Introduction
Gut microbiota dysbiosis is associated with the
development and progression of chronic kidney disease (CKD)
(1,2), cardiovascular complications (CVC)
(2–4), obesity (5) and type 2 diabetes mellitus (T2DM)
(5–8). High levels of serum lipopolysaccharide
(LPS), a potent endotoxin produced by Gram-negative bacteria, have
been associated with pathological processes, including diabetes,
the progression of kidney disease, obesity and inflammation
(9,10). The release of LPS from Gram-negative
bacteria in the gut and its passage to the blood causes
LPS-associated toxicity (11). LPS
induces inflammation via a cascade of inflammatory responses
following the recognition of lipid A of LPS by immune cells
(12,13). Lipid A is the toxic component of LPS
and serves as the microbe-specific molecular signal that binds to
the surface receptor complexes of immune cells, which comprise
Toll-like receptor 4 (TLR4) and myeloid differentiation factor 2
(MD2) (14,15). The formation of the signaling complex
can induce the activation of nuclear factor-κB (NF-κB). NF-κB
serves key role in signaling for the production of proinflammatory
cytokines. Therefore, the detection of low quantities of
circulating LPS by the TLR4 receptor system initiates the cascade
of protein-protein interactions, leading to the production of
proinflammatory cytokines (16,17).
A number of factors associated with microbial
alterations in the gut microbiota can lead to changes in host
inflammatory responses (18). Diet
and T2DM-associated medications are among the reported factors that
alter the composition of the gut microbiome (19,20). The
consumption of foods high in fat increases the abundance of
LPS-containing Gram-negative bacteria in the gut at the expense of
Gram-positive bacteria (20,21). Metformin, a blood glucose regulator,
was reported to increase the relative abundance of LPS-producing
Gram-negative gut bacteria (22).
Numerous studies characterized the composition of gut microbiota in
CKD (23,24), and reported the correlation between
LPS, increased intestinal permeability (11,25) and
inflammation in patients with CKD (1,26).
Metabolic syndrome and obesity are influenced by interactions
between the gut microbiota and host genetics (27). Collectively, diet, the host genome
and T2DM medication may be associated with imbalances in the gut
microbiome. The multifactorial network between gut microbiome
dysbiosis and the health of patients suggests an association
between LPS, impaired gut permeability, cardiovascular diseases,
CKD, and patients with T2DM and CKD (T2DM-CKD).
Of note, the aforementioned studies did not analyze
patients with T2DM and advanced CKD (stages 4 and 5, not on
dialysis). The present study investigated the link between gut
Gram-negative bacteria and the derived LPS endotoxin, and the
consequent inflammatory responses in patients with T2DM-CKD. We
evaluated gut microbiome dysbiosis associated with LPS-producing
bacteria in patients with T2DM-CKD compared with healthy control
subjects. In addition, whether elevated serum LPS levels correlated
with chronic inflammation was determined. Improved understanding in
this research field is important for the development of effective
therapeutic and diagnostic approaches for LPS-associated
diseases.
Patients and methods
Study design
Healthy (n=20) and T2DM-CKD (stages 4 and 5, not on
dialysis) (n=20) subjects were ≥18-years-old at enrollment into the
present study. Healthy participants were referred by primary care
physicians, and had no notable medical histories and were not on
any medication. Patients with T2DM-CKD were recruited from the
outpatient clinic of the Texas Tech University Health Sciences
Center (Amarillo, TX, USA). Patients with T2DM-CKD with a
glomerular filtration rate (GFR) of <30 ml/min/1.72
m2 and not on dialysis were enrolled in the present
study. The exclusion criteria included patients treated with
antibiotics for ≥3 days during the month prior to the initiation of
analysis, those diagnosed with end-stage liver disease or chronic
gut-related diseases other than diabetes, pregnant, patients who
previously had bariatric surgeries, and those with a mental status
that restricted informed consent for enrollment. Criteria from
Electronic Medical Records and Genomics (eMERGE) Network were used
in categorizing patients for the study (28). The GFR was calculated using the
CKD-EPI formula (29) and staging
corresponded to the Kidney Disease Outcomes Quality Initiative
guidelines (30). The Institutional
Review Board of Texas Tech University Health Sciences Center
approved the present study. Patients provided informed consent
prior to study participation. The study design included analyses of
the dysbiosis of Gram-negative bacteria in the gut microbiome,
serum inflammatory markers, and endothelial dysfunction biomarkers
in healthy patients and those with T2DM-CKD (stages 4 and 5, not on
dialysis).
Fecal sample collection and DNA
analysis
Stool samples were collected from subjects within 24
h prior to a clinical visit. The samples were processed for DNA
extraction within 24 h using PSP Spin Stool DNA PLUS Kits (Invitek
Biotechnology and Biodesign LTD, Berlin, Germany) according to the
manufacturer's protocols. DNA sequencing and gut microbiota
classification were conducted as previously described (31–33).
Briefly, the hypervariable sub-regions of 16S ribosomal RNA genes
were amplified by polymerase chain reaction with the bacterial
universal primer sets, 341F and 805R, which contained Illumina
adaptors. The obtained libraries were sequenced by MiSeq sequencer
(Illumina Inc., San Diego, CA, USA) using a 600 cycle v3 sequencing
kit. The Human 16S rRNA database was used to remove matching DNA
sequences in the human genome. The microbiome operational taxonomic
units (OTUs) were classified and compiled into the taxonomic levels
as ‘counts’ and ‘percentage’ files.
Biomarkers assays
ELISA kits were employed to assay biomarkers in the
serum from the blood of fasting patients and healthy controls.
ELISA kits (R&D Systems, Minneapolis, MN, USA) were used to
assay endothelin-1 (ET-1), C-reactive protein (CRP), tumor necrosis
factor-α (TNFα), and interleukin-6 (IL6). An ELISA kit obtained
from TSZ ELISA (Waltham, MA, USA) was used to LPS levels.
