Introduction
Thyroid cancer (TC) is a common malignancy of the
endocrine system. According to the pathological types, >80% of
TC is papillary thyroid carcinoma (PTC) (1). A total of 15–30% thyroid nodules
evaluated using fine-needle aspiration (FNA) are not clearly benign
or malignant (2). Genomic
alterations have been fully interpreted by studies in recent years.
The Cancer Genome Atlas (TCGA) Research Network performed the
largest cohort study of PTC, and created a comprehensive
multiplatform analysis (3). A
previous study by Ye et al (4) drew the genetic landscape of benign
thyroid nodules and thyroid cancer, and suggested that PTC and
benign nodules have independent origins (4).
BRAF V600E is the most common mutation observed in
PTC (3), and was recommended for the
evaluation of thyroid nodules by the NCCN Guidelines of Thyroid
Carcinoma (5). However, tumor tissue
DNA sequencing has limitations. Tumors are heterogeneous, and the
results of sequencing of a tumor section do not represent the whole
tumor (6). Additionally, a surplus
of tissues following FNA cytology is difficult to obtain, or the
amount obtained may not be sufficient for sequencing to be
performed. Circulating tumor DNA (ctDNA) is the cell-free fragment
of DNA, which is released into the bloodstream by tumor cells, and
the ctDNA load is associated with tumor staging and prognosis
(7). Therefore, the detection of
these known somatic gene mutations in tumors and the corresponding
plasma has the potential to aid in the diagnosis of thyroid tumors.
Previous studies have confirmed that there is controversy regarding
ctDNA detection in patients with thyroid tumors. Although some
studies have clarified the clinical significance of ctDNA (8–10),
others have raised the issue that the analysis of ctDNA cannot
improve the clinical management of patients with thyroid carcinoma,
due to plasma ctDNA being undetectable (11,12).
In the present study, whole-exome sequencing (WES)
was performed on tumor and matched normal tissues from 10 patients
with PTC from Quanzhou, China, and a genomic alteration profile was
created. The validation cohort, including 59 patients with PTC, was
recruited to detect hotspot mutations in tumor DNA and cell-free
DNA using QuantStudio™ 3D digital PCR. The accuracy of ctDNA
detection was analyzed according to the sequencing results of the
tumor DNA.
Materials and methods
Patients and samples
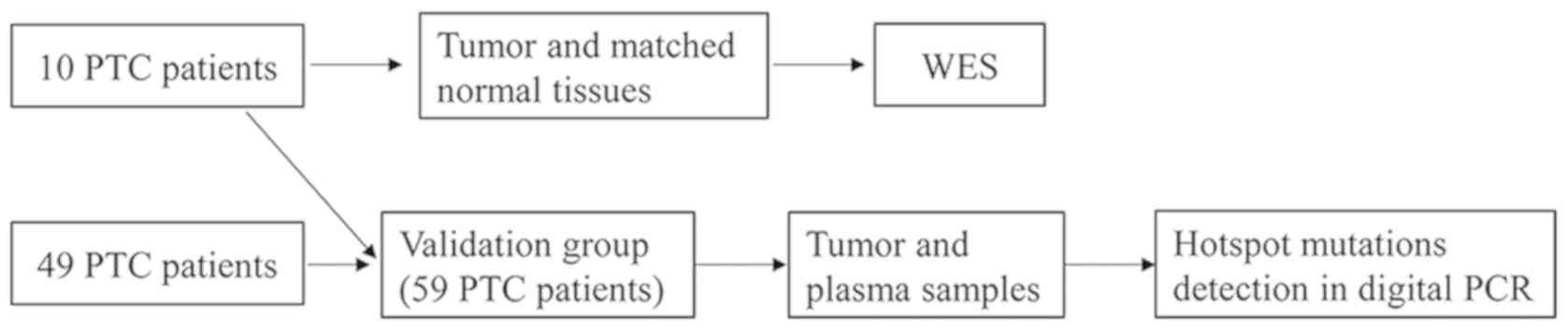
A total of 59 patients (Fig. 1) with PTC were recruited from
Quanzhou First Hospital Affiliated to Fujian Medical University
(Quanzhou, China) between August 2017 and June 2018. None of them
had received therapeutic procedures such as chemotherapy. For 10 of
the 59 patients, tumor and matched normal tissues from these
patients were sampled during surgery, stored at −80°C then
subjected to WES (Table I). The
matched normal tissues were collected which were at least 3 cm
distance from cancer-foci. Tumor tissue and plasma were collected
from all 59 patients, whose clinical characteristics are presented
in Table II. The TMN Classification
of Malignant Tumors (TNM) staging refers to AJCC Cancer Staging
Manual Eighth Edition (13). All
surgical specimens were pathologically diagnosed as PTC, and
patients with other malignant tumors were excluded. Fresh tissues
for WES were snap-frozen in liquid nitrogen immediately following
surgical resection and subsequently stored at −80°C. Tumor tissues
for PCR, from 49 patients, were formalin-fixed and paraffin
embedded (FFPE) and collected from the pathology department
following surgery. Tissues were fixed in 10% neutral formalin
overnight at room temperature before embedding in paraffin. Prior
to surgery, 10 ml peripheral blood was collected in a Cell-Free DNA
BCT blood collection tube (Streck, Inc.), and the plasma was
separated immediately and subjected to 800 × g centrifugation for
10 min at 4°C and a 2500 × g centrifugation for 10 min at 4°C.
 | Table I.Clinical characteristics and
whole-exome sequencing quality control of patients with papillary
thyroid carcinoma. |
Table I.
