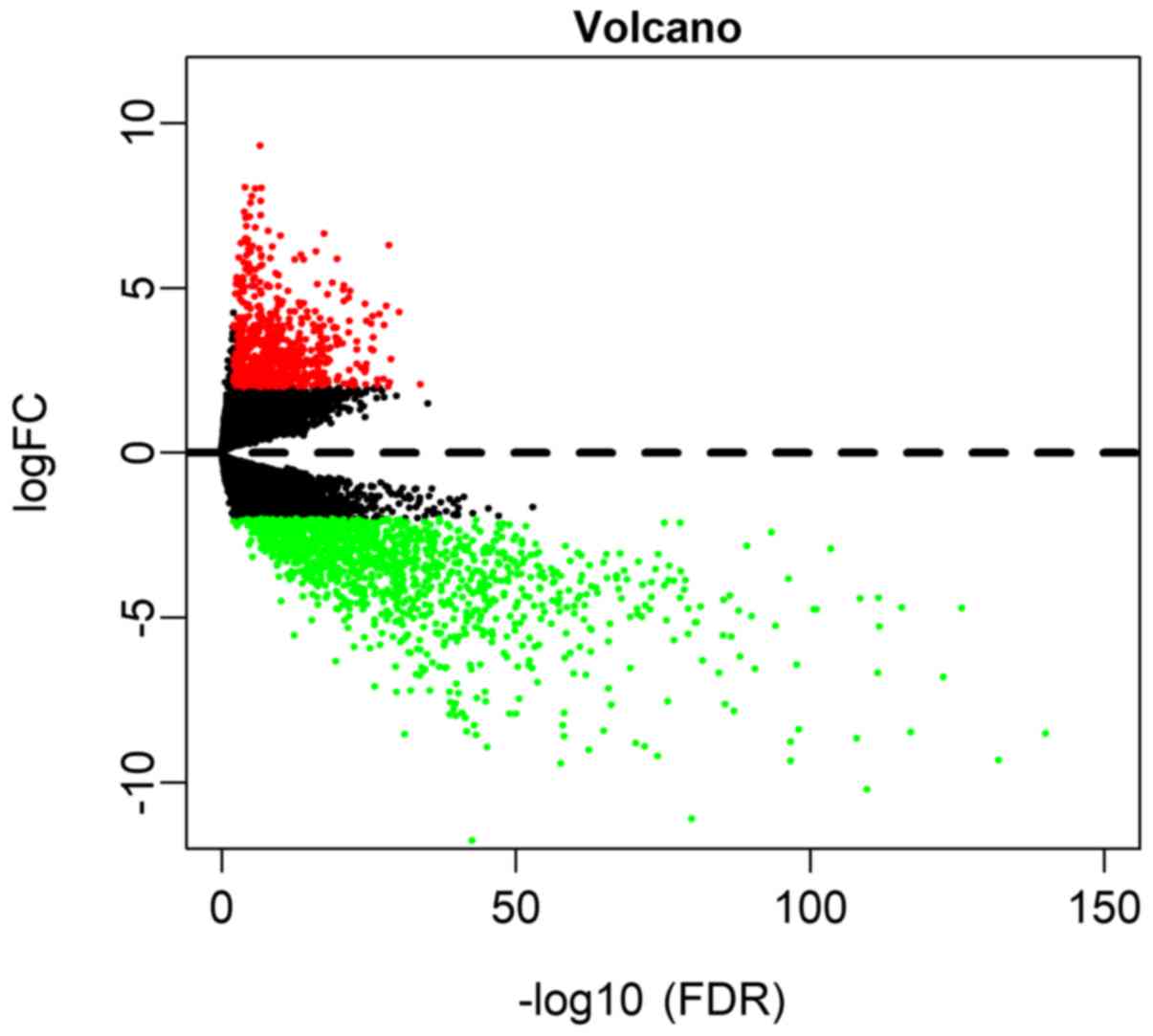
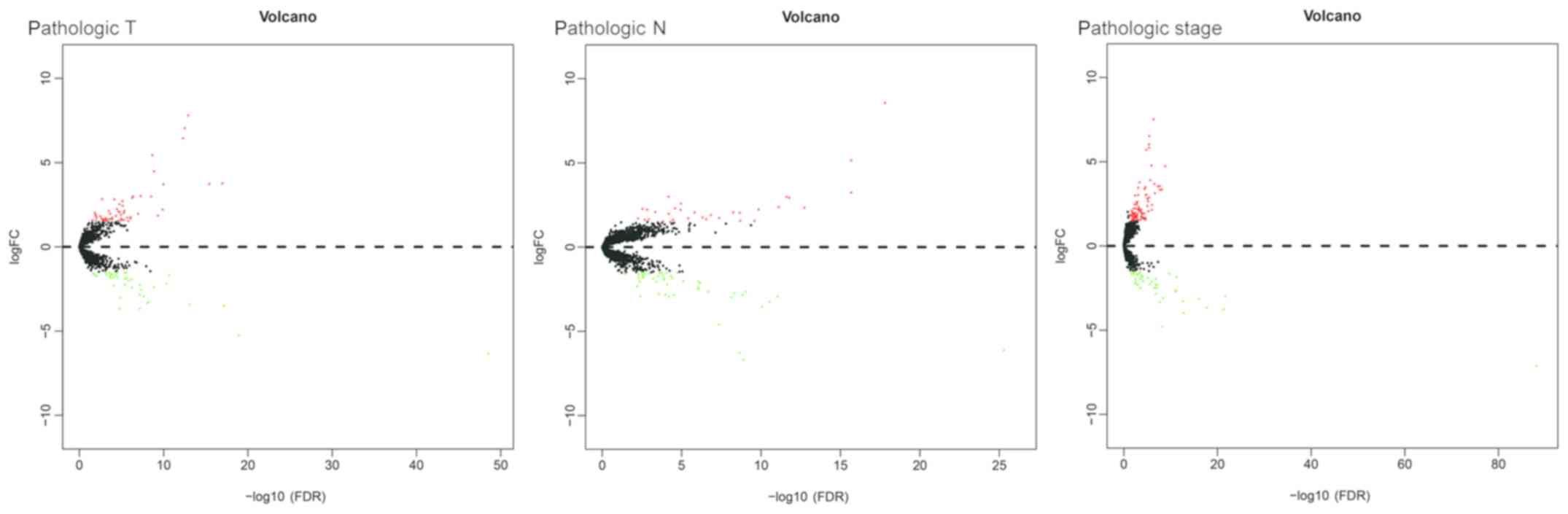