Statistical analysis
Excel Data Analysis Toolpack and GraphPad Prism 7.01
(GraphPad Software, Inc., La Jolla, CA, USA) were used for the
statistical analysis of various parameters via paired or unpaired
t-tests, and two-way analysis of variance (ANOVA) with Bonferroni's
post hoc test. A Mann-Whitney test was used to compare data between
patients with T2DM-CKD and healthy controls. Regression and
Spearman correlation analyses were used to assess the strength and
direction of correlations between LPS and serum biomarkers.
P<0.05 was considered to indicate a statistically significant
difference. To demonstrate the significance of the source of
variations in the abundance of identified OTUs between the two
groups, two-way ANOVA with an adjusted P-value (multiplicity test)
was applied for each comparison. Multiplicity tests revealed three
adjusted P-values, interaction [p(I)], column factor [p(CF)] and
row factor [p(RF)].
Results
Clinical characteristics
The healthy controls and patients with T2DM-CKD
exhibited no significant differences in gender, age and body mass
index values (Table I). Significant
differences were observed between the control and T2DM-CKD groups
for total cholesterol, triglycerides, and low- and high-density
lipoprotein. Insulin dependence among the diabetic group was 40% at
enrollment. The remaining patients of the group had received
sitagliptin or glipizide. None of the patients with T2MD-CKD were
administered metformin. Patients in the T2DM-CKD group used
ranitidine (10% of the group) and proton pump inhibitors (20% of
the group). Antihypertensive medications were prescribed to 90% of
the recruited patients with T2MD-CKD (mono or combined therapy),
with 40% on angiotensin-converting enzyme inhibitors and 20% on
angiotensin II receptor blockers. A total of 40% of the T2MD-CKD
group were taking statins.
 | Table I.Clinical parameters of patients with
T2DM-CKD and control participants recruited in the present
study. |
Table I.
Clinical parameters of patients with
T2DM-CKD and control participants recruited in the present
study.
| Parameter | T2DM-CKD | Control | P-value |
|---|
| Sex | 9 male, 11
female | 11 male, 9
female | 0.75 |
| Age (years) | 62.8±3.6 | 58.5±4.1 | 0.23 |
| BMI
(kg/m2) | 32.7±3.8 | 29.4±2.6 | 0.18 |
| eGFR (ml/min/1.72
m2) | 16.54±3.01 | >60 | N/A |
| Proteinuria (g/24
h) | 3.58±2.29 | N/A | N/A |
| HDL (mg/dl) | 38.9±6.2 | 58.5±8.4 | 0.01 |
