Introduction
Neuroblastoma is a childhood cancer that affects
~1/7,000 children (1). Neuroblastoma
is the most common pediatric extracranial solid tumor worldwide
that originates from the sympatoadrenal lineage, which resides in
the neural crest (2–4). Unique features of neuroblastoma include
the early age of onset, the high frequency of metastatic disease at
diagnosis and the tendency for spontaneous regression of tumours in
infancy (5). Currently, there are
many methods for treating neuroblastoma including chemotherapy,
radiation and autologous transplantation (6), but these treatments are unsatisfactory.
The five-year survival rate of neuroblastoma in children aged 1–14
years is 68% (7). Given such a high
mortality rate, it is necessary to find an effective treatment for
neuroblastoma.
MicroRNAs (miRNAs or miRs) are small non-coding RNA
molecules that are generally 19–26 nucleotides in length, which
serve to regulate the expression of genes at the
posttranscriptional level (8–10).
miRNAs do not encode protein and function to inhibit the expression
of multiple target genes by binding to the target mRNAs 3
untranslated region (3-UTR) (11–13).
Previous studies have indicated that miRNAs, which are encoded by
various plants, animals and viruses, can regulate many diverse
biological and physiological processes including organ development,
apoptosis, tumorogenesis, proliferation, stress response and fat
metabolism (14,15). A previous study has revealed that
miR-3934 is highly expressed in cervical cancer (16). It has also been reported that
miR-3934 is associated with lung adenocarcinoma (17). However, the role of miR-3934-5p in
neuroblastoma remains unclear.
Tumor protein 53-induced nuclear protein 1
(TP53INP1) is a potential target gene for many miRNAs, including
miR-221 (18), miR-30a (19) and miR-205 (20). Additionally, it has been reported
that TP53INP1 is a regulator of autophagy and may interact with
autophagy-associated molecules, including light chain 3 and
autophagy related protein 8-family proteins, which indicates that
it may not only regulate, but also promote autophagy (21,22).
Researchers have also suggested that TP53INP1 may serve an
important role in certain types of cancer, including hepatocellular
carcinoma (23), breast cancer
(24) and human osteosarcoma
(25).
In the present study, the expression of miR-3934-5p
in neuroblastoma tissues and cell lines was investigated and the
underlying mechanisms of miR-3934-5p in neuroblastoma were assessed
in more detail. The association between miR-3934-5p and TP53INP1
was also investigated. The results of the present study may provide
promising targets for the development of novel approaches in the
management of neuroblastoma.
Materials and methods
Clinical samples
A total of 30 (Female, 12; Male, 18; age range, 3
months to 14 years) neuroblastoma tissues and normal matched
adjacent tissues (>2 cm from the tumor site) were obtained from
Jianou Municipal Hospital (Jianou, China) between June 2015 and
December 2017. Individuals with severe concomitant diseases,
including cardiovascular disease, malabsorption syndrome, liver or
kidney disease, another tumor and immunodepression were excluded
from the study. No patients had received any radiotherapy or
chemotherapy prior to surgery. The present study was approved by
the Ethical Committee of Jianou Municipal Hospital (Jianou, China).
Written informed consent was obtained from each patient and their
parent or guardians.
Cell culture and cell
transfection
Human neuroblastoma cell lines CHLA-20, CHLA-15 and
SMS-KAN (Children's Oncology Group, Cell Culture and Xenograft
Repository, Texas Tech University Health Science Centre; Lubbock,
TX, USA) were cultured in Iscoves Modified Dulbeccos Medium
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). SK-N-SH (cat. no.
ATCC® HTB-11™; American Type Culture Collection,
Manassas, VA, USA) were cultured in Eagles Minimum Essential medium
(EMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C for 24 h. Human
umbilical vein endothelial cells (HUVECs) were also cultured in
M199 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 20% FBS (Gibco; Thermo Fisher Scientific, Inc.).
On the day prior to cell transfection, SK-N-SH cells
were plated into a six-well plate at a density of 1×106
cells per well and cultured at 37°C with 5% CO2. SK-N-SH
cells were transfected with 50 nM inhibitor control [cat. no.