Clinical characteristics and
whole-exome sequencing quality control of patients with papillary
thyroid carcinoma.
| Case no. | Age, years | Sex | Multifocality | TNM stage | Tumor sequencing
depth (x) | Normal sequencing
depth (x) | TMB
(mutations/Mb) | BRAF V600E |
|---|
| 1 | 40 | Female | Yes | T1aN0M0 | 187.32 | 99.89 | 0.29 | − |
| 2 | 29 | Female | No | T1aN0M0 | 179.27 | 98.48 | 0.35 | − |
| 3 | 43 | Female | No | T1aN0M0 | 169.37 | 86.95 | 0.38 | − |
| 4 | 65 | Male | Yes | T1aN0M0 | 82.39 | 56.64 | 0.29 | − |
| 5 | 38 | Female | Yes | T1aN1aM0 | 67.98 | 31.87 | 0.16 | − |
| 6 | 45 | Female | No | T1aN1aM0 | 159.76 | 39.63 | 1.35 | + |
| 7 | 34 | Female | No | T1aN1aM0 | 174.48 | 47.25 | 1.06 | + |
| 8 | 37 | Female | No | T1aN1aM0 | 81.04 | 46.23 | 0.32 | + |
| 9 | 29 | Female | No | T1aN1aM0 | 133.27 | 37.27 | 1.07 | + |
| 10 | 55 | Male | No | T1aN1aM0 | 150.61 | 79.92 | 0.38 | + |
 | Table II.Clinical characteristics of the
validation cohort, and association with BRAF V600E status in
tumors. |
Table II.
Clinical characteristics of the
validation cohort, and association with BRAF V600E status in
tumors.
|
|
| BRAF V600E in
tumors |
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristic | No. patients | Positive
(n=26) | Negative
(n=33) | P-value |
|---|
| Age (median,
range) | 41 (22–65) | 39 (22–64) | 41 (22–65) | 0.283 |
| Sex |
|
|
|
|
| Male
(%) | 17 (28.81%) | 7 | 10 | 0.078 |
| Female
(%) | 42 (71.19%) | 19 | 23 |
|
| Tumor size, cm;
median (range) | 1.8 (0.2–5.4) | 1.7 (0.2–4.6) | 2.3 (0.6–5.4) | 0.658 |
| LNM |
|
|
|
|
|
Positive (%) | 20 (33.90) | 15 | 5 | 0.001b |
|
Negative (%) | 39 (66.10) | 11 | 28 |
|
| Capsule
invasion |
|
|
|
|
|
Positive (%) | 20 (33.90) | 11 | 9 | 0.226 |
|
Negative (%) | 39 (66.10) | 15 | 24 |
|
| Stage |
|
|
|
|
| I+II
(%) | 38 (64.41) | 12 | 26 | 0.009a |
| III+IV
(%) | 21 (35.59) | 14 | 7 |
|
DNA extraction
Genomic DNA (gDNA) from fresh tissues was extracted
using a Hipure Tissue DNA Mini kit (Magen). gDNA was extracted from
FFPE tissues using a GeneRead DNA FFPE kit (Qiagen China Co., Ltd.)
and plasma cfDNA was extracted using a HiPure Circulating DNA Midi
kit (Pharmaceutical Product Development, Inc.), according to the
manufacturers' protocols. The quantity and purity of gDNA were
assessed using a Qubit® 3.0 Fluorometer (Invitrogen;
Thermo Fisher Scientific, Inc.) and a NanoDrop ND-1000 device
(Thermo Fisher Scientific, Inc.).
WES
A total of 300 ng of each gDNA sample, based on the
Qubit quantification, were mechanically fragmented (duty factor
10%; peak incident power 175 W; cycles per burst 150; treatment
time 150 sec; bath temperature 4–8°C) on a Covaris E220 focused
ultrasonicator (Covaris, Inc.). Sheared gDNA (200 ng) was used to
perform the end repairs, and A-tailing and adapter ligation was
performed with KAPA Hyper Prep kit (cat. no. KK8504; Kapa
Biosystems, Inc.), according to the manufacturer's protocol.
Libraries were then captured using Agilent SureSelect Human All
Exon (version 6; Agilent Technologies, Inc.) probes, and
subsequently amplified. Libraries were sequenced using an Illumina
HiSeq 2500 platform (Illumina, Inc.) on high output mode, and 2×150
cycles were performed using TruSeq SBS chemistry (version 3;
Illumina, Inc.).
Bioinformatics analysis
Clean data were obtained from filtering out the
adapter, low-quality reads and reads with a proportion >10%.
Reads were aligned to the reference human genome (UCSC hg19;
http://genome.ucsc.edu/) using the
Burrows-Wheeler Aligner (version 0.7.12) (14). Local realignment, PCR duplicate
marking, base-quality recalibration, and calculation of coverage
metrics were performed using GATK v.3.2 (15) and Picard tools (http://broadinstitute.github.io/picard/).