| LDL (mg/dl) | 112±12.7 | 81±9.6 | 0.01 |
| Total cholesterol
(mg/dl) | 161.7±15.2 | 189.0±10.7 | 0.01 |
| Triglycerides
(mg/dl) | 193.1±22.7 | 82.0±17.1 | 0.01 |
Diversity of LPS producing gut
Gram-negative bacteria in the T2DM-CKD and control groups
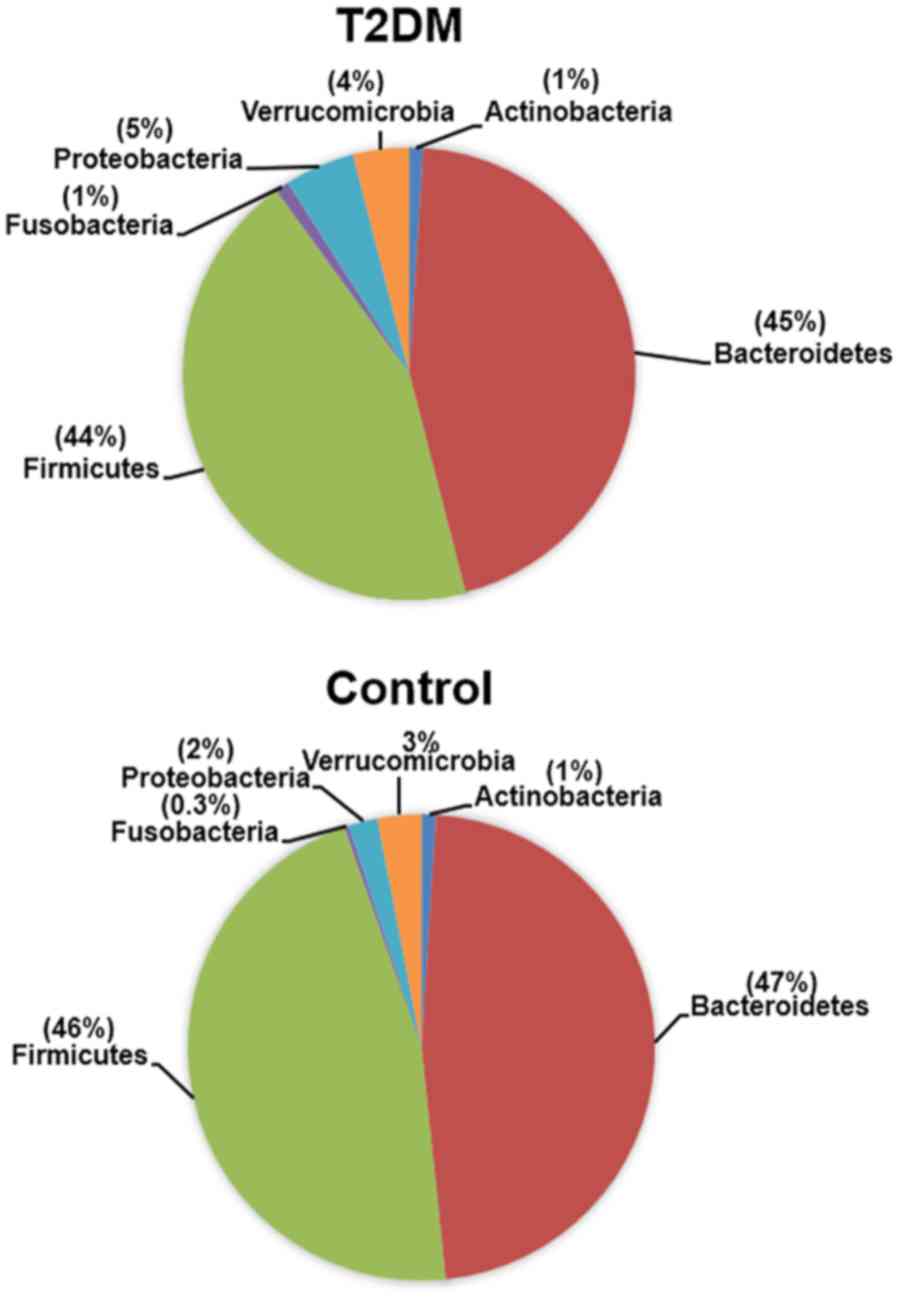
We identified four Gram-negative phyla in the gut
microbiota of the T2DM-CKD and control groups, including
Bacteroidetes, Proteobacteria, Verrucomicrobia and Fusobacteria. Of
the Gram-positive phyla, Firmicutes and Actinobacteria were
identified; the phyla exhibited a mean percentage of ≥0.3%
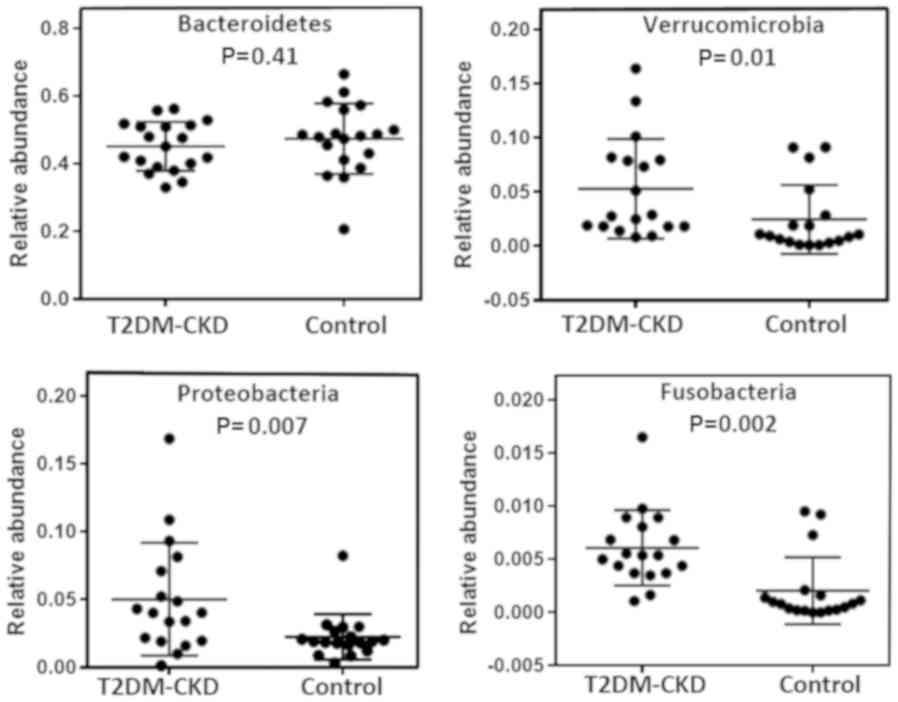
(Fig. 1). The relative abundance of
each phylum in the two study groups, T2DM-CKD and control, revealed
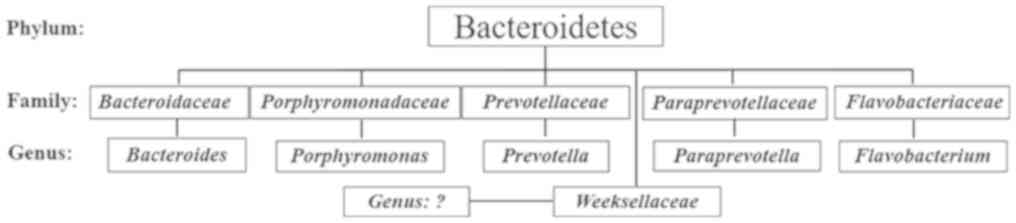
no significant difference for possessing Bacteroidetes (Fig. 2). Within the Bacteroidetes, five
families were identified, including Bacteroidaceae,
Porphyromonadaceae, Prevotellaceae, Paraprevotellaceae and
Flavobacteriaceae; five genera (Bacteroides, Porphyromonas,
Prevotella, Paraprevotella and Flavobacterium) were also
reported in the human gut (Fig. 3).
In addition, bacteria of a sixth family, Weeksellaceae in
the phylum Bacteroidetes, were detected (Fig. 3). To the best our knowledge,
Weeksellaceae has not been reported in the human gut
microbiota, but has been identified in human saliva. Furthermore,
the results of the present study revealed that members of the
Bacteroides genus of the Bacteroidetes phylum were dominant
in the gut microbiota of patients with T2DM-CKD (36%) and controls
(34%); however, no significant difference in the mean percentages
was observed.
The three Gram-negative phyla (Proteobacteria,
Verrucomicrobia and Fusobacteria) exhibited lower mean percentages
(0.3–5%) than Bacteroidetes in the T2DM-CKD and control groups
(Fig. 1). As presented in Fig. 2, Mann-Whitney analysis demonstrated
significantly increased relative abundance of the three
aforementioned Gram-negative phyla in the gut of patients with
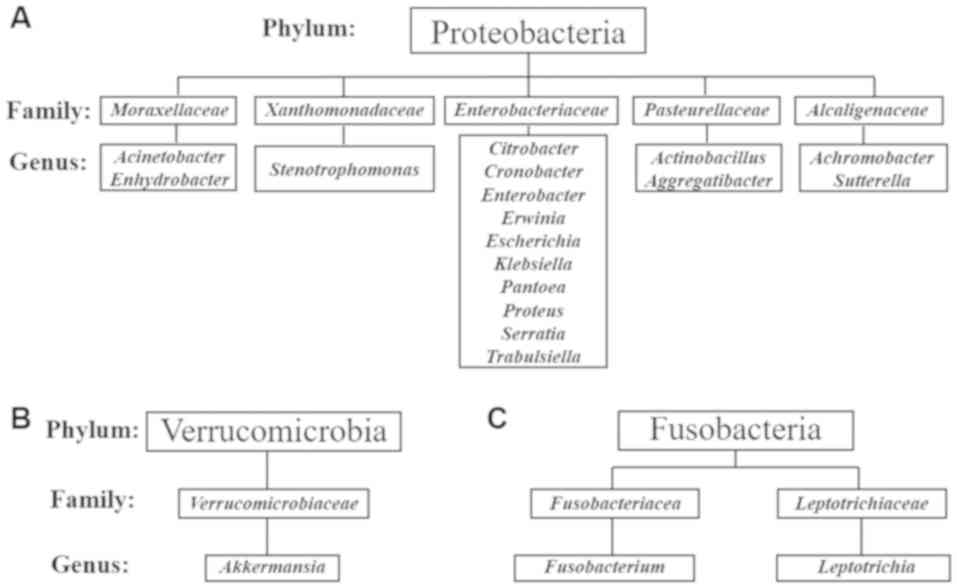
T2DM-CKD compared with the control. A total of 17 genera in the
Gram-negative phylum Proteobacteria and 10 genera within the family
Enterobacteriaceae were reported (Fig. 4A). Akkermansia was identified
in the Gram-negative phylum Verrucomicrobia (Fig. 4B) and two genera, Leptotrichia
and Fusobacterium, were detected in the Gram-negative phylum
Fusobacteria (Fig. 4C).