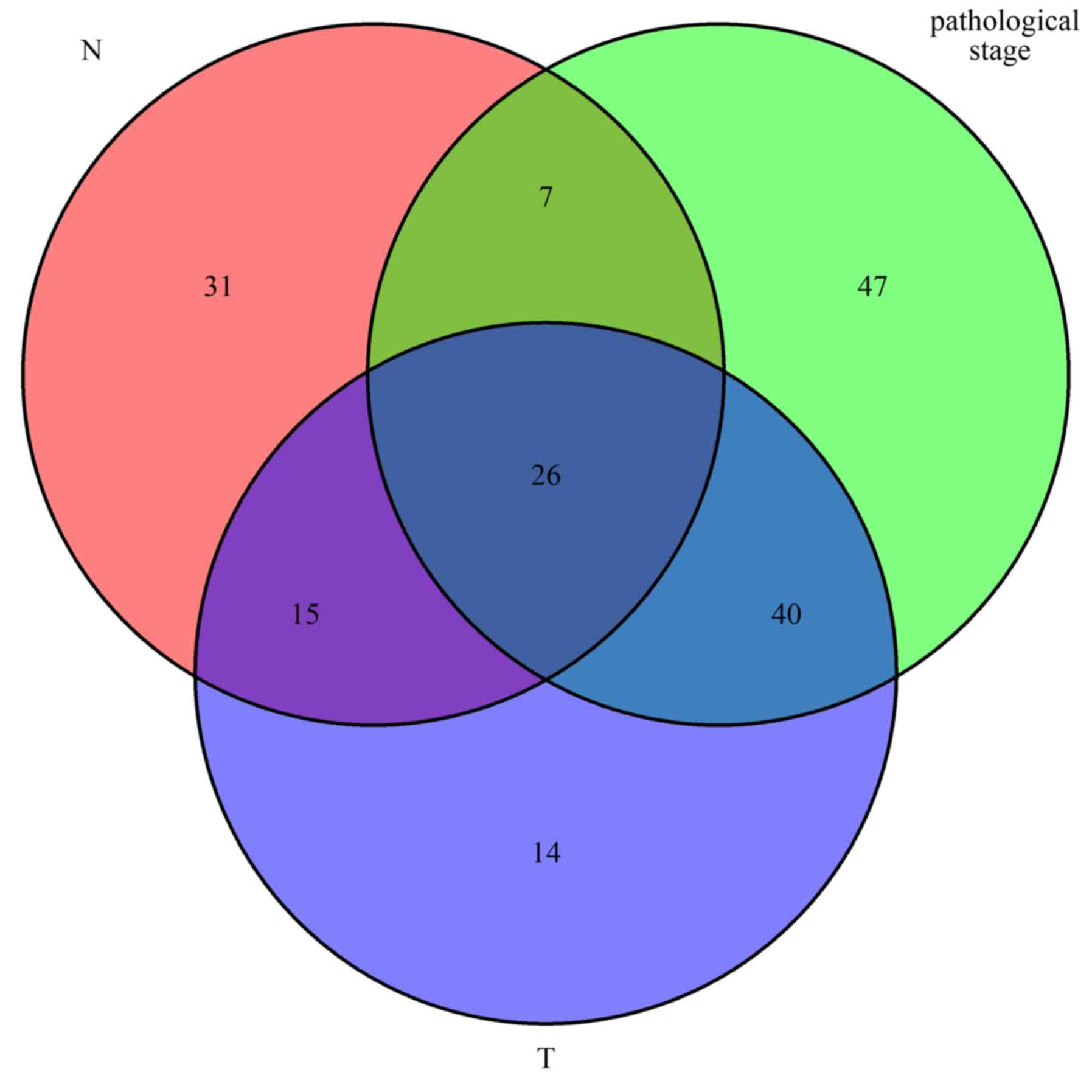
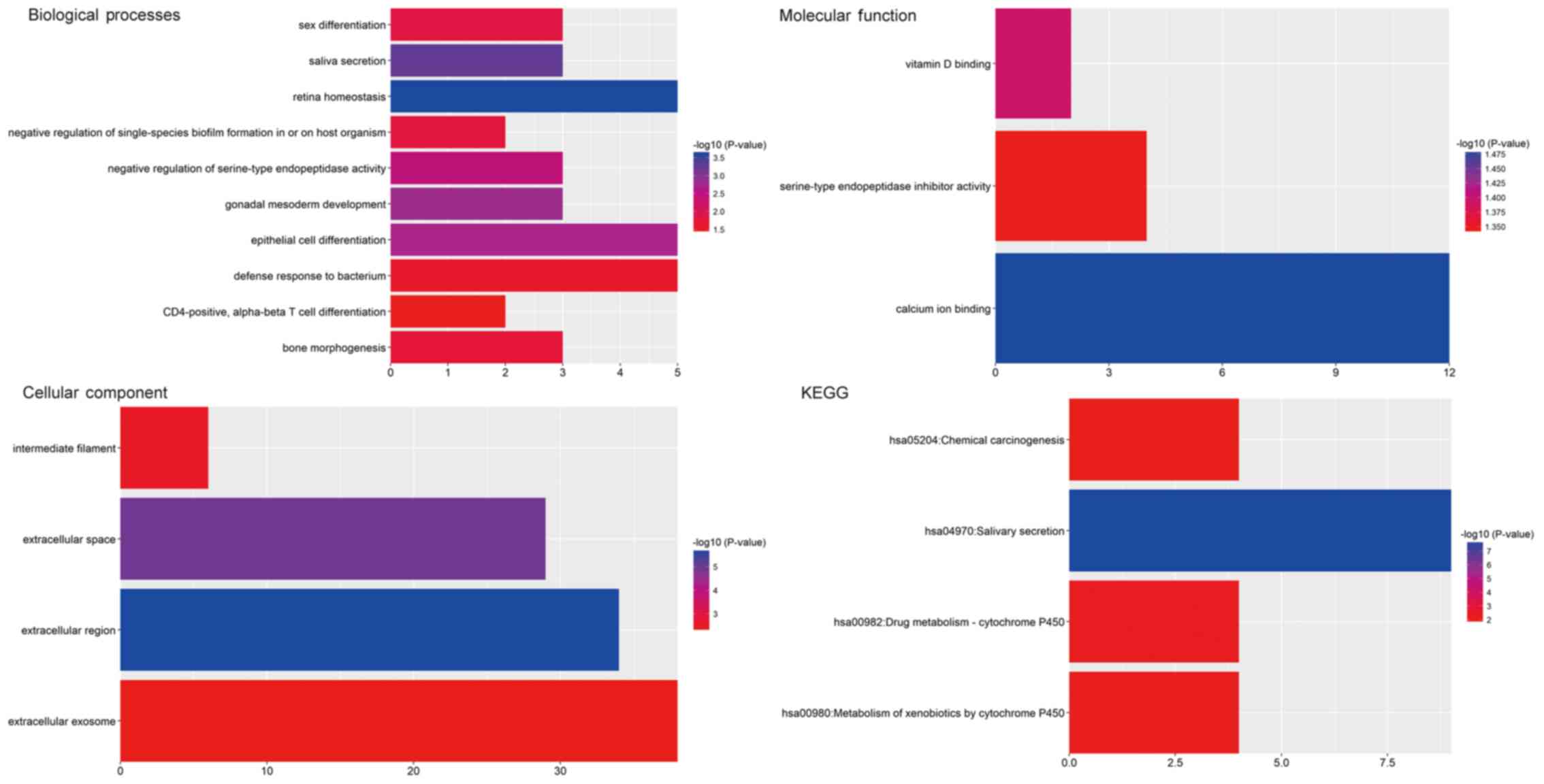