Somatic single-nucleotide variants and InDels of
tumors compared with matched normal tissues were annotated using
MuTect software (version 1.1.4) (16). The mutations with variant allele
frequencies >1% were filtered and the variants in non-coding
regions and synonymous variants were excluded from analysis. Gene
ontology (GO; http://www.geneontology.org/) and Kyoto Encyclopedia
of Genes and Genomes (KEGG; http://www.kegg.jp/) enrichment analyses were
performed in order to investigate the biological importance of the
somatic mutated genes using ClusterProfiler (17) in R software (version 3.5.1) (18).
Hotspot mutations detected by
QuantStudio™ 3D digital PCR
BRAF V600E and NRAS p.Q61R were detected using
QuantStudio™ 3D Digital PCR Master Mix v2 (cat. no. A26358) in a
QuantStudio™ 3D digital PCR system (both Thermo Fisher Scientific,
Inc.). The primer sequences of BRAF V600E were as follows: Forward,
5′-CTACTGTTTTCCTTTACTTACTACACCTCAGA-3′ and reverse,
5′-ATCCAGACAACTGTTCAAACTGATG-3′. The primer sequences of NRAS
p.Q61R were as follows: Forward, 5′-CACACCCCCAGGATTCTTACA-3′ and
reverse, 5′-TGTATTGGTCTCTCATGGCACTGT-3′. A solution containing 20
µl PCR reaction solution was prepared. The QuantStudio™ 3D Digital
PCR 20 K Chip kit v2 (cat. no. A26316; Thermo Fisher Scientific,
Inc.) was used to prepare the PCR reaction with 14.5 µl sealed
sample oil, which was then placed in the sample slot of the PreFlex
PCR system for PCR amplification. Procedures were performed at 95°C
for 10 min; 98°C for 30 sec; and 60°C for 2 min, for a total of 45
cycles. The amplified chip was placed in the chip analyzer to read
the data, and the QuantStudio™ 3D Analysis Suite software was used
to quantify the number of mutations in the samples from the
collected data in the QuantStudio™ 3D Digital PCR System.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 19.0; IBM Corp.). Differences in distributions
between the clinicopathological characteristics and molecular
status were assessed using a χ2 or Fisher's exact test,
as appropriate. Linear regression analysis was performed and
graphical plots were generated using GraphPad Prism software
(version 6.0; GraphPad Software, Inc.), OriginPro 8.1 (OriginLab,
Corp.) and R. The OncoPrint function of ComplexHeatmap package
(19) in R was used to visualize
genomic alterations. P<0.05 was considered to indicate a
statistically significant difference.
Results
Clinical characteristics and
whole-exome sequencing data
The clinical characteristics of the WES cohort and
all patients with PTC are presented in Tables I and II, respectively. The median patient age
was 41 years (range, 22–65 years). Of the 59 patients with PTC, 42