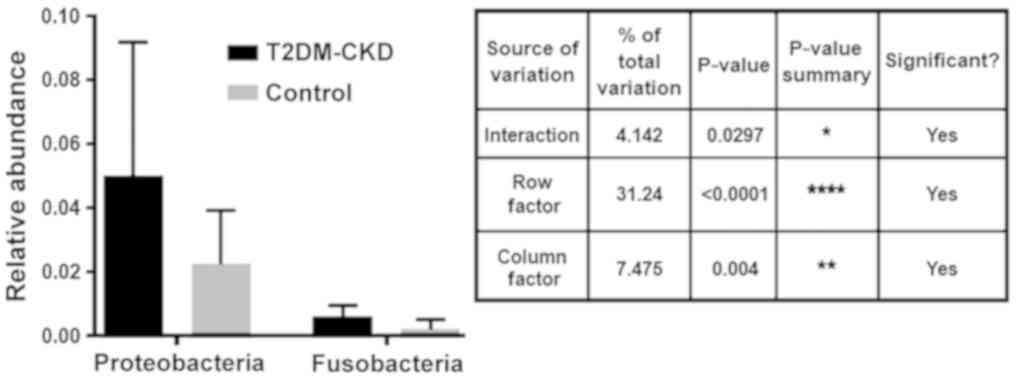
Multiplicity tests with adjusted P-values revealed
the abundance of the identified OTUs between the two study groups,
T2DM-CKD and controls. Analysis demonstrated a significant
difference in the specified source of variation for the relative
abundance of Proteobacteria and Fusobacteria between the T2DM-CKD
and control groups [p(I)=0.0226, p(RF)<0.0001 and p(CF)=0.0184;
Fig. 5]. On the contrary, the
multiplicity tests indicated no significant differences in the
abundance of Proteobacteria and Verrucomicrobia, or Verrucomicrobia
and Fusobacteria between the T2DM-CKD and control groups.
Correlation between elevated levels of
serum LPS and the assayed biomarkers in the T2DM-CKD group
The significant difference observed in the relative
abundance of gut Gram-negative bacteria in patients with T2DM-CKD
compared with the control group may be associated with increased
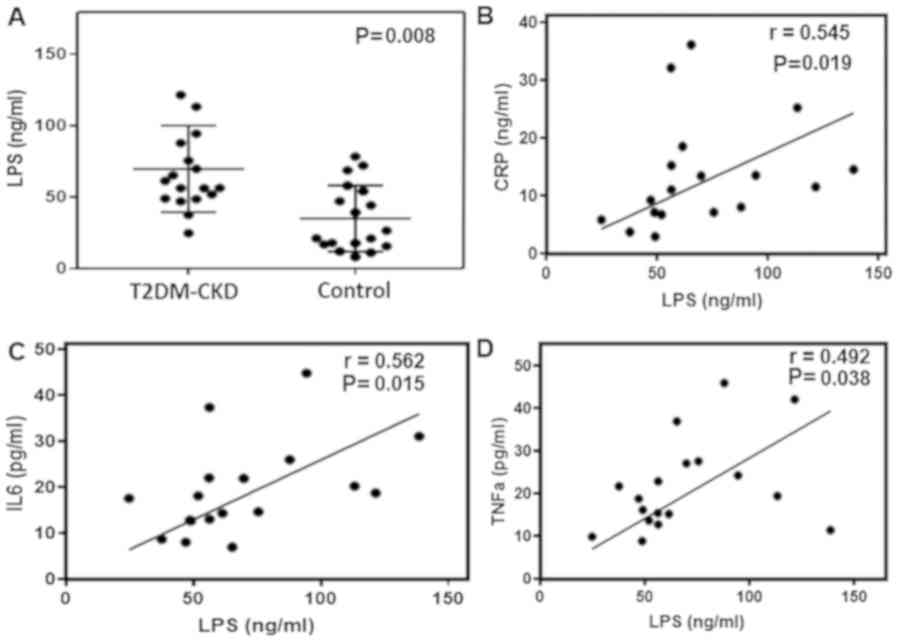
levels of serum LPS. ELISA demonstrated that serum LPS levels were
significantly higher in patients with T2DM-CKD compared with the
controls (P≤0.05; Fig. 6A). Then,
the correlation between elevated serum LPS levels and inflammatory
biomarkers was determined by linear regression and Spearman
correlation analyses. Linear regression was performed using
circulating inflammatory biomarkers as a dependent variable and LPS
as an independent variable. The results demonstrated that elevated
LPS levels were associated with increased levels of CRP, IL6 and
TNFα in the blood of patients with T2DM-CKD (P≤0.05; Fig. 6B-D). Whereas, LPS and endothelial
dysfunction biomarker, ET-1, exhibited a non-significant
correlation. The levels of inflammatory biomarkers were
significantly decreased in healthy individuals compared with
patients with T2DM-CKD and exhibited a non-significant correlation
with LPS.