CS8005; Biomics Biotechnologies (Nantong) Co., Ltd.], 50 nM
miR-3934-5p inhibitor [cat. no. hsa-miR-3934-5p; Biomics
Biotechnologies (Nantong) Co., Ltd], 2 µl control-small interfering
RNA (siRNA; cat. no. sc-36869; Santa Cruz Biotechnology, Inc.), 2
µl TP53INP1-siRNA (cat. no. sc-76715; Santa Cruz Biotechnology,
Inc.) or 50 nM miR-3934-5p inhibitor+2 µl TP53INP1-siRNA using
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturers protocol. 48 h later, the
transfection efficiency was detected using reverse
transcription-quantitative (RT-q) PCR.
RT-qPCR
Total RNA from tissues and cells was extracted using
the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturers protocol. Total RNA concentration
was then detected via Nanodrop2000 (Thermo Fisher Scientific, Inc.)
and stored at −80°C until further use. The synthesis of cDNA was
performed using the RevertAid™ First Strand cDNA Synthesis kit
(Thermo Fisher Scientific, Inc.) according to the manufacturers
protocol. qPCR was subsequently performed using the SYBR Premix Ex
TaqTM II (TliRNaseH Plus) kit (Takara Bio, Inc.). The amplification
conditions for qPCR were as follows: 10 min at 95°C followed by 35
cycles of 15 sec at 95°C, 40 sec at 55°C and 72°C for 30 sec. The
primer sequences used for qPCR were as follows: TP53INP1 forward,
5′-GACTCACGGGCACAGAAGTGGAAGC-3′ and reverse,
5′-CCACTGGGAAGGGCGAAAG-3′; GAPDH forward,
5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse,
5′-GTAGAGGCAGGGATGATGTTCT-3′; U6 forward,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; Bcl-2 forward,
5′-TTGGATCAGGGAGTTGGAAG-3′ and reverse, 5′-TGTCCCTACCAACCAGAAGG-3′;
Bax forward, 5′-CGTCCACCAAGAAGCTGAGCG-3′ and reverse,
5′-CGTCCACCAAAGCTGAGCG3-3′; Cyclin dependent kinase inhibitor 1A
(p21) forward, 5′-TGAGCCGCGACTGTGATG-3′ and reverse,
5′-GTCTCGGTGACAAAGTCGAAGTT-3′. The relative expression of genes
were calculated using the 2−ΔΔCq method (26) following normalization with reference
to the expression of GAPDH or U6. All experiments were performed in
triplicate to ensure for minimum deviation.
Western blotting
Cells were washed three times with cold PBS and
total cellular proteins were extracted using
Radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime
Institute of Biotechnology) at 4°C for 30 min. The bicinchoninic
acid protein assay kit (Beyotime Institute of Biotechnology) was
used for protein quantification. Equal quantities of protein (30 µg
per lane) were separated on 10% SDS-PAGE gel and transferred to
PVDF membranes. The membranes were then blocked with 5% non-fat
milk at room temperature for 2 h. This was then followed by
incubation with primary antibodies against TP53INP1 (1:1,000; cat.
no. A04229; Wuhan Boster Biological Technology, Ltd.), Bcl-2
(1:1,000; cat. no. 4223; Cell Signaling Technology, Inc.), Bax
(1:1,000; cat. no. 5023; Cell Signaling Technology, Inc.), p21
(1:1,000; cat. no. 2947; Cell Signaling Technology, Inc.) or
β-actin (1:1,000; cat. no. 4970; Cell Signaling Technology, Inc.)
overnight at 4°C. The membranes were then incubated with
horseradish peroxidase-conjugated secondary anti-rabbit IgG
antibody (1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.),
for 2 h at room temperature. Finally, protein bands were visualized
using the Western Blotting Luminol Reagent (cat. no. sc-2048; Santa
Cruz Biotechnology, Inc.) according to the manufacturers
protocol.
Cell counting kit-8 (CCK-8) assay
A CCK-8 assay was performed to measure cell
viability. Logarithmic phase cells were seeded in a 96-well plate
with 1×104 cells per well and incubated in 37°C with 5%
CO2 for 12 h, after which 10 µl CCK-8 solution (Beyotime
Institute of Biotechnology) was added to each well and cells were
incubated for a further 2 h at 37°C with 5% CO2.