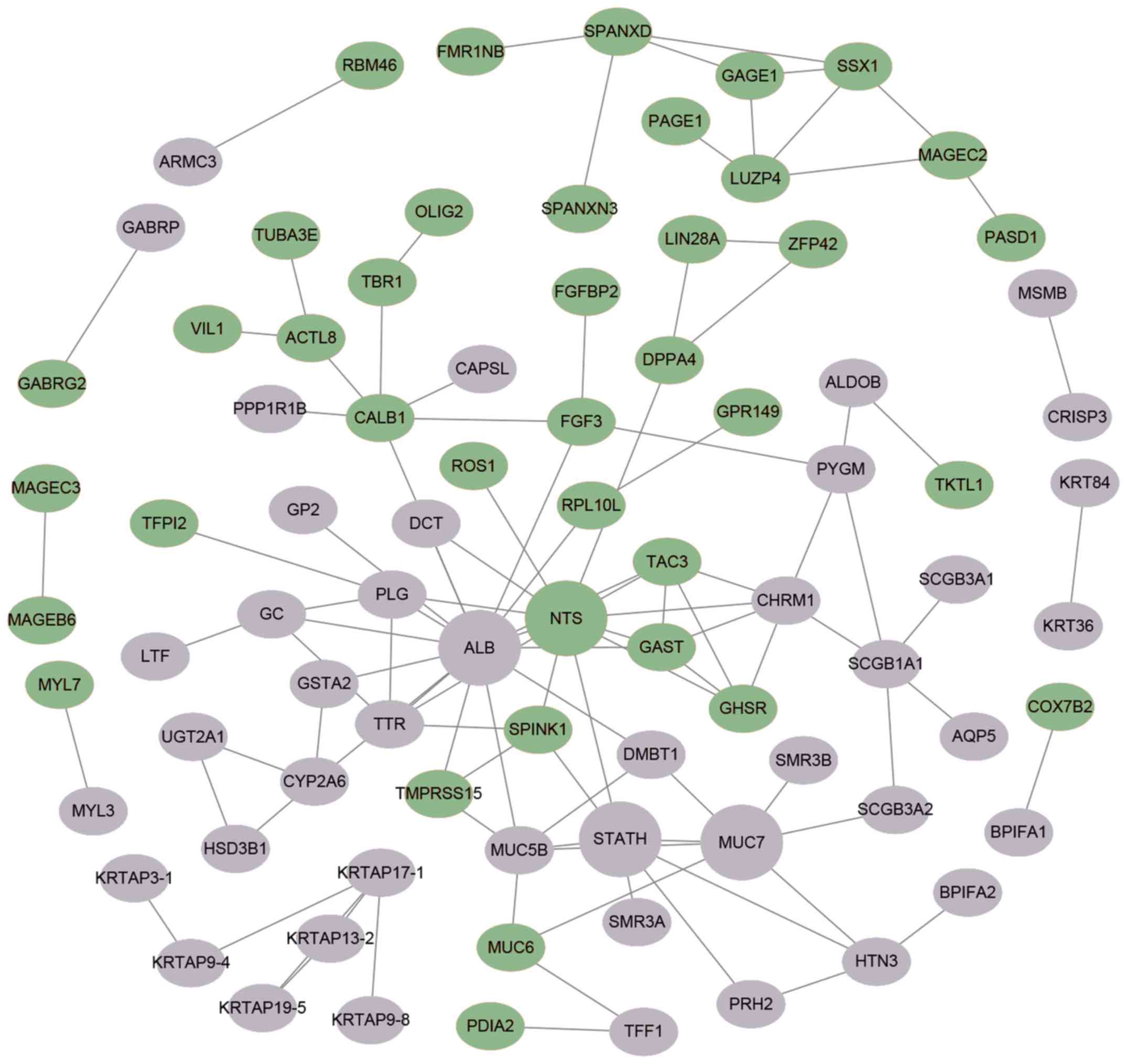
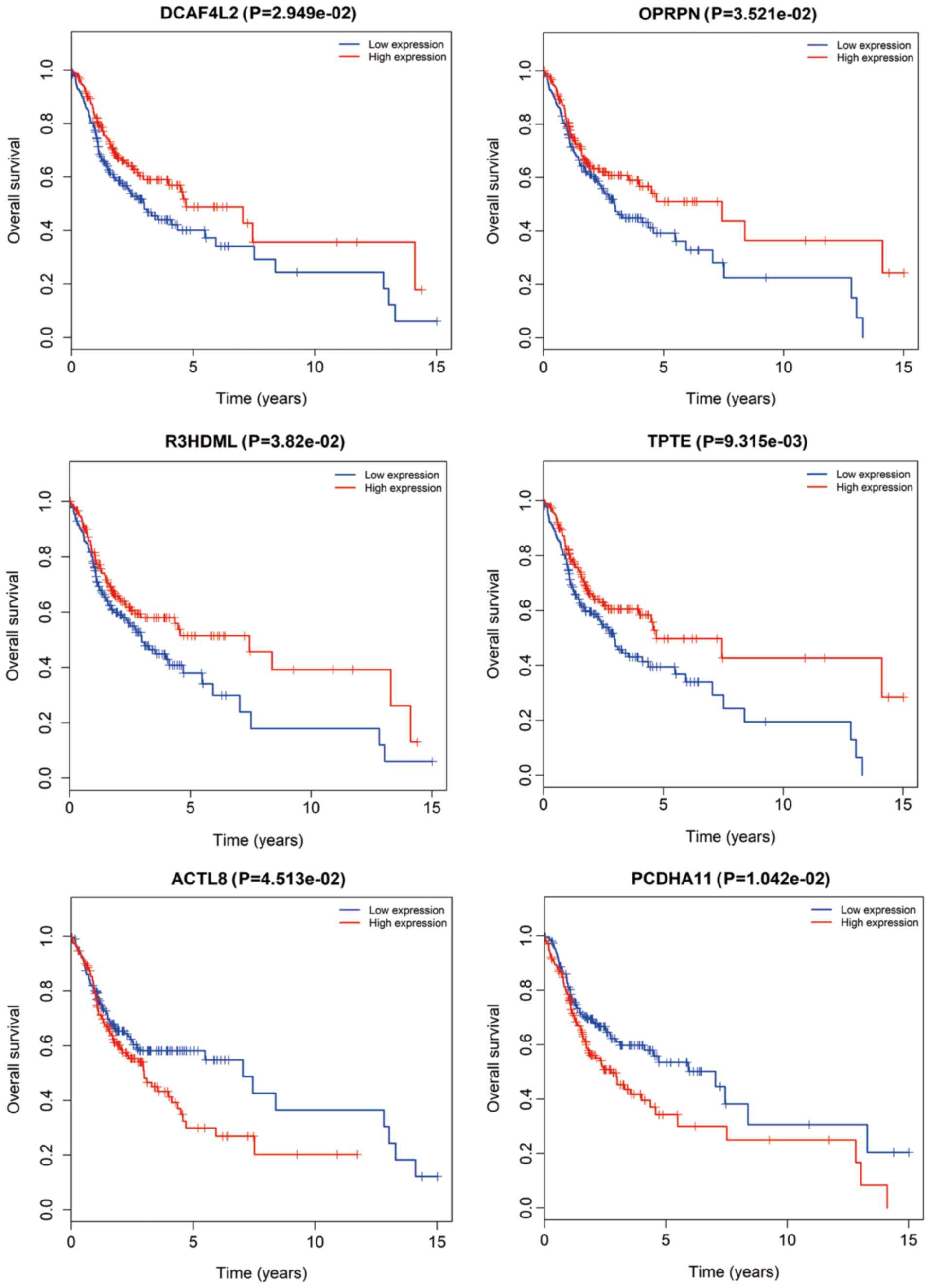
|
1
|
Shimomura H, Sasahira T, Nakashima C,
Shimomura-Kurihara M and Kirita T: Downregulation of DHRS9 is
associated with poor prognosis in oral squamous cell carcinoma.