were female, and the remaining 17 were male. Tumors of 20 (33.90%)
patients exhibited lymph node metastasis (LNM), and capsule
invasion also occurred in 20 (33.90%) tumors.
For the sequencing depth of 10 patients with PTC,
WES achieved an average of 138.55× coverage of tumor genomes and
62.41× coverage of the germline genomes (Table I). The average tumor mutation burden
was 0.565 (range, 0.29–1.35) mutations/Mb. PTC presents a low
frequency of somatic alterations compared with other types of
carcinoma (3), which may contribute
to the good prognosis exhibited by PTC.
Somatic gene mutations in PTC as
identified using WES
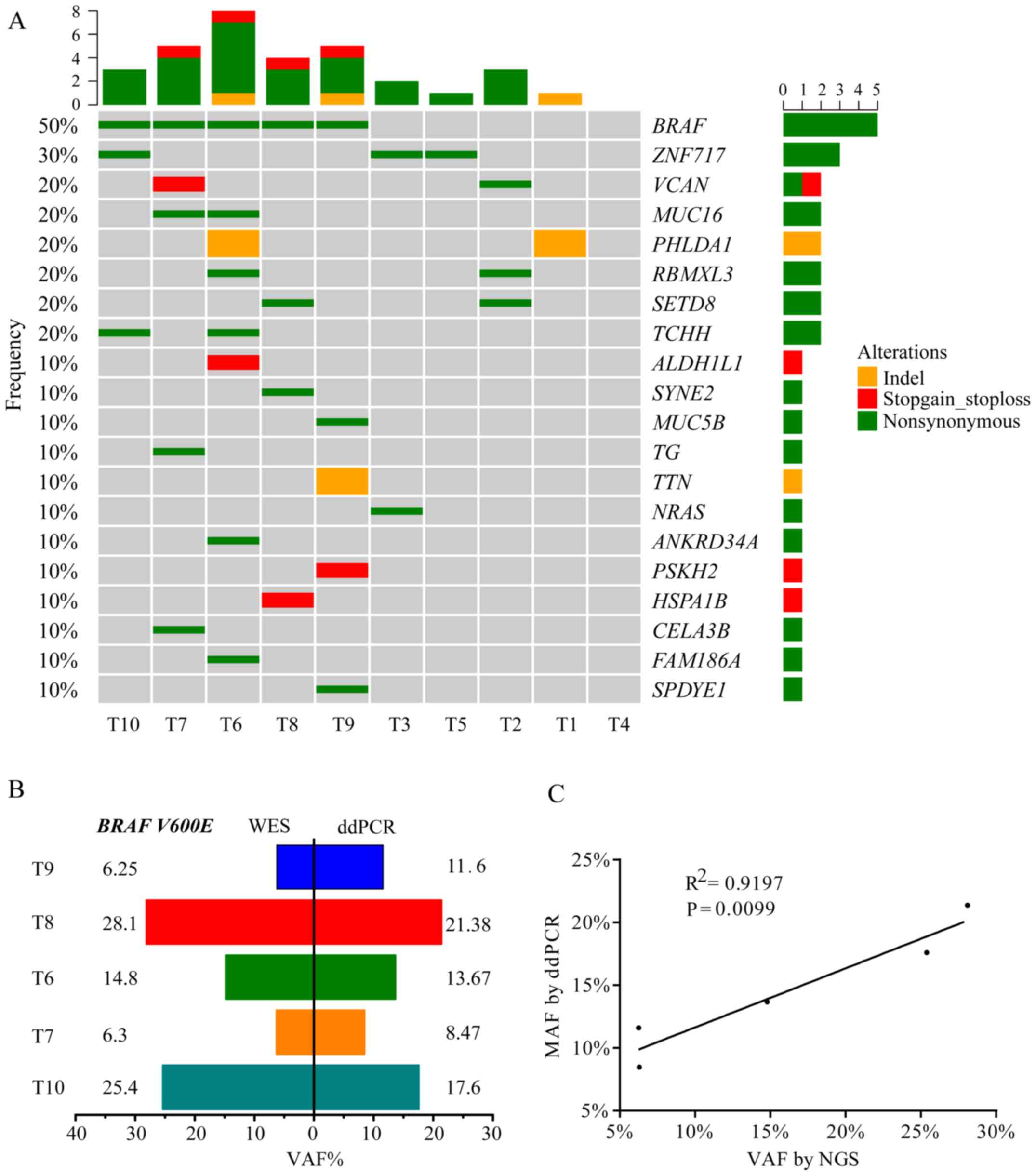
Fig. 2A presents the
WES results of 10 patients with PTC, including the top 20 mutated
genes, mutation type and frequency. BRAF V600E was the hotspot
mutation in PTC and exhibited a 50.0% frequency, which was
comparable to a TCGA network study of PTCs (59.7% mutated frequency
of BRAF V600E) and to other previous studies (3,20–22). The
results also revealed that five tumors with the BRAF V600E mutation
were all from patients with PTC who exhibited lymph node
metastasis. Digital PCR was used to validate the BRAF
V600E-positive tumors that were detected using WES. The results of
this analysis indicated a good degree of consistency in BRAF status
(Fig. 2B) and variation allele
frequency (VAF; Fig. 2C) between WES
and digital PCR. VAF of BRAF V600E detected by WES had a positive
linear correlation (R2=0.9197; P=0.0099) with digital
PCR (Fig. 2C). A number of novel
gene mutations were identified, including zinc finger protein
(ZNF)717, pleckstrin homology like domain family A member 1
(PHLDA1), RBMX like 3 (RBMXL3), lysine methyltransferase 5A (SETD8)
and trichohyalin (TCHH), along with reported genes, including BRAF,
NRAS and mucin 16 (MUC16).
Functional enrichment analysis of
mutated genes
To further determine the biological function of all
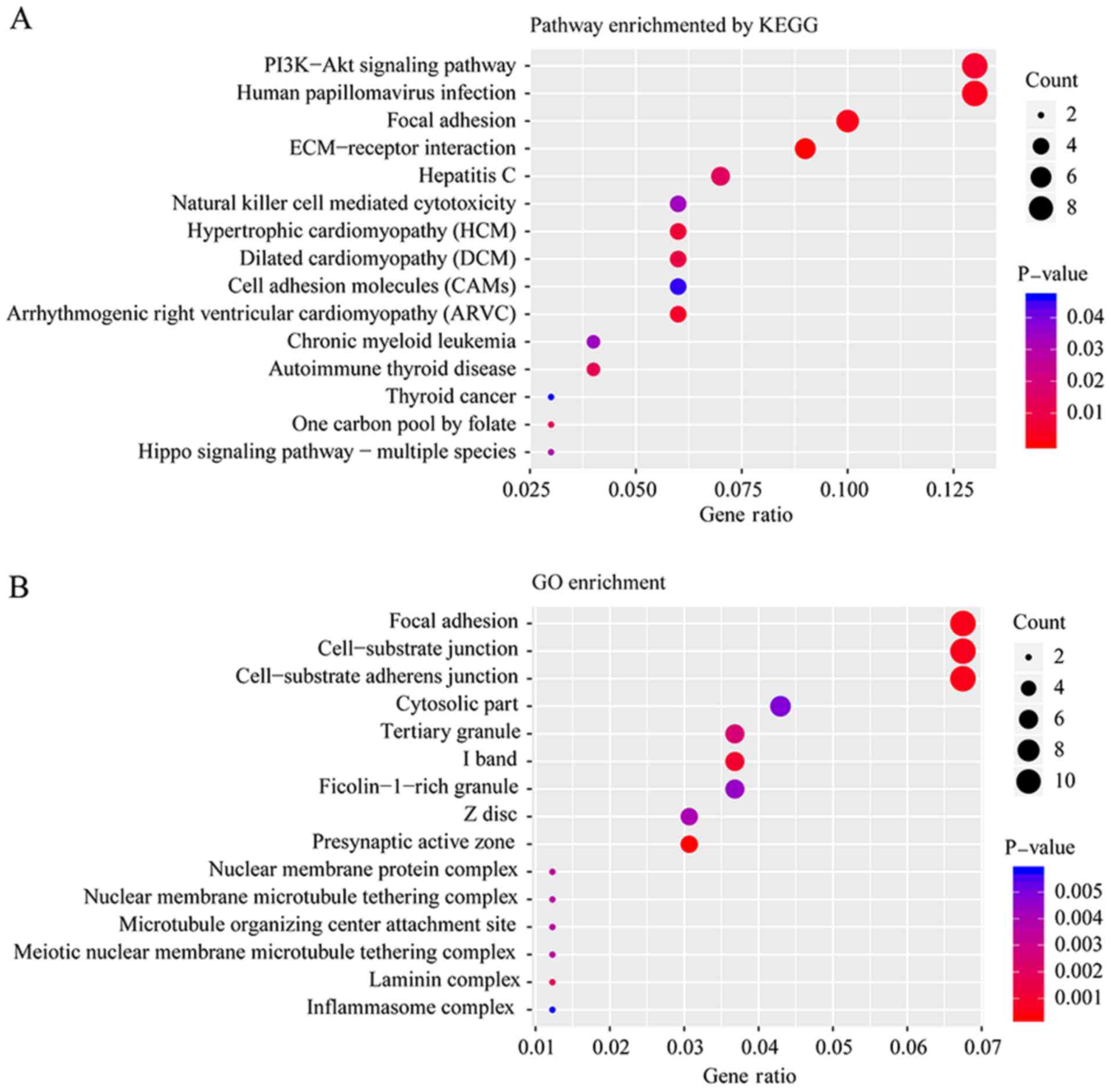
mutated genes, KEGG and GO enrichment were performed. Fig. 3A and B presents the top 15 pathways
and biological functions enriched by KEGG and GO analysis,
respectively, according to their P-values. ‘Focal adhesion’ was a
pathway that was significantly altered according to both the KEGG
and GO enrichment analyses. Adhesion to the substrate via specific
focal adhesion points has previously been considered an essential
step in cell migration (23). In the
present study, 11 mutated genes were enriched in three biological
functions: ‘Focal adhesion’, ‘cell-substrate junction’ and
‘cell-substrate adherens junction’, including cadherin 2, heat
shock family A member 1B, PTPRF interacting protein alpha 1,