Absorbance was measured at a wavelength of 450 nm using a
FLUOstar® Omega Microplate Reader to assess cell
viability (BMG Labtech GmbH).
Flow cytometry assay
Cells were collected in the logarithmic growth phase
via trypsinization, washed three times with PBS and then
trypsinized into single cell suspensions. Apoptotic cells were
detected using the Annexin V-(FITC)/propidium iodide (PI) apoptosis
detection kit [cat. no. 70-AP101-100; Hangzhou MultiSciences
(Lianke) Biotech Co., Ltd.] according to the manufacturers
instructions. Cells were stained with 5 µl Annexin V-FITC and 5 µl
PI for 30 min for 15 min in darkness at room temperature. Flow
cytometry was performed (BD Biosciences) according to the
manufacturers protocol to detect cell apoptosis. The apoptotic rate
was determined using FlowJo software version 7.6.1 (FlowJo
LLC).
Dual-luciferase reporter assay
To predict the targets of miR-3934-5p, TargetScan
bioinformatics software (www.targetscan.org/vert_71) was applied. To
investigate the association between miR-3934-5p and TP53INP1, the
wild type (WT-TP53INP1) and mutant (MUT-TP53INP1) 3-UTRs of
TP53INP1 were cloned into a pmiR-RB-ReportTM dual luciferase