Pathology. 50:642–647. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sandulache VC, Michikawa C, Kataria P,
Gleber-Netto FO, Bell D, Trivedi S, Rao X, Wang J, Zhao M, Jasser
S, et al: High-risk TP53 mutations are associated with extranodal
extension in oral cavity squamous cell carcinoma. Clin Cancer Res.
24:1727–1733. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng Q, Deng Z, Pan H, Gu L, Liu O and
Tang Z: Mitogen-activated protein kinase signaling pathway in oral
cancer (Review). Oncol Lett. 15:1379–1388. 2018.PubMed/NCBI
|
|
4
|
Sahingur SE and Yeudall WA: Chemokine
function in periodontal disease and oral cavity cancer. Front
Immunol. 6:2142015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao X, Sun S, Zeng X and Cui L:
Expression profiles analysis identifies a novel three-mRNA
signature to predict overall survival in oral squamous cell
carcinoma. Am J Cancer Res. 8:450–461. 2018.PubMed/NCBI
|
|
6
|
Chen PK, Hua CH, Hsu HT, Kuo TM, Chung CM,
Lee CP, Tsai MH, Yeh KT and Ko YC: ALPK1 expression is associated
with lymph node metastasis and tumor growth in oral squamous cell
carcinoma patients. Am J Pathol. 189:190–199. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang G, Tang Q, Jia L, Xia S, Li J, Chen
Y, Li H, Ding X, Wang F, Hou D, et al: High expression of TROP2 is
correlated with poor prognosis of oral squamous cell carcinoma.
Pathol Res Pract. 214:1606–1612. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang H, Liu J, Fu X and Yang A:
Identification of key genes and pathways in tongue squamous cell
carcinoma using bioinformatics analysis. Med Sci Monit.
23:5924–5932. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang H, Niu L, Jiang S, Zhai J, Wang P,
Kong F and Jin X: Comprehensive analysis of aberrantly expressed
profiles of lncRNAs and miRNAs with associated ceRNA network in
muscle-invasive bladder cancer. Oncotarget. 7:86174–86185. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lydiatt WM, Patel SG, O'Sullivan B,
Brandwein MS, Ridge JA, Migliacci JC, Loomis AM and Shah JP: Head
and Neck cancers-major changes in the American Joint Committee on
cancer eighth edition cancer staging manual. CA Cancer J Clin.
67:122–137. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gao C, Zhuang J, Zhou C, Ma K, Zhao M, Liu
C, Liu L, Li H, Feng F and Sun C: Prognostic value of aberrantly
expressed methylation gene profiles in lung squamous cell
carcinoma: A study based on The cancer genome atlas. J Cell
Physiol. 234:6519–6528. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sarode SC, Sarode GS and Patil S: Role of
statherin in oral carcinogenesis. Oral Oncol. 50:e55–e56. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Sousa-Pereira P, Amado F, Abrantes J,
Ferreira R, Esteves PJ and Vitorino R: An evolutionary perspective
of mammal salivary peptide families: Cystatins, histatins,
statherin and PRPs. Arch Oral Biol. 58:451–458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sabatini LM, Carlock LR, Johnson GW and
Azen EA: cDNA cloning and chromosomal localization (4q11-13) of a
gene for statherin, a regulator of calcium in saliva. Am J Hum
Genet. 41:1048–1060. 1987.PubMed/NCBI
|
|
15
|
Contucci AM, Inzitari R, Agostino S,
Vitali A, Fiorita A, Cabras T, Scarano E and Messana I: Statherin
levels in saliva of patients with precancerous and cancerous
lesions of the oral cavity: A preliminary report. Oral Dis.