spectrin repeat containing nuclear envelope protein 2,
transcriptional adaptor 1, basigin, integrin subunit-α (ITGA)4,
trio and f-actin binding protein, ZNF185, ITGA8 and NHS actin
remodeling regulator.
Hotspot mutations detected by digital
PCR in the tumor and plasma tissue of the validation cohort
The clinical significance of BRAF and RAS mutations
has been previously studied (20),
and it has been recommended that the diagnosis of thyroid nodules
should occur using indeterminate FNA results according to the NCCN
guidelines for thyroid carcinoma (5). To evaluate the accuracy of ctDNA
detection compared with tumor tissues, BRAF and NRAS hotspot
mutations were detected in the plasma and tumor tissues in the
validation group using digital PCR. For the tumor tissues of the 59
patients with PTC, the BRAF V600E mutation frequency was 44.07%
(Table II). Among the 26 patients
with BRAF V600E tumors, BRAF V600E mutation was detected in the
cell-free DNA of the plasma of 16 patients, while BRAF mutations in
cell-free DNA were detected in 3 patients from the 33 with BRAF
wild-type tumors. The sensitivity and specificity of BRAF V600E in
cell-free DNA of patients with negative and positive results,
detected by digital PCR, were 61.54 and 90.91%, respectively
(Table III).
 | Table III.Sensitivity and specificity of BRAF
V600E and NRAS Q61R detection in plasma in papillary thyroid
carcinoma, according to the results gained from tumor tissue
analysis. |
Table III.
Sensitivity and specificity of BRAF
V600E and NRAS Q61R detection in plasma in papillary thyroid
carcinoma, according to the results gained from tumor tissue
analysis.
|
|
| Mutation in
plasma |
|
|---|
|
|
|
|
|
|---|
| Tumor mutation | Negative | Positive | Sensitivity
(%) | Specificity
(%) |
|---|
| BRAF
V600E |
|
Negative | 30 | 3 | 61.54 | 90.91 |
|
Positive | 10 | 16 |
|
|
| NRAS
p.Q61R |
|
Negative | 54 | 1 | 50 | 98.18 |
|
Positive | 2 | 2 |
|
|
As presented in Table
II, BRAF V600E in patients' tumor tissues was significantly
associated with advanced stage (P=0.009) and LNM (P=0.001).
However, no significant association was observed with age, sex,
tumor size or capsule invasion. The characteristics of the tumors
(including stage, tumor size and LNM) were also investigated to
determine whether these influenced the sensitivity of BRAF V600E in
plasma. Table IV demonstrated that
there was no significant correlation between the sensitivity of
BRAF V600E in the plasma and clinical parameters, despite there
being a marked trend.
 | Table IV.Association between cell-free BRAF
V600E status and tumor characteristics in 26 patients with BRAF
V600E-positive tumors. |
Table IV.
