
Control, ocular trauma group; Primary, primary pterygium group;
Recurrent, recurrent pterygium; SPRR2A, small proline-rich protein
2A; KRT24, keratin 24; SPRR2F, small proline-rich protein 2F;
DEFB4A, defensin β 4A; SERPINA3, serpin family A member 3; S100A7,
S100 calcium-binding protein A7; FCGBP, Fc fragment of IgG binding
protein; BPIFA1, BPI Fold Containing Family A Member 1; H19, H19
imprinted maternally expressed transcript; SCGB2A1, secretoglobin
family 2A member 1. |
S100A7 regulates the immune defense
mechanism and mitogen-activated protein kinase (MAPK)
activation
As S100A7 was significantly upregulated in recurrent
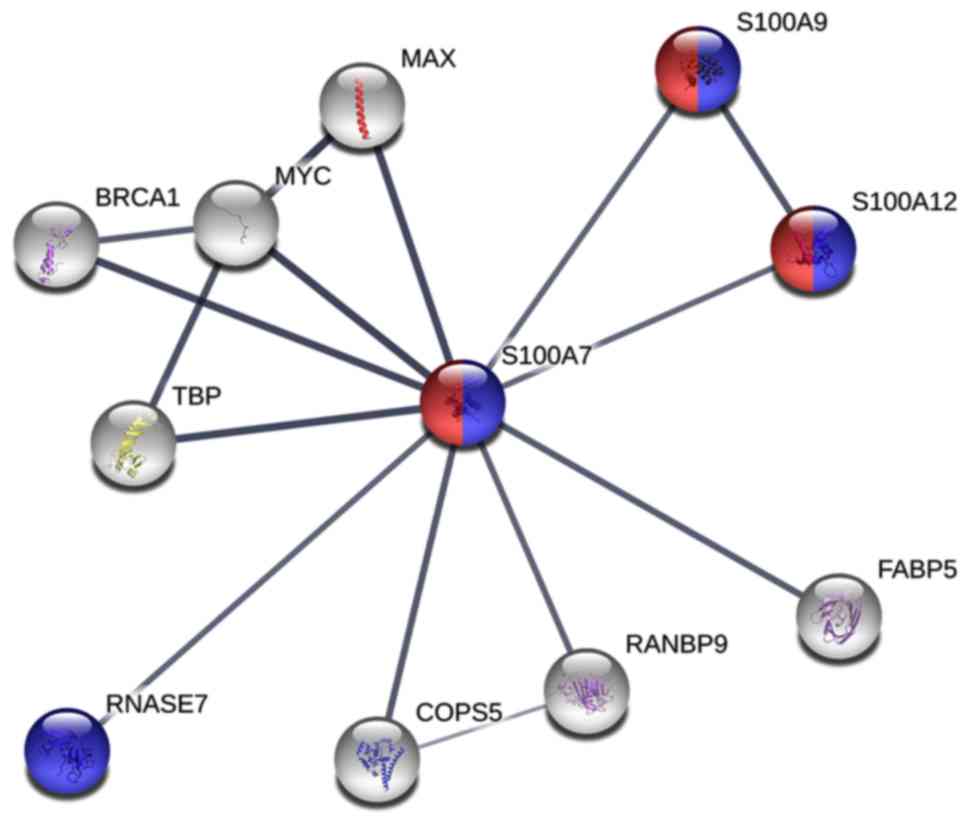
pterygium tissues, function enrichment was performed by PPI and GO
analysis. PPI analysis suggested that S100A7 interacts with ten
genes TATA-box binding protein, breast cancer type 1 susceptibility
protein, MYC, protein max, S100A9, S100A12, ribonuclease 7, COP9
signalosome complex subunit 5, importin-5 and fatty acid-binding
protein 5 (Fig. 3). Furthermore, GO
analysis revealed that these genes were mainly involved in
regulating defense mechanisms against bacteria, MAPK cascade
activation (Table II) and receptor
for advanced glycation end-products (RAGE) receptor binding
(Table III).
 | Table II.Biological processes of genes that
interact with S100A7. |
Table II.