Discussion
The results demonstrated a shift in the gut
microbial community in patients with T2DM-CKD towards increased
relative abundances of Gram-negative bacteria. The dysbiosis of
Gram-negative gut microbiota is a health risk associated with
elevated LPS levels and correlated with increased levels of
inflammatory biomarkers that can cause chronic inflammation in
patients with T2DM-CKD. LPS forms the outer monolayer of the outer
membranes of the majority of Gram-negative bacteria with profound
effects on the immune system (10,34,35).
Nicholson et al (18)
reported the involvement of bacterial LPS in metabolic and numerous
inflammatory disorders (18).
A total of four Gram-negative phyla, including
Bacteroidetes, Proteobacteria, Verrucomicrobia and Fusobacteria
were identified in the gut microbiota of T2DM-CKD and control
subjects. Previous studies proposed Bacteroidetes as the primary
Gram-negative microbiota in the gastrointestinal tract (36,37). The
present study identified five previously reported genera in the
phylum Bacteroidetes, including Porphyromonas, Prevotella,
Paraprevotella and Flavobacterium (38,39).
Members of the Weeksellaceae family in the Bacteroidetes
phylum had been detected in human saliva but were reported not to
inhabit the human gut (40). In the
present study, members of the genus Bacteroides were
reported to be dominant in the gut microbiota of the T2DM-CKD
(stages 4 and 5, not on dialysis) and control groups; no
significant differences were observed. Jiang et al (41) revealed that members of the genus
Bacteroides were prevalent in samples collected from
patients with end-stage renal disease.
The other three Gram-negative phyla, Proteobacteria,
Verrucomicrobia and Fusobacteria exhibited reduced abundances in
the T2DM-CKD group compared with that of Bacteroidetes. The mean
percentages for Proteobacteria (5%), Verrucomicrobia (4%) and
Fusobacteria (1%) in patients with T2DM-CKD were significantly
higher than the relative abundance in the controls (2, 3 and 0.3%,
respectively). A total of 17 genera in five families in the phylum
Proteobacteria were identified. Several of the identified genera
were within the family Enterobacteriaceae. Of note, the
activity of LPS in Bacteroidetes is less effective compared with
that of other Gram-negative bacteria. Numerous studies reported
that Escherichia coli and other bacteria in the
Enterobacteriaceae family possess markedly increased LPS endotoxin
activity than LPS extracted from the envelope of Bacteroides
fragilis of the Bacteroidetes phylum (42,43). LPS
is structurally similar to lipid A of E. coli; however,
lipid A of B. fragilis differs by the lack of a phosphate
group on C4 of the non-reducing amino sugar, and by the presence of
fewer and various fatty acids (42).
These differences may account for the low endotoxic activity of
B. fragilis-derived LPS. In addition, Bacteroides
fragilis endotoxins are a notable cause of anaerobic bacteremia
and sepsis (44). The present study
also reported the three genera, Akkermansia, Fusobacterium
and Leptotrichia which belong to two phyla, Verrucomicrobia
and Fusobacteria, are a part of the human gut microbiota. Recent
studies reported the genera identified in the present study,
including, Leptotrichia (45), Akkermansia (46) and Fusobacterium (45,47) to
be a part of the human gut flora.
Additionally, the present study revealed a
significant correlation between elevated levels of LPS, and
inflammatory biomarkers, including CRP, TNFα and IL-6 in patients
with T2DM-CKD compared with the controls. Increased levels of
circulating LPS have been reported to promote systemic inflammation
(42,43). The inflammatory response is initiated
following the recognition of lipid A by TLR4 and MD2 of immune
cells (12–15). The formation of the TLR4-MD-2 complex
activates the signaling pathway controlling the expression of
various inflammatory genes (48–51).
Thus, when the levels of LPS in the blood exceed a threshold
concentration, myeloid cells are systemically activated to produce
excessive quantities of pro-inflammatory cytokines, including
IL-1β, IL-6 and TNFα (52,53).
An additional factor contributing to elevated LPS in
the blood is diet. Consuming foods high in fat promotes
Gram-negative bacteria in the gut (20,21). In
the present study, it was observed that patients with T2DM-CKD
consumed more fat than control participants. Furthermore, several
studies revealed the effects of the gut microbiota in regulating
the integrity of the intestine permeability; gut permeability is
critical for health (54,55). Gut bacteria can induce alterations in
zonulin, an essential component of the intercellular tight junction
and an important factor associated with gut permeability (56,57). The
loss of integrity of intestinal permeability allows the
translocation of LPS from the gut into the blood (58–61).
Thus, the loss of gut wall integrity associated with gut microbiota
dysbiosis could increase the levels of circulating LPS in patients
with T2DM-CKD.