Association between cell-free BRAF
V600E status and tumor characteristics in 26 patients with BRAF
V600E-positive tumors.
|
| BRAF V600E in
plasma |
|
|---|
|
|
|
|
|---|
| Characteristic | Negative | Positive | P-value |
|---|
| Stage, no.
patients |
|
| 0.422 |
|
I+II | 6 | 6 |
|
|
III+IV | 4 | 10 |
|
| LNM, no.
patients |
|
| 1.000 |
|
Negative | 4 | 7 |
|
|
Positive | 6 | 9 |
|
| Tumor size, cm;
median (range) | 1.55 (0.2–3.6) | 1.7 (0.4–4.6) | 0.766 |
NRAS p.Q61R was also detected in the tumor tissues
and plasma in the validation cohort. NRAS p.Q61R was present in
four tumors (6.78%). The sensitivity of NRAS p.Q61R in ctDNA was
50.00% and the specificity was 98.18% (Table III). Low frequency of NRAS p.Q61R
in tumors limits its independent application for liquid biopsy
whereas combined detection of BRAF V600E and NRAS p.Q61R in plasma
is feasible.
Discussion
Cellular pathology following FNA remains the most
reliable method of distinguishing benign and malignant thyroid
tumors prior to surgery (24).
However, the features of 10–30% of thyroid nodules are still unable
to be confirmed by FNA (24), which
leads to these patients receiving unnecessary surgical treatment.
For these patients, the detection of genetic mutations in their
tumor tissues, via fine needle puncture, can be used to screen
high-risk patients with thyroid cancer (25), and help guide further treatment.
In the present study, WES was used to investigate
the genomic alteration profile of 10 patients with PTC from
Quanzhou, China. All 59 patients with PTC formed the validation
cohort to verify the results. BRAF V600E was the most frequent
mutation in the WES cohort (5/10) and the mutation frequency was
44.07% in the validation cohort (26/59), which was consistent with
previous studies (26,27). BRAF gene mutations have been reported
to be associated with tumor progression, sensitivity to iodine
treatment and poor prognosis (26,28,29). The
results from the present study revealed that BRAF V600E was
significantly associated with an advanced disease stage (P=0.009)
and LNM (P=0.001). Additionally, consistency was observed in the
identification of the BRAF genotype and VAF between WES and digital
detection, which provided evidence for the reliability of WES.
Currently, the BRAF status, based on surgical pathology specimens,
is used by clinicians to evaluate patients at risk of recurrence
(20).
Mutated genes, including ZNF717, PHLDA1, RBMXL3,
SETD8 and TCHH, were also identified in the present study. These
genes were novel and had not been reported in the TCGA study
(3). For example, ZNF717 mutated in
three samples, which encodes a Kruppel-associated box zinc-finger
protein (30). Duan et al
(31) reported that ZNF717 was a
potential driver gene in hepatocellular carcinoma, as a tumor
suppressor acting through the regulation of the
interleukin-6/signal transducer and activator of transcription 3
pathway. Further evaluation of these genes should be performed in
future studies, with validation of these mutated genes in a larger
cohort, and determination of their pathogenic mechanisms.
Liquid biopsy is beneficial in diagnosis and
treatment of cancers (32). ctDNA
can be tested without accessing tumor tissues. Additionally, the
short half-life of ctDNA and its wide dynamic range (33) may allow for the rapid assessment of
therapeutic responses for early- and late-stage disease. Previous
studies have indicated that the detection of ctDNA in plasma can
aid in cancer diagnosis and can guide treatment course (34), including liver cancer (35), colorectal cancer (36) and melanoma (37). However, controversy exists regarding
the detection of ctDNA in the plasma of patients with thyroid
cancer. Cradic et al (38)
and Zane et al (9)
demonstrated that the mutation rate of BRAF in the plasma of
patients with thyroid cancer was low, and that this was therefore
not associated with thyroid cancer. However, a study performed by
Kim et al (22), which
included 72 patients with thyroid cancer, revealed that gene
mutations in cell-free DNA were positive in three patients with LNM
and lung metastasis, and this study concluded that there was a
positive correlation between ctDNA and lung metastasis in patients
with thyroid carcinoma.
In the present study, BRAF V600E and NRAS p.Q61R
were analyzed in the tumor tissue and plasma of 59 patients with
PTC in the validation cohort, and the accuracy of ctDNA detection
was analyzed according to the results from the QuantStudio™ 3D PCR.
The sensitivity of BRAF V600E and NRAS p.Q61R in ctDNA was 61.54
and 50.00%, respectively, and the specificity was 90.91 and 98.18%,
respectively.
In the present study, for BRAF V600E, the ctDNA
samples of 38.46% patients with positive BRAF V600E tumors were
negative. In a study performed by Pupilli et al (10), the corresponding ctDNA was negative
in 22 PTC patients whose tumors were BRAF V600E positive, using
reverse transcription-PCR and digital PCR. In the aforementioned
study, the release efficiency of DNA from the tumor to the
peripheral blood was affected by a variety of factors, such as
capsule invasion, size and tumor stage. ctDNA was not detected when
its concentration was lower than the sensitivity limitation of
QuantStudio™ 3D PCR. Additionally, there exists a high risk of
degradation for cell-free DNA due to its short half-life (39). Few tumors in this aforementioned
study were wild-type, and the sequencing results of the
corresponding plasma were mutated, which may be accounted for by
this. Tumors possess heterogeneity, and the sequencing results of a
small section of tumor tissue cannot represent the entire tumor
(6). Additionally, this potential
false-sensitive result could be caused by the ultrahigh sensitivity
of digital PCR.