11:95–99. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Souazé F, Dupouy S, Viardot-Foucault V,
Bruyneel E, Attoub S, Gespach C, Gompel A and Forgez P: Expression
of neurotensin and NT1 receptor in human breast cancer: A potential
role in tumor progression. Cancer Res. 66:6243–6249. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu Z, Galmiche A, Liu J, Stadler N, Wendum
D, Segal-Bendirdjian E, Paradis V and Forgez P: Neurotensin
regulation induces overexpression and activation of EGFR in HCC and
restores response to erlotinib and sorafenib. Cancer Lett.
388:73–84. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morgat C, Mishra AK, Varshney R, Allard M,
Fernandez P and Hindié E: Targeting neuropeptide receptors for
cancer imaging and therapy: Perspectives with bombesin,
neurotensin, and neuropeptide-Y receptors. J Nucl Med.
55:1650–1657. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dupouy S, Viardot-Foucault V, Alifano M,
Souazé F, Plu-Bureau G, Chaouat M, Lavaur A, Hugol D, Gespach C,
Gompel A and Forgez P: The neurotensin receptor-1 pathway
contributes to human ductal breast cancer progression. PLoS One.
4:e42232009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Younes M, Wu Z, Dupouy S, Lupo AM, Mourra
N, Takahashi T, Fléjou JF, Trédaniel J, Régnard JF, Damotte D, et
al: Neurotensin (NTS) and its receptor (NTSR1) causes EGFR, HER2
and HER3 over-expression and their autocrine/paracrine activation
in lung tumors, confirming responsiveness to erlotinib. Oncotarget.
5:8252–8269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ye Y, Long X, Zhang L, Chen J, Liu P, Li
H, Wei F, Yu W, Ren X and Yu J: NTS/NTR1 co-expression enhances
epithelial-to-mesenchymal transition and promotes tumor metastasis
by activating the Wnt/β-catenin signaling pathway in hepatocellular
carcinoma. Oncotarget. 7:70303–70322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiao P, Long X, Zhang L, Ye Y, Guo J, Liu
P, Zhang R, Ning J, Yu W, Wei F and Yu J: Neurotensin/IL-8 pathway
orchestrates local inflammatory response and tumor invasion by
inducing M2 polarization of Tumor-Associated macrophages and
epithelial-mesenchymal transition of hepatocellular carcinoma
cells. Oncoimmunology. 7:e14401662018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alos L, Lujan B, Castillo M, Nadal A,
Carreras M, Caballero M, de Bolos C and Cardesa A: Expression of
membrane-bound mucins (MUC1 and MUC4) and secreted mucins (MUC2,
MUC5AC, MUC5B, MUC6 and MUC7) in mucoepidermoid carcinomas of
salivary glands. Am J Surg Pathol. 29:806–813. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
NguyenHoang S, Liu Y, Xu L, Chang Y, Zhou
L, Liu Z, Lin Z and Xu J: High mucin-7 expression is an independent
predictor of adverse clinical outcomes in patients with clear-cell
renal cell carcinoma. Tumour Biol. 37:15193–15201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu D, Pavlidis P, Thamadilok S, Redwood E,
Fox S, Blekhman R, Ruhl S and Gokcumen O: Recent evolution of the
salivary mucin MUC7. Sci Rep. 6:317912016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mahomed F: Recent advances in mucin
immunohistochemistry in salivary gland tumors and head and neck
squamous cell carcinoma. Oral Oncol. 47:797–803. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagashio R, Ueda J, Ryuge S, Nakashima H,
Jiang SX, Kobayashi M, Yanagita K, Katono K, Satoh Y, Masuda N, et
al: Diagnostic and prognostic significances of MUC5B and TTF-1
expressions in resected non-small cell lung cancer. Sci Rep.