Numerous cross-sectional studies reported that
insulin resistance and T2DM are associated with increased levels of
CRP, IL-6 and TNFα, markers of subclinical systemic inflammation
(62). The precise mechanisms
underlying the upregulation of these inflammatory markers require
further investigation; however, the inflammatory response under the
conditions of T2DM may be multifactorial. The present study aimed
to demonstrate LPS as a major factor in the onset of inflammation;
however, other factors, including age, quantity of adipose tissue
and advanced glycation end products may serve a role. Furthermore,
the host genome and drugs have been associated with gut microbiome
imbalances (19,20,27). The
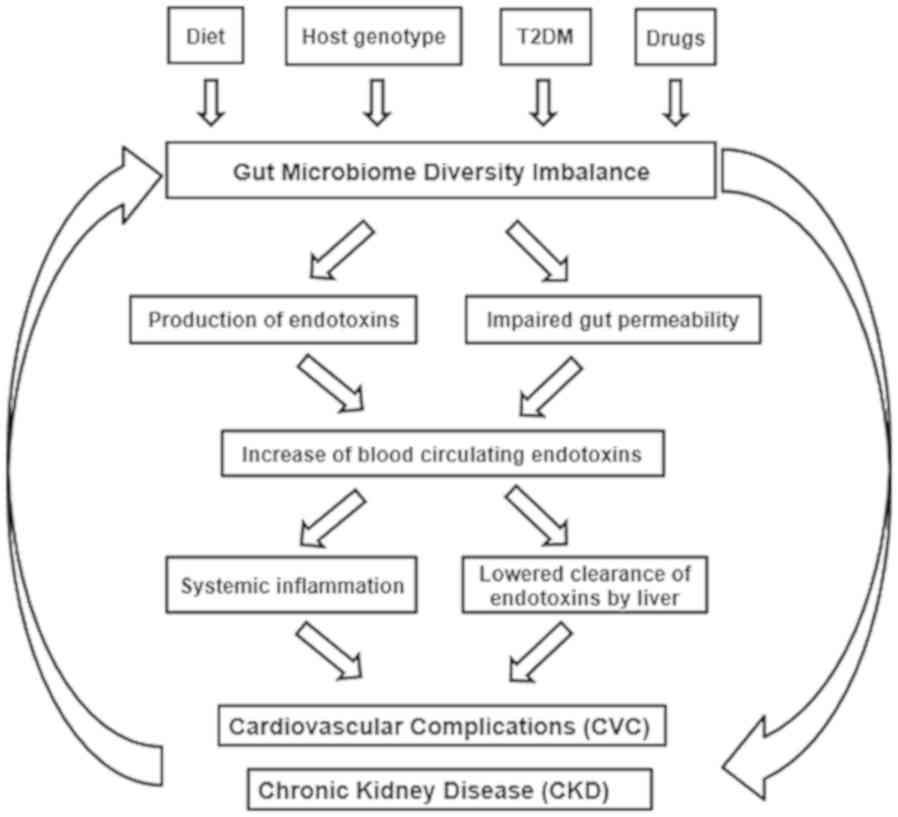
present study proposed a multifactorial association between the
composition of the gut microbiome and the health of patients with
T2DM-CKD (Fig. 7), including the
contribution of T2DM, LPS endotoxin and impaired gut permeability
in the development of CVC, CKD and T2DM-CKD.
However, there are two limitations to this study.
First, using a small number of subjects is not to quantify the
general performance of dysbiosis within T2DM -CKD patients but to
document the existence of an effect (63). The second limitation is the analysis
of the potential association of only four Gram-negative phyla with
LPS. These phyla predominantly exhibited a mean percentage of
greater than or equal to 0.3%.
In conclusion, the present study revealed the
dysbiosis of three Gram-negative phyla, including Proteobacteria,
Verrucomicrobia and Fusobacteria in the gut microbiota of patients
with T2DM-CKD, which was associated with elevated levels of LPS
endotoxin in the blood. Direct therapeutic interventions are
required to reduce the relative abundance of LPS-producing bacteria
in patients with T2DM-CKD.
Acknowledgements
This work is a part of MVS master's thesis of the
Master's Program in Clinical Research, Center for Clinical Research
and Management Education, Division of Health Care Sciences, Dresden
International University, Dresden, Germany. The authors would like
to acknowledge the assistance of Mrs Ibtisam Al-Obaidi (School of
Medicine, Texas Tech University Health Sciences Center, Amarillo,
Texas, USA) for technical services, Dr Kameswara Rao Kottapalli
(Center for Biotechnology and Genomics, Texas Tech University,
Lubbock, Texas, USA) for DNA sequencing, and Dr Candace A. Myers
and Ms Noel Howard (School of Medicine, Texas Tech University
Health Sciences Center, Amarillo, Texas, USA) for editing the
manuscript.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TS and TLV conceived, designed and supervised the
study. MVS, MAIAO and RS performed the experiments and analyzed the
data. MVS, MAIAO, TS and TLV wrote and revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The Institutional Review Board of Texas Tech
University Health Sciences Center/Amarillo, Texas approved the
protocol, which was performed in accordance with the Declaration of
Helsinki. Written informed consent was obtained from each
participant prior to participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chaves LD, McSkimming DI, Bryniarski MA,
Honan AM, Abyad S, Thomas SA, Wells S, Buck M, Sun Y, Genco RJ, et
al: Chronic kidney disease, uremic milieu, and its effects on gut
bacterial microbiota dysbiosis. Am J Physiol Renal Physiol.
315:F487–F502. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mafra D, Lobo JC, Barros AF, Koppe L,
Vaziri ND and Fouque D: Role of altered intestinal microbiota in
systemic inflammation and cardiovascular disease in chronic kidney
disease. Future Microbiol. 9:399–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bu J and Wang Z: Cross-Talk between gut
microbiota and heart via the routes of metabolite and immunity.
Gastroenterol Res Pract. 2018:64580942018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang WW and Hazen SL: The contributory
role of gut microbiota in cardiovascular disease. J Clin Invest.
124:4204–4211. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Harsch IA and Konturek PC: The role of gut
microbiota in obesity and type 2 and type 1 diabetes mellitus: New
insights into ‘old’ diseases. Med Sci (Basel). 6(pii):
E322018.PubMed/NCBI
|
|
6
|
Gomes JMG, Costa JA and Alfenas RCG:
Metabolic endotoxemia and diabetes mellitus: A systematic review.
Metabolism. 68:133–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sabatino A, Regolisti G, Cosola C,
Gesualdo L and Fiaccadori E: Intestinal microbiota in type 2
diabetes and chronic kidney disease. Curr Diab Rep. 17:162017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fallucca F, Porrata C, Fallucca S and
Pianesi M: Influence of diet on gut microbiota, inflammation and
type 2 diabetes mellitus. First experience with macrobiotic Ma-Pi 2
diet. Diabetes Metab Res Rev. 30 (Suppl 1):S48–S54. 2014.
View Article : Google Scholar
|
|
9
|
Harley IT and Karp CL: Obesity and the gut
microbiome: Striving for causality. Mol Metab. 1:21–31. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Raetz CR and Whitfield C:
Lipopolysaccharide endotoxins. Annu Rev Biochem. 71:635–700. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo S, Al-Sadi R, Said HM and Ma TY:
Lipopolysaccharide causes an increase in intestinal tight junction
permeability in vitro and in vitro and in vivo by inducing
enterocyte membrane expression and localization of TLR-4 and CD14.
Am J Pathol. 182:375–387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tidswell M, Tillis W, Larosa SP, Lynn M,
Wittek AE, Kao R, Wheeler J, Gogate J and Opal SM; Eritoran Sepsis
Study Group, : Phase 2 trial of eritoran tetrasodium (E5564), a
toll-like receptor 4 antagonist, in patients with severe sepsis.