In conclusion, the present study drew a genomic
alteration profile of patients with PTC from southeast China, and
provided evidence for the accuracy of ctDNA analysis in patients
with PTC. ctDNA is an important tool for assisting in the diagnosis
of thyroid nodules, predicting prognosis and tracking the progress
of PTC when tumor tissue cannot be obtained.
Acknowledgements
Not applicable.
Funding
This work was supported by the Natural Science
Foundation of Fujian Province (grant no. 2014J01435), Quanzhou
Science and Technology Project (grant no. 2013Z58), and Projects of
Special Development Funds in Zhangjiang National Independent
Innovation Demonstration Zone, Shanghai (grant no.
ZJ2017-ZD-012).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QP and HL wrote the manuscript; HL, ZS and SW
contributed to the study design and manuscript revision; JZhan, ML
and CW collected samples and performed PCR; SW performed
whole-exome sequencing; JZhao analyzed the whole-exome sequencing
data; QP collected clinical data, analyzed the PCR data and
developed the figures and tables. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Ethical approval for the recruitment of human
subjects was obtained from the Ethics Committee of Affiliated
Quanzhou First Hospital of Fujian Medical University and was
consistent with ethical guidelines provided by the Declaration of
Helsinki (1975). Written informed consent was obtained from each
patient.
Patient consent for publication
All individuals whose data were included in the
current study provided informed consent for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Prescott JD and Zeiger MA: The RET
oncogene in papillary thyroid carcinoma. Cancer. 121:2137–2146.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alexander EK, Kennedy GC, Baloch ZW, Cibas
ES, Chudova D, Diggans J, Friedman L, Kloos RT, LiVolsi VA, Mandel
SJ, et al: Preoperative diagnosis of benign thyroid nodules with
indeterminate cytology. N Engl J Med. 367:705–715. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas Research Network, .
Integrated genomic characterization of papillary thyroid carcinoma.
Cell. 159:676–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ye L, Zhou X, Huang F, Wang W, Qi Y, Xu H,
Yang S, Shen L, Fei X, Xie J, et al: The genetic landscape of
benign thyroid nodules revealed by whole exome and transcriptome
sequencing. Nat Commun. 8:155332017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
National Comprehensive Cancer Network
(NCCN), . NCCN Clinical Practice Guidelines in Oncology. Thyroid
Carcinoma (Version 1.2019). https://www.nccn.org/professionals/physician_gls/default.aspx#thyroidMarch
28–2019
|
|
6
|
Burrell RA, McGranahan N, Bartek J and
Swanton C: The causes and consequences of genetic heterogeneity in
cancer evolution. Nature. 501:338–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Diaz LA Jr and Bardelli A: Liquid
biopsies: Genotyping circulating tumor DNA. J Clin Oncol.
32:579–586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allin DM, Shaikh R, Carter P, Thway K,
Sharabiani MTA, Gonzales-de-Castro D, O'Leary B, Garcia-Murillas I,
Bhide S, Hubank M, et al: Circulating tumour DNA is a potential
biomarker for disease progression and response to targeted therapy
in advanced thyroid cancer. Eur J Cancer. 103:165–175. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zane M, Agostini M, Enzo MV, Casal Ide E,
Del Bianco P, Torresan F, Merante Boschin I, Pennelli G, Saccani A,
Rubello D, et al: Circulating cell-free DNA, SLC5A8 and SLC26A4
hypermethylation, BRAF(V600E): A non-invasive tool panel for early
detection of thyroid cancer. Biomed Pharmacother. 67:723–730. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pupilli C, Pinzani P, Salvianti F, Fibbi
B, Rossi M, Petrone L, Perigli G, De Feo ML, Vezzosi V, Pazzagli M,
et al: Circulating BRAFV600E in the diagnosis and follow-up of
differentiated papillary thyroid carcinoma. J Clin Endocrinol
Metab. 98:3359–3365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Condello V, Macerola E, Ugolini C, De
Napoli L, Romei C, Materazzi G, Elisei R and Basolo F: Analysis of
circulating tumor DNA does not improve the clinical management of
patients with locally advanced and metastatic papillary thyroid
carcinoma. Head Neck. 40:1752–1758. 2018.PubMed/NCBI
|
|
12
|
Lupo M, Guttler R, Geck Z, Tonozzi TR,
Kammesheidt A and Braunstein GD: Is measurement of circulating
tumor DNA of diagnostic use in patients with thyroid nodules?
Endocr Pract. 24:453–459. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
AJCC Cancer Staging Manual. 8th. Springer;
New York, NY: 2017
|
|
14
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The Genome Analysis Toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cibulskis K, Lawrence MS, Carter SL,
Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES
and Getz G: Sensitive detection of somatic point mutations in
impure and heterogeneous cancer samples. Nat Biotechnol.