reporter gene plasmid vector (Guangzhou RiboBio Co., Ltd.) as per
the manufacturers instructions. Cells seeded in 24-well plates
(5×104 cells per well) were then co-transfected with
miR-3934-5p mimics or mimic controls and the MUT or WT of TP53INP1
using the Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h, together with the Renilla
luciferase pRL-TK vector as a control. After transfection for 48 h,
cells were lysed with RIPA buffer (Beyotime Institute of
Biotechnology). Relative luciferase activity was detected using the
dual-luciferase reporter assay system (Promega Corporation)
according to the manufacturers instructions. Luciferase activity
was normalized to that of Renilla luciferase activity.
Statistical analysis
Each experiment was performed at least three times.
All data was presented as the mean ± standard deviation.
Significant differences multiple groups were measured using one-way
ANOVA with a Tukeys post hoc test, and the Students t-test was used
to perform comparisons between two groups. P<0.05 was considered
to indicate a statistically significant difference. Data analyses
were performed using SPSS software version 17.0 (SPSS, Inc.).
Results
Expression of miR-3934-5p in
neuroblastoma tissues and cell lines
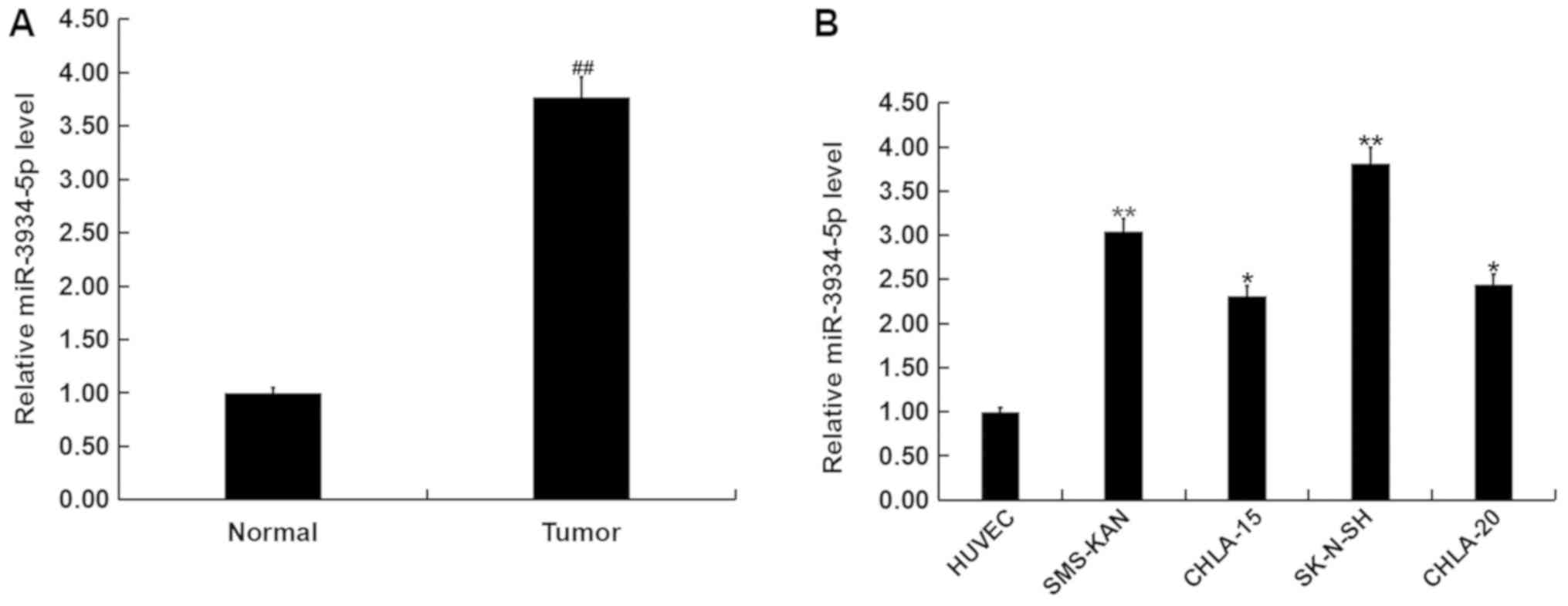
To assess the role of miR-3934-5p in neuroblastoma,
the level of miR-3934-5p in neuroblastoma and normal matched
adjacent tissues were detected using RT-qPCR. The results indicated
that the expression of miR-3934-5p was upregulated in neuroblastoma
tissues compared with normal matched adjacent tissues (Fig. 1A). miR-3934-5p levels in different
neuroblastoma cell lines (CHLA-20, CHLA-15, SMS-KAN and SK-N-SH)
and HUVECs were also measured. The current study demonstrated that
the expression of miR-3934-5p was significantly upregulated in
CHLA-20, CHLA-15, SMS-KAN and SK-N-SH cells in comparison with
HUVEC. The highest level of miR-3934-5p was observed in SK-N-SH
cells (Fig. 1B).
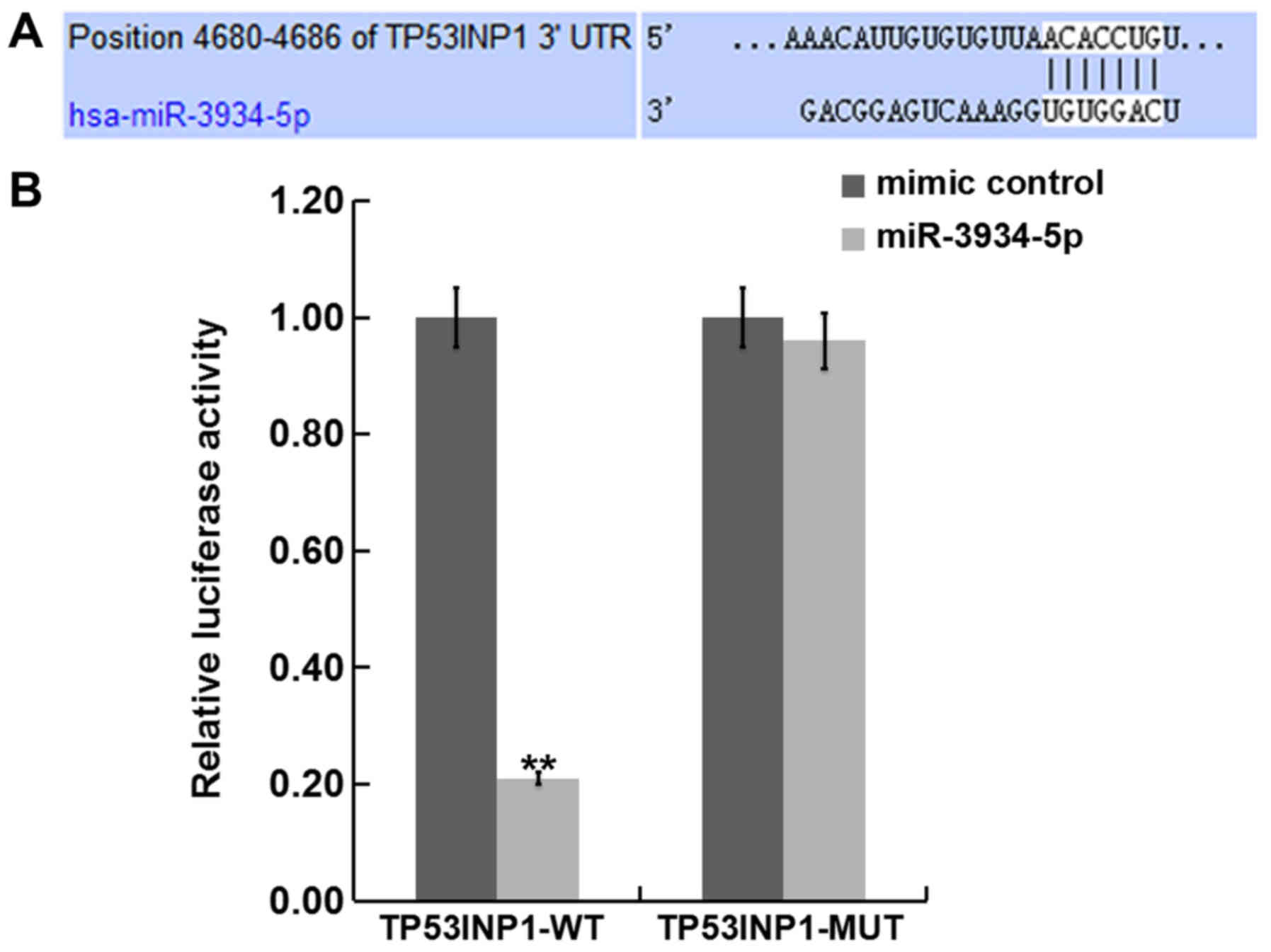
TP53INP1 is a direct gene of
miR-3934-5p
In order to determine the interaction between miRNAs
and their target genes, TargetScan was used to predict the target
gene of miR-3934-5p. The software revealed that the TP53INP1 3UTR
of mRNA contains a putative site that is partially complementary to
miR-3934-5p (Fig. 2A). A luciferase