Crit Care Med. 38:72–83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bohannon JK, Hernandez A, Enkhbaatar P,
Adams WL and Sherwood ER: The immunobiology of toll-like receptor 4
agonists: From endotoxin tolerance to immunoadjuvants. Shock.
40:451–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dauphinee SM and Karsan A:
Lipopolysaccharide signaling in endothelial cells. Lab Invest.
86:9–22. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carpenter S and O'Neill LA: Recent
insights into the structure of Toll-like receptors and
post-translational modifications of their associated signalling.
Biochem J. 422:1–10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kuzmich NN, Sivak KV, Chubarev VN, Porozov
YB, Savateeva-Lyubimova TN and Peri F: TLR4 signaling pathway
modulators as potential therapeutics in inflammation and sepsis.
Vaccines (Basel). 5(pii): E342017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang F, Liu J, Weng T, Shen K, Chen Z, Yu
Y, Huang Q, Wang G, Liu Z and Jin S: The inflammation induced by
lipopolysaccharide can be mitigated by short-chain fatty acid,
butyrate, through upregulation of IL-10 in septic shock. Scand J
Immunol. 85:258–263. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nicholson JK, Holmes E, Kinross J,
Burcelin R, Gibson G, Jia W and Pettersson S: Host-gut microbiota
metabolic interactions. Science. 336:1262–1267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu H, Esteve E, Tremaroli V, Khan MT,
Caesar R, Mannerås-Holm L, Ståhlman M, Olsson LM, Serino M,
Planas-Fèlix M, et al: Metformin alters the gut microbiome of
individuals with treatment-naive type 2 diabetes, contributing to
the therapeutic effects of the drug. Nat Med. 23:850–858. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ahola AJ, Lassenius MI, Forsblom C,
Harjutsalo V, Lehto M and Groop PH: Dietary patterns reflecting
healthy food choices are associated with lower serum LPS activity.
Sci Rep. 7:65112017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cani PD, Neyrinck AM, Fava F, Knauf C,
Burcelin RG Tuohy KM, Gibson GR and Delzenne NM: Selective
increases of bifidobacteria in gut microflora improve
high-fat-diet-induced diabetes in mice through a mechanism
associated with endotoxaemia. Diabetologia. 50:2374–2383. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Montandon SA and Jornayvaz FR: Effects of
antidiabetic drugs on gut microbiota composition. Genes (Basel).
8(pii): E2502017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Al Khodor S and Shatat IF: Gut microbiome
and kidney disease: A bidirectional relationship. Pediatr Nephrol.
32:921–931. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan
J, DeSantis TZ, Ni Z, Nguyen TH and Andersen GL: Chronic kidney
disease alters intestinal microbial flora. Kidney Int. 83:308–315.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Forsyth CB, Shannon KM, Kordower JH, Voigt
RM, Shaikh M, Jaglin JA, Estes JD, Dodiya HB and Keshavarzian A:
Increased intestinal permeability correlates with sigmoid mucosa
alpha-synuclein staining and endotoxin exposure markers in early
Parkinson's disease. PLoS One. 6:e280322011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Terawaki H, Yokoyama K, Yamada Y, Maruyama
Y, Iida R, Hanaoka K, Yamamoto H, Obata T and Hosoya T: Low-grade
endotoxemia contributes to chronic inflammation in hemodialysis
patients: Examination with a novel lipopolysaccharide detection
method. Ther Apher Dial. 14:477–482. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ussar S, Griffin NW, Bezy O, Fujisaka S,
Vienberg S, Softic S, Deng L, Bry L, Gordon JI and Kahn CR:
Interactions between gut microbiota, host genetics and diet
modulate the predisposition to obesity and metabolic syndrome. Cell
Metab. 22:516–530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Richesson RL, Rusincovitch SA, Wixted D,
Batch BC, Feinglos MN, MirandaM L, Hammond WE, Califf RM and Spratt
SE: A comparison of phenotype definitions for diabetes mellitus. J
Am Med Inform Assoc. 20:e319–e326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
National Kidney Foundation: GFR
calculator. https://www.kidney.org/professionals/kdoqi/gfr_calculatorAugust
9–2017PubMed/NCBI
|
|
30
|
National Kidney Foundation: K/DOQI
clinical practice guidelines for chronic kidney disease:
Evaluation, classification, and stratification. Am J Kidney Dis. 39
(Suppl 1):S1–S266. 2002.PubMed/NCBI
|
|
31
|
Al-Obaide MAI, Singh R, Datta P,
Rewers-Felkins KA, Salguero MV, Al-Obaidi I, Kottapalli KR and
Vasylyeva TL: Gut microbiota-dependent trimethylamine-N-oxide and
serum biomarkers in patients with T2DM and advanced CKD. J Clin
Med. 6(pii): E862017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hiergeist A, Reischl U; Priority Program
1656 Intestinal Microbiota Consortium/quality assessment
participants, ; Gessner A: Multicenter quality assessment of 16S
ribosomal DNA-sequencing for microbiome analyses reveals high
inter-center variability. Int J Med Microbiol. 306:334–342. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Balvočiūtė M and Huson DH: SILVA, RDP,
greengenes, NCBI and OTT-how do these taxonomies compare? BMC
Genomics. 18 (Suppl 2):S1142017. View Article : Google Scholar
|
|
34
|
Raetz CR, Ulevitch RJ, Wright SD, Sibley
CH, Ding A and Nathan CF: Gram-negative endotoxin: An extraordinary
lipid with profound effects on eukaryotic signal transduction.
FASEB J. 5:2652–2660. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Raetz CR, Guan Z, Ingram BO, Six DA, Song
F, Wang X and Zhao J: Discovery of new biosynthetic pathways: The
lipid a story. J Lipid Res. 50 (Suppl):S103–S108. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thomas F, Hehemann JH, Rebuffet E, Czjzek
M and Michel G: Environmental and gut bacteroidetes: The food
connection. Front Microbiol. 2:932011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
d'Hennezel E, Abubucker S, Murphy LO and
Cullen TW: Total lipopolysaccharide from the human gut microbiome
silences Toll-like receptor signaling. mSystems. 2(pii): e00046–17.