Biological processes of genes that
interact with S100A7.
| Pathway ID | Pathway
description | FDR | Proteins in
network |
|---|
| GO.0042742 | Defense response to
bacterium | 0.028 | RNASE7, S100A12,
S100A7, S100A9 |
| GO.0043408 | Regulation of MAPK
cascade | 0.0423 | COPS5, MYC, RANBP9,
S100A12, S100A7 |
| GO.0045893 | Positive regulation
of transcription, DNA-template | 0.0423 | BRCA1, COPS5, MYC,
S100A12, S100A9, TBP |
| GO.0051238 | Sequestering of metal
ion | 0.0423 | S100A7, S100A9 |
| GO.1902531 | Regulation of
intracellular signal transduction | 0.0423 | COPS5, MYC, RANBP9,
S100A12, S100A7, S100A9 |
| GO.0009966 | Regulation of signal
transduction | 0.0464 | BRCA1, COPS5, MYC,
RANBP9, S100A12, S100A7, S100A9 |
 | Table III.Molecular function (GO) of genes that
interact with S100A7. |
Table III.
Molecular function (GO) of genes that
interact with S100A7.
| Pathway ID | Pathway
description | FDR | Proteins in
network |
|---|
| GO.0050786 | RAGE receptor
binding |
3.99×10−5 | S100A12, S100A7,
S100A9 |
Discussion
In the present study, an integrated analysis of
differential gene expression was performed in primary and recurrent
pterygium tissues by microarray analysis. It was identified then
verified that S100A7 was significantly upregulated in recurrent
pterygium tissues compared with primary pterygium and normal
conjunctival tissues. Bioinformatics further demonstrated that
S100A7 was associated with defense mechanisms against bacteria,
MAPK cascade activation and RAGE receptor binding.
By reviewing worldwide academic research on
pterygium recurrence following primary surgery, it was determined
that age, sunlight exposure, limbal stem cell deficiency and a
large pterygium extent were relevant to pterygium recurrence
(6). At the molecular level,
vascular endothelial growth factor (VEGF) mRNA was observed in
recurrent pterygium tissues (7) with
abnormal expression of heat shock protein 90 also associated with
pterygium recurrence (8). However,
studies investigating the molecular mechanisms of pterygium remain
limited. In the present study, it was demonstrated that the level
of S100A7 was obviously increased in patients with recurrent
pterygium, which suggested that S100A7 may be a potential target
for the treatment of recurrent pterygium.
The S100A7 protein is overexpressed in hyper
proliferative skin diseases, exhibits anti-microbial activities
against bacteria and induces immunomodulatory activities (9). In the present study, elevation of
S100A7 was observed in patients with recurrent pterygium; however,
it could not be determined with certainty whether the upregulation
of S100A7 was a cause or consequence (or both) of recurrent
pterygium. According to previous studies, S100A7 is mainly
associated with the occurrence and development of tumors and tissue
hyperplasia (10,11). Additionally, in psoriasis patients,
S100A7 was also determined to be overexpressed in the epidermis of
psoriatic skin lesions (11). In
addition, S100A7 is closely associated with autoimmune diseases
(12), chronic inflammatory and
angiogenesis (12,13). Pterygium is considered a chronic
inflammatory disease caused by external stimulation and is
manifested as degeneration, hypertrophy, hyperplasia and
angiogenesis of conjunctival and sub-conjunctival tissues (14). Hence, the present study hypothesized
that S100A7-mediated inflammation may be the main cause of
pterygium recurrence. To investigate the potential underlying
mechanism, PPI and GO analyses were performed, and the results
suggested that all genes interacting with S100A7 were also involved
in the immune defense against bacteria and the MAPK inflammatory
response pathway. Furthermore, molecular function analysis
identified that RAGE receptor binding is a major molecular function
of S100A7. RAGE, a nucleic acid receptor, can promote the
inflammatory response to DNA (15).
S100A7 reportedly upregulates the expression of VEGF and promotes
endothelial cell proliferation, which also involves RAGE receptor
binding (16). Therefore, the
aforementioned results revealed that S100A7-mediated inflammation
may be one of the main causes of the recurrence of pterygium. In
fact, S100 proteins, such as S100A8 and S100A9, are all regulatory
proteins that mediate cell growth, apoptosis, differentiation and
inflammation (17); however,
abnormal expression of other S100 family members was not
investigated in this study.
For S100A7 to be established as a biomarker of
pterygium recurrence and a potential therapeutic target, advanced
research utilizing larger datasets is required. The present study
had certain limitations and can be expanded in several ways. First,
it will be interesting to compare the expression of S100A7 with
S100A6, S100A8 and S100A10 then analyze their correlation with
other differentially expressed genes. Second, different S100A
proteins should be investigated to determine whether they might be
superior markers and if the expression of these genes is regulated
by S100A7. Third, investigation into whether S100A7 is correlated
with the size and thickness of primary and recurrent pterygium
tissues should be performed. Finally, the link between the
recurrence of pterygium and the inflammatory response or MAPK
activation needs to be investigated in future studies.