reporter assay was performed to examine if miR-3934-5p interacts
directly with the target gene TP53INP1. Compared with
co-transfection of MUT-TP53INP1 and miR-3934-5p mimics, luciferase
activity was significantly decreased following co-transfection with
WT-TP53INP1 and miR-3934-5p mimics (Fig.
2B). These results suggest that TP53INP1 may be a direct target
gene of miR-3934-5p.
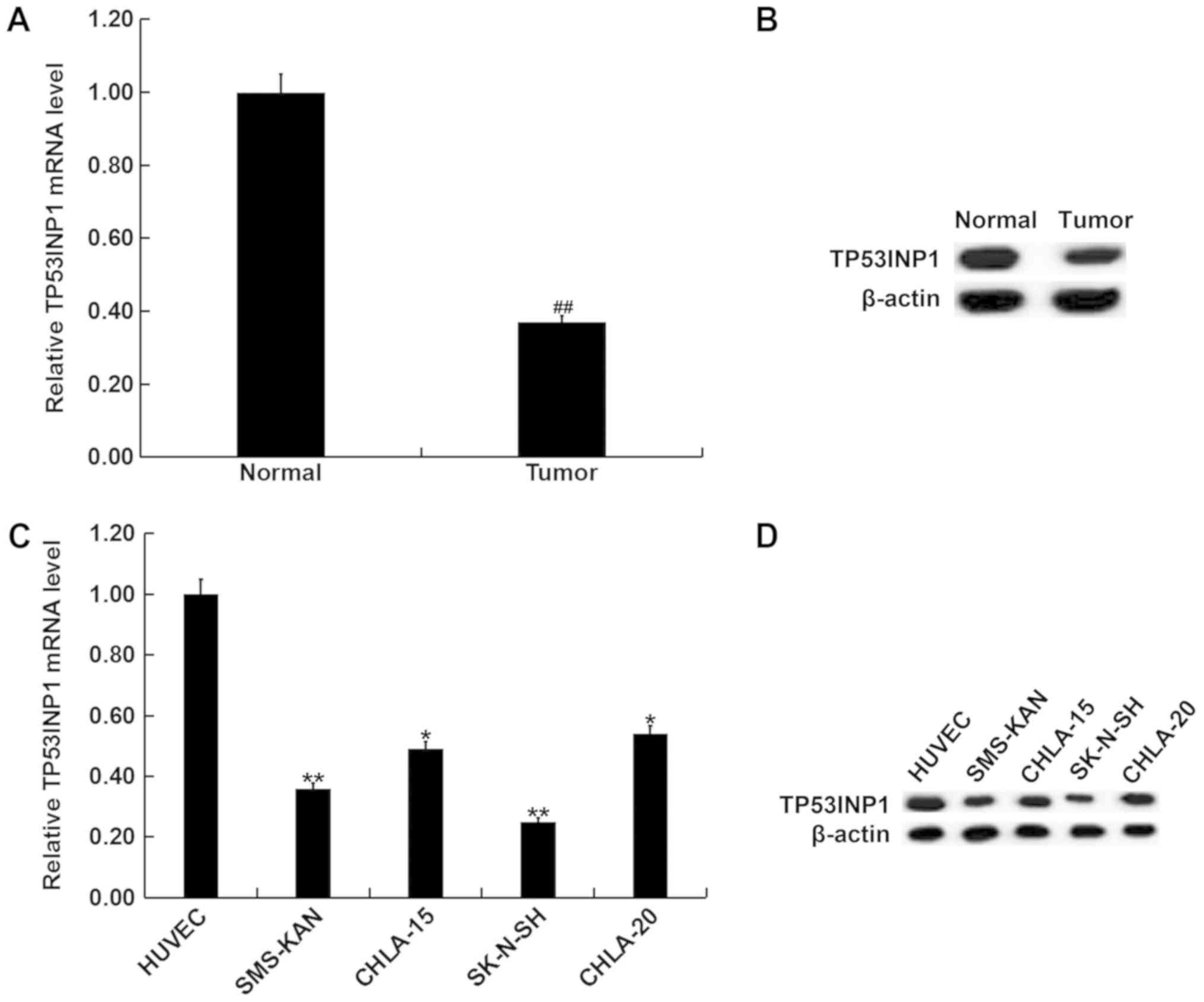
Expression of TP53INP1 in
neuroblastoma tissues and cell lines
The differences in expression of TP53INP1 between
neuroblastoma tissues and normal matched adjacent tissues were
determined using RT-qPCR and western blotting. The data revealed
that TP53INP1 was downregulated in neuroblastoma tissues compared
with normal matched adjacent tissues (Fig. 3A and B). In addition, RT-qPCR and
western blotting revealed that compared with HUVEC, TP53INP1 was
significantly downregulated in neuroblastoma cell lines, including
CHLA-20, CHLA-15, SMS-KAN and SK-N-SH at both mRNA and protein
levels. The lowest expression was observed in SK-N-SH cells
(Fig. 3C and D). The results
indicate that there is an inverse association between the
expression of miR-3934-5p and TP53INP1 in neuroblastoma.