2017.PubMed/NCBI
|
|
38
|
Santoru ML, Piras C, Murgia A, Palmas V,
Camboni T, Liggi S, Ibba I, Lai MA, Orrù S, Blois S, et al: Cross
sectional evaluation of the gut-microbiome metabolome axis in an
Italian cohort of IBD patients. Sci Rep. 7:95232017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun J and Kato I: Gut microbiota,
inflammation and colorectal cancer. Genes Dis. 3:130–143. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guerrero-Preston R, Godoy-Vitorino F,
Jedlicka A, Rodríguez-Hilario A, González H, Bondy J, Lawson F,
Folawiyo O, Michailidi C, Dziedzic A, et al: 16S rRNA amplicon
sequencing identifies microbiota associated with oral cancer, human
papilloma virus infection and surgical treatment. Oncotarget.
7:51320–51334. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jiang S, Xie S, Lv D, Wang P, He H, Zhang
T, Zhou Y, Lin Q, Zhou H, Jiang J, et al: Alteration of the gut
microbiota in Chinese population with chronic kidney disease. Sci
Rep. 7:28702017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lindberg AA, Weintraub A, Zähringer U and
Rietschel ET: Structure-activity relationships in
lipopolysaccharides of Bacteroides fragilis. Rev Infect Dis.
12 (Suppl 2):S133–S241. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ramachandran G: Gram-positive and
gram-negative bacterial toxins in sepsis: A brief review.
Virulence. 5:213–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lukiw WJ: Bacteroides fragilis
lipopolysaccharide and inflammatory signaling in alzheimer's
disease. Front Microbiol. 7:15442016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Couturier MR, Slechta ES, Goulston C,
Fisher MA and Hanson KE: Leptotrichia bacteremia in patients
receiving high-dose chemotherapy. J Clin Microbiol. 50:1228–1232.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cekanaviciute E, Yoo BB, Runia TF,
Debelius JW, Singh S, Nelson CA, Kanner R, Bencosme Y, Lee YK,
Hauser SL, et al: Gut bacteria from multiple sclerosis patients
modulate human T cells and exacerbate symptoms in mouse models.
Proc Natl Acad Sci USA. 114:10713–10718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hsieh YY, Tung SY, Pan HY, Yen CW, Xu HW,
Lin YJ, Deng YF, Hsu WT, Wu CS and Li C: Increased abundance of
Clostridium and Fusobacterium in gastric microbiota
of patients with gastric cancer in Taiwan. Sci Rep. 8:1582018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Apte RN, Pluznik DH and Galanos C: Lipid
A, the active part of bacterial endotoxins in inducing serum colony
stimulating activity and proliferation of splenic
granulocyte/macrophage progenitor cells. J Cell Physiol. 87:71–78.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu YC, Yeh WC and Ohashi PS: LPS/TLR4
signal transduction pathway. Cytokine. 42:145–151. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Park BS, Song DH, Kim HM, Choi BS, Lee H
and Lee JO: The structural basis of lipopolysaccharide recognition
by the TLR4-MD-2 complex. Nature. 458:1191–1195. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
McGettrick AF and O'Neill LA: Regulators
of TLR4 signaling by endotoxins. Subcell Biochem. 53:153–171. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Oberholzer A, Oberholzer C and Moldawer
LL: Sepsis syndromes: Understanding the role of innate and acquired
immunity. Shock. 16:83–96. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Grasset E, Puel A, Charpentier J, Collet
X, Christensen JE, Tercé F and Burcelin R: A specific gut
microbiota dysbiosis of type 2 diabetic mice induces GLP-1
resistance through an enteric NO-dependent and gut-brain axis
mechanism. Cell Metab. 25:1075–1090.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Farhadi A, Banan A, Fields J and
Keshavarzian A: Intestinal barrier: An interface between health and
disease. J Gastroenterol Hepatol. 18:479–497. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Camilleri M, Madsen K, Spiller R,
Greenwood-Van Meerveld B and Verne GN: Intestinal barrier function
in health and gastrointestinal disease. Neurogastroenterol Motil.
24:503–512. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tripathi A, Lammers KM, Goldblum S,
Shea-Donohue T, Netzel-Arnett S, Buzza MS, Antalis TM, Vogel SN,
Zhao A, Yang S, et al: Identification of human zonulin, a
physiological modulator of tight junctions, as prehaptoglobin-2.
Proc Natl Acad Sci USA. 106:16799–16804. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li C, Gao M, Zhang W, Chen C, Zhou F, Hu Z
and Zeng C: Zonulin regulates intestinal permeability and
facilitates enteric bacteria permeation in coronary artery disease.
Sci Rep. 6:291422016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kelly JR, Kennedy PJ, Cryan JF, Dinan TG,
Clarke G and Hyland NP: Breaking down the barriers: The gut
microbiome, intestinal permeability and stress-related psychiatric
disorders. Front Cell Neurosci. 9:3922015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bischoff SC, Barbara G, Buurman W,
Ockhuizen T, Schulzke JD, Serino M, Tilg H, Watson A and Wells JM:
Intestinal permeability-a new target for disease prevention and
therapy. BMC Gastroenterol. 14:1892014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wang L, Llorente C, Hartmann P, Yang AM,
Chen P and Schnabl B: Methods to determine intestinal permeability
and bacterial translocation during liver disease. J Immunol
Methods. 421:44–53. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Fukui H, Brauner B, Bode JC and Bode C:
Plasma endotoxin concentrations in patients with alcoholic and
non-alcoholic liver disease: Reevaluation with an improved
chromogenic assay. J Hepatol. 12:162–169. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
de Rekeneire N, Peila R, Ding J, Colbert
LH, Visser M, Shorr RI, Kritchevsky SB, Kuller LH, Strotmeyer ES,
Schwartz AV, et al: Diabetes, hyperglycemia, and inflammation in
older individuals: The health, aging and body composition study.
Diabetes Care. 29:1902–1908. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Anderson AJ and Vingrys AJ: Small samples:
Does size matter? Invest Ophthalmol Vis Sci. 42:1411–1413.
2001.PubMed/NCBI
|