The present study determined that elevation of
S100A7 expression was observed in patients with recurrent
pterygium, therefore, the gene may have potential as a therapeutic
target for treatment of pterygium recurrence.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ performed the experiments, analysis and
interpretation of data. FL designed the study and wrote the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Yantai Affiliated Hospital of Binzhou Medical
University. Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu T, Liu Y, Xie L, He X and Bai J:
Progress in the pathogenesis of pterygium. Curr Eye Res.
38:1191–1197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Monden Y, Hotokezaka F and Yamakawa R:
Recurrent pterygium treatment using mitomycin C, double amniotic
membrane transplantation, and a large conjunctival flap. Int Med
Case Rep J. 11:47–52. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Malozhen SA, Trufanov SV and Krakhmaleva
DA: Pterygium: Etiology, pathogenesis, treatment. Vestn Oftalmol.
133:76–83. 2017.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Raffaele N and Federico T: How to minimize
pterygium recurrence rates: Clinical perspectives. Clin Ophthalmol.
12:2347–2362. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Masters JS and Harris DJ Jr: Low
recurrence rate of pterygium after excision with conjunctival
limbal autograft: A retrospective study with long-term follow-up.
Cornea. 34:1569–1572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Poenaru Sava MG, Raica ML and Cimpean AM:
VEGF mRNA assessment in human pterygium: A new ‘scope’ for a future
hope. Ophthalmic Res. 52:130–135. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sebastiá R, Ventura MP, Solari HP, Antecka
E, Orellana ME and Burnier MN Jr: Immunohistochemical detection of
Hsp90 and Ki-67 in pterygium. Diagn Pathol. 8:322013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wittersheim M, Cordes J, Meyer-Hoffert U,
Harder J, Hedderich J and Gläser R: Differential expression and in
vivo secretion of the antimicrobial peptides psoriasin (S100A7),
RNase 7, human beta-defensin-2 and −3 in healthy human skin. Exp
Dermatol. 22:364–366. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dey KK, Bharti R, Dey G, Pal I, Rajesh Y,
Chavan S, Das S, Das CK, Jena BC, Halder P, et al: S100A7 has an
oncogenic role in oral squamous cell carcinoma by activating
p38/MAPK and RAB2A signaling pathway. Cancer Gene Ther. 23:382–391.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kong F, Li Y, Hu E, Wang R, Wang J, Liu J,
Zhang J, He D and Xiao X: The characteristic of S100A7 induction by
the Hippo-YAP pathway in cervical and glossopharyngeal squamous
cell carcinoma. PLoS One. 11:e01670802016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Batycka-Baran A, Hattinger E, Zwicker S,
Summer B, Zack Howard OM, Thomas P, Szepietowski JC, Ruzicka T,
Prinz JC and Wolf R: Leukocyte-derived koebnerisin (S100A15) and
psoriasin (S100A7) are systemic mediators of inflammation in
psoriasis. J Dermatol Sci. 79:214–221. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mishra S, Ahirwar DK and Ganju RK:
Psoriasin (S100A7): A novel mediator of angiogenesis. Br J
Dermatol. 175:1141–1142. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hsu MY, Lee HN, Liang CY, Wei LC, Wang CY,
Lin KH and Shen YC: Pterygium is related to a decrease in corneal
endothelial cell density. Cornea. 33:712–715. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sirois CM, Jin T, Miller AL, Bertheloot D,
Nakamura H, Horvath GL, Mian A, Jiang J, Schrum J, Bossaller L, et
al: RAGE is a nucleic acid receptor that promotes inflammatory
responses to DNA. J Exp Med. 210:2447–2463. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shubbar E, Vegfors J, Carlström M,
Petersson S and Enerbäck C: Psoriasin (S100A7) increases the
expression of ROS and VEGF and acts through RAGE to promote
endothelial cell proliferation. Breast Cancer Res Treat. 134:71–80.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Charoenpong H, Osathanon T, Pavasant P,
Limjeerajarus N, Keawprachum B, Limjeerajarus CN,
Cheewinthamrongrod V, Palaga T, Lertchirakarn V and Ritprajak P:
Mechanical stress induced S100A7 expression in human dental pulp
cells to augment osteoclast differentiation. Oral Dis. 25:812–821.
2019. View Article : Google Scholar : PubMed/NCBI
|