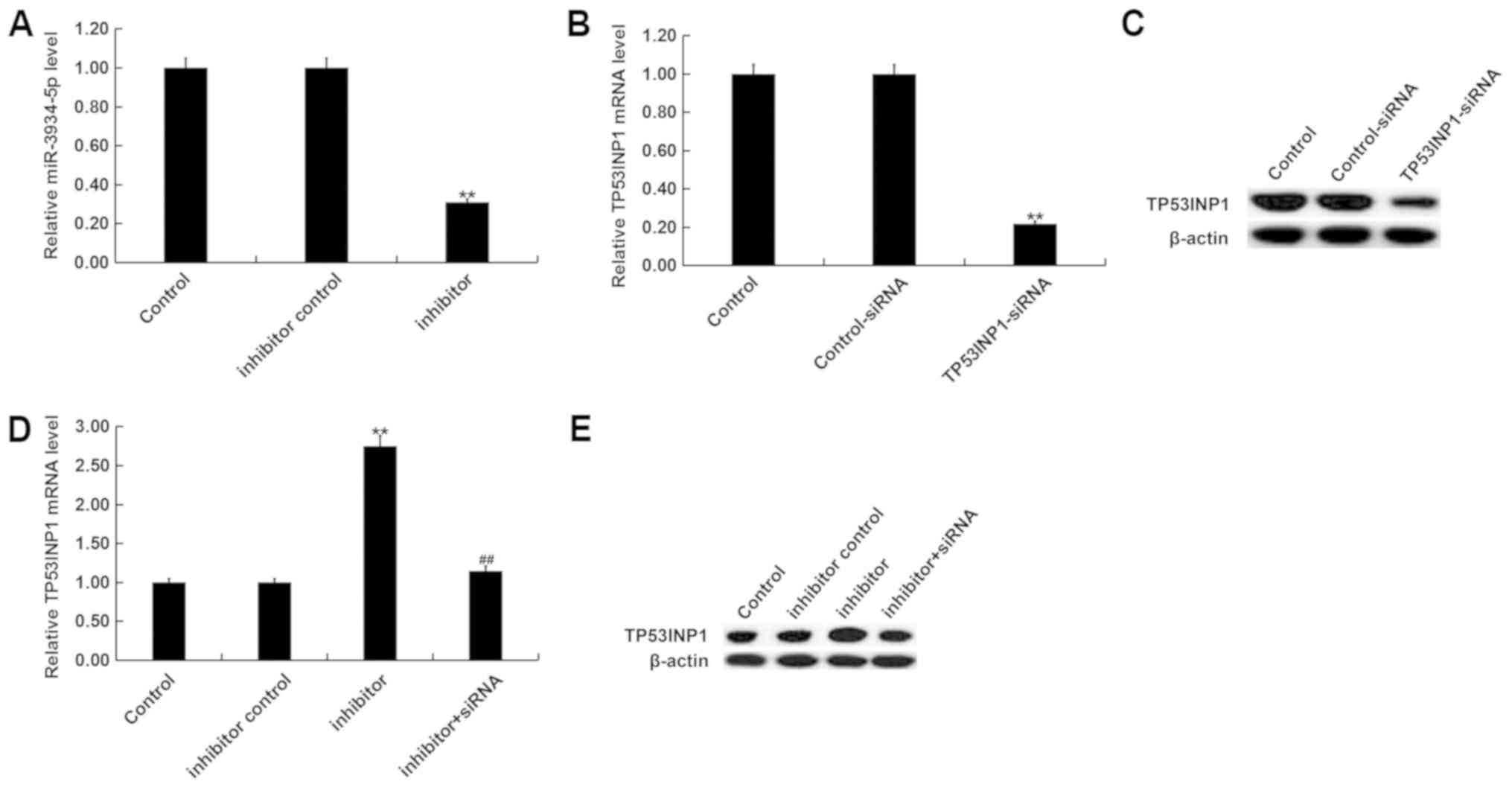
miR-3934-5p negatively regulates the
expression of TP53INP1 in SK-N-SH cells
SK-N-SH cells were transfected with inhibitor
controls, miR-3934-5p inhibitors, control-siRNA, TP53INP1-siRNA, or
miR-3934-5p inhibitor+TP53INP1-siRNA for 48 h. RT-qPCR or western
blot analysis was then performed to detect transfection efficiency.
The RT-qPCR assay revealed that the miR-3934-5p inhibitor
significantly reduced the expression of miR-3934-5p in SK-N-SH
cells (Fig. 4A). RT-qPCR and western
blot analysis also indicated that TP53INP1-siRNA decreased the mRNA
and protein expression of TP53INP1 in SK-N-SH cells (Fig. 4B and C). The results also indicated
that the miR-3934-5p inhibitor significantly increased the mRNA and
protein expression of TP53INP1. However, this effect was reversed
via TP53INP1-siRNA treatment (Fig. 4D
and E).