31:213–219. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
R Core Team: R, . A language and
environment for statistical computing. R Foundation for Statistical
Computing; Vienna, Austria: 2018, https://www.R-project.org
|
|
19
|
Gu Z, Eils R and Schlesner M: Complex
heatmaps reveal patterns and correlations in multidimensional
genomic data. Bioinformatics. 32:2847–2849. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xing M, Alzahrani AS, Carson KA, Shong YK,
Kim TY, Viola D, Elisei R, Bendlová B, Yip L, Mian C, et al:
Association between BRAF V600E mutation and recurrence of papillary
thyroid cancer. J Clin Oncol. 33:42–50. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim TH, Park YJ, Lim JA, Ahn HY, Lee EK,
Lee YJ, Kim KW, Hahn SK, Youn YK, Kim KH, et al: The association of
the BRAF(V600E) mutation with prognostic factors and poor clinical
outcome in papillary thyroid cancer: A meta-analysis. Cancer.
118:1764–1773. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim BH, Kim IJ, Lee BJ, Lee JC, Kim IS,
Kim SJ, Kim WJ, Jeon YK, Kim SS and Kim YK: Detection of plasma
BRAF(V600) mutation is associated with lung metastasis in papillary
thyroid carcinomas. Yonsei Med J. 56:634–640. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Paluch EK, Aspalter IM and Sixt M: Focal
adhesion-independent cell migration. Annu Rev Cell Dev Biol.
32:469–490. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cibas ES and Ali SZ: The 2017 Bethesda
system for reporting thyroid cytopathology. Thyroid. 27:1341–1346.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Haugen BR, Alexander EK, Bible KC, Doherty
GM, Mandel SJ, Nikiforov YE, Pacini F, Randolph GW, Sawka AM,
Schlumberger M, et al: 2015 American thyroid association management
guidelines for adult patients with thyroid nodules and
differentiated thyroid cancer: The American thyroid association
guidelines task force on thyroid nodules and differentiated thyroid
cancer. Thyroid. 26:1–133. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xing M, Alzahrani AS, Carson KA, Viola D,
Elisei R, Bendlova B, Yip L, Mian C, Vianello F, Tuttle RM, et al:
Association between BRAF V600E mutation and mortality in patients
with papillary thyroid cancer. JAMA. 309:1493–1501. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin KL ZX and Dai XX: Detection of BRAF
gene mutations in patients with high risk papillary thyroid
carcinoma before the operation. Chin J Oncol. 33:130–131. 2011.
|
|
28
|
Prescott JD, Sadow PM, Hodin RA, Le LP,
Gaz RD, Randolph GW, Stephen AE, Parangi S, Daniels GH and Lubitz
CC: BRAFV600E status adds incremental value to current
risk classification systems in predicting papillary thyroid
carcinoma recurrence. Surgery. 152:984–990. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Riesco-Eizaguirre G, Gutiérrez-Martínez P,
García-Cabezas MA, Nistal M and Santisteban P: The oncogene BRAF
V600E is associated with a high risk of recurrence and less
differentiated papillary thyroid carcinoma due to the impairment of
Na+/I- targeting to the membrane. Endocr Relat Cancer. 13:257–269.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Muzny DM, Scherer SE, Kaul R, Wang J, Yu
J, Sudbrak R, Buhay CJ, Chen R, Cree A, Ding Y, et al: The DNA
sequence, annotation and analysis of human chromosome 3. Nature.
440:1194–1198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Duan M, Hao J, Cui S, Worthley DL, Zhang
S, Wang Z, Shi J, Liu L, Wang X, Ke A, et al: Diverse modes of
clonal evolution in HBV-related hepatocellular carcinoma revealed
by single-cell genome sequencing. Cell Res. 28:359–373. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fiala C and Diamandis EP: Utility of
circulating tumor DNA in cancer diagnostics with emphasis on early
detection. BMC Med. 16:1662018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mengual L, Lozano JJ, Ingelmo-Torres M,
Gazquez C, Ribal MJ and Alcaraz A: Using microRNA profiling in
urine samples to develop a non-invasive test for bladder cancer.
Int J Cancer. 133:2631–2641. 2013.PubMed/NCBI
|
|
34
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ye Q, Ling S, Zheng S and Xu X: Liquid
biopsy in hepatocellular carcinoma: Circulating tumor cells and
circulating tumor DNA. Mol Cancer. 18:1142019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Garlan F, Laurent-Puig P, Sefrioui D,
Siauve N, Didelot A, Sarafan-Vasseur N, Michel P, Perkins G, Mulot
C, Blons H, et al: Early evaluation of circulating tumor DNA as
marker of therapeutic efficacy in metastatic colorectal cancer
patients (PLACOL study). Clin Cancer Res. 23:5416–5425. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tan L, Sandhu S, Lee RJ, Li J, Callahan J,
Ftouni S, Dhomen N, Middlehurst P, Wallace A, Raleigh J, et al:
Prediction and monitoring of relapse in stage III melanoma using
circulating tumor DNA. Ann Oncol. 30:804–814. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cradic KW, Milosevic D, Rosenberg AM,
Erickson LA, McIver B and Grebe SK: Mutant BRAF(T1799A) can be
detected in the blood of papillary thyroid carcinoma patients and
correlates with disease status. J Clin Endocrinol Metab.
94:5001–5009. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Khier S and Lohan L: Kinetics of
circulating cell-free DNA for biomedical applications: Critical
appraisal of the literature. Future Sci OA. 4:FSO2952018.
View Article : Google Scholar : PubMed/NCBI
|