5:86492015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kumar MH, Sanjai K, Kumarswamy J,
Keshavaiah R, Papaiah L and Divya S: Expression of MUC1 mucin in
potentially malignant disorders, oral squamous cell carcinoma and
normal oral mucosa: An immunohistochemical study. J Oral Maxillofac
Pathol. 20:214–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thakur A, Tupkari JV, Joy T, Kende PP,
Siwach P and Ahire MS: Expression of mucin-1 in oral squamous cell
carcinoma and normal oral mucosa: An immunohistochemical study. J
Oral Maxillofac Pathol. 22:210–215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Micel LN, Tentler JJ, Smith PG and
Eckhardt GS: Role of ubiquitin ligases and the proteasome in
oncogenesis: Novel targets for anticancer therapies. J Clin Oncol.
31:1231–1238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Beaty TH, Taub MA, Scott AF, Murray JC,
Marazita ML, Schwender H, Parker MM, Hetmanski JB, Balakrishnan P,
Mansilla MA, et al: Confirming genes influencing risk to cleft lip
with/without cleft palate in a case-parent trio study. Hum Genet.
132:771–781. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang H, Chen Y, Han J, Meng Q, Xi Q, Wu G
and Zhang B: DCAF4L2 promotes colorectal cancer invasion and
metastasis via mediating degradation of NFκB negative regulator
PPM1B. Am J Transl Res. 8:405–418. 2016.PubMed/NCBI
|
|
33
|
Miller RE, Uwamahoro N and Park JH: PPM1B
depletion in U2OS cells supresses cell growth through RB1-E2F1
pathway and stimulates bleomycin-induced cell death. Biochem
Biophys Res Commun. 500:391–397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Saitoh E, Taniguchi M, Ochiai A, Kato T,
Imai A and Isemura S: Bioactive peptides hidden in human salivary
proteins. J Oral Biosciences. 59:71–79. 2017. View Article : Google Scholar
|
|
35
|
Saitoh E, Sega T, Imai A, Isemura S, Kato
T, Ochiai A and Taniguchi M: The PBII gene of the human salivary
proline-rich protein P-B produces another protein, Q504X8, with an
opiorphin homolog, QRGPR. Arch Oral Biol. 88:10–18. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lang Z, Wu Y, Pan X, Qu G and Zhang T:
Study of differential gene expression between invasive
multifocal/multicentric and unifocal breast cancer. J BUON.
23:134–142. 2018.PubMed/NCBI
|
|
37
|
Kuemmel A, Simon P, Breitkreuz A, Röhlig
J, Luxemburger U, Elsässer A, Schmidt LH, Sebastian M, Sahin U,
Türeci Ö and Buhl R: Humoral immune responses of lung cancer
patients against the Transmembrane phosphatase with TEnsin homology
(TPTE). Lung Cancer. 90:334–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yao J, Caballero OL, Yung WK, Weinstein
JN, Riggins GJ, Strausberg RL and Zhao Q: Tumor subtype-specific
cancer-testis antigens as potential biomarkers and
immunotherapeutic targets for cancers. Cancer Immunol Res.
2:371–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cuffel C, Rivals JP, Zaugg Y, Salvi S,
Seelentag W, Speiser DE, Liénard D, Monnier P, Romero P, Bron L and
Rimoldi D: Pattern and clinical significance of cancer-testis gene
expression in head and neck squamous cell carcinoma. Int J Cancer.
128:2625–2634. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Piotti KC, Scognamiglio T, Chiu R and Chen
YT: Expression of cancer/testis (CT) antigens in squamous cell
carcinoma of the head and neck: Evaluation as markers of squamous
dysplasia. Pathol Res Pract. 209:721–726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li B, Zhu J and Meng L: High expression of
ACTL8 is poor prognosis and accelerates cell progression in head
and neck squamous cell carcinoma. Mol Med Rep. 19:877–884.
2019.PubMed/NCBI
|
|
42
|
Freitas M, Malheiros S, Stávale JN, Biassi
TP, Zamunér FT, de Souza Begnami M, Soares FA and Vettore AL:
Expression of cancer/testis antigens is correlated with improved
survival in glioblastoma. Oncotarget. 4:636–646. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mahmoud AM: Cancer testis antigens as
immunogenic and oncogenic targets in breast cancer. Immunotherapy.
10:769–778. 2018. View Article : Google Scholar : PubMed/NCBI
|