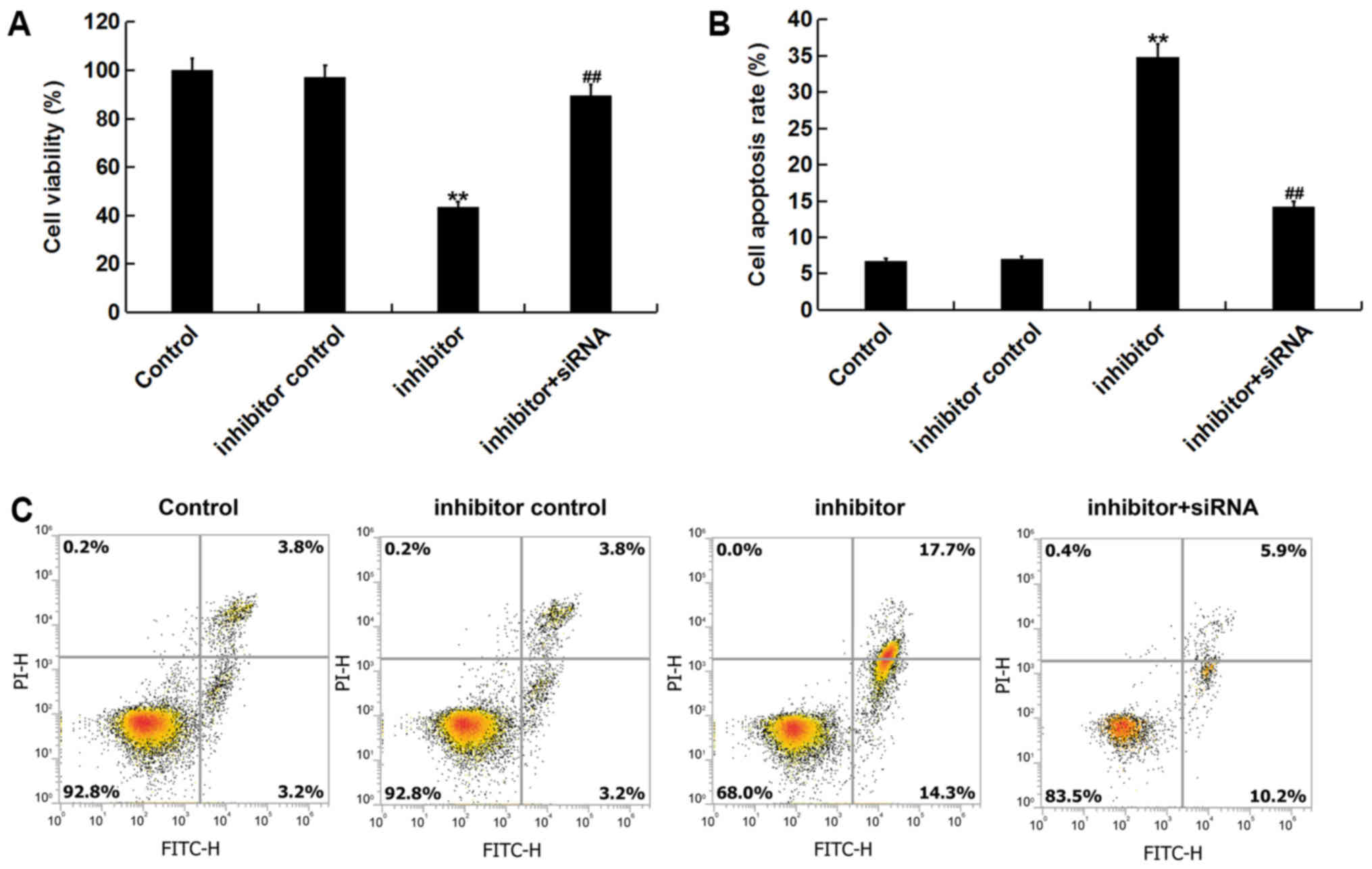
Effect of miR-3934-5p on viability and
apoptosis of neuroblastoma cells
To assess the role of miR-3934-5p in neuroblastoma
cells, the effect of miR-3934-5p on the viability of SK-N-SH cells
was assessed. The result of the CCK-8 assay indicated that compared
with the control group, the miR-3934-5p inhibitor significantly
decreased cell viability. This decrease was subsequently reversed
by TP53INP1-siRNA treatment (Fig.
5A). To determine the apoptotic effect of miR-3934-5p, flow
cytometry was performed. Analysis revealed that transfection with
the miR-3934-5p inhibitor significantly induced cell apoptosis.
This apoptotic effect was then reversed via TP53INP1-siRNA
treatment (Fig. 5B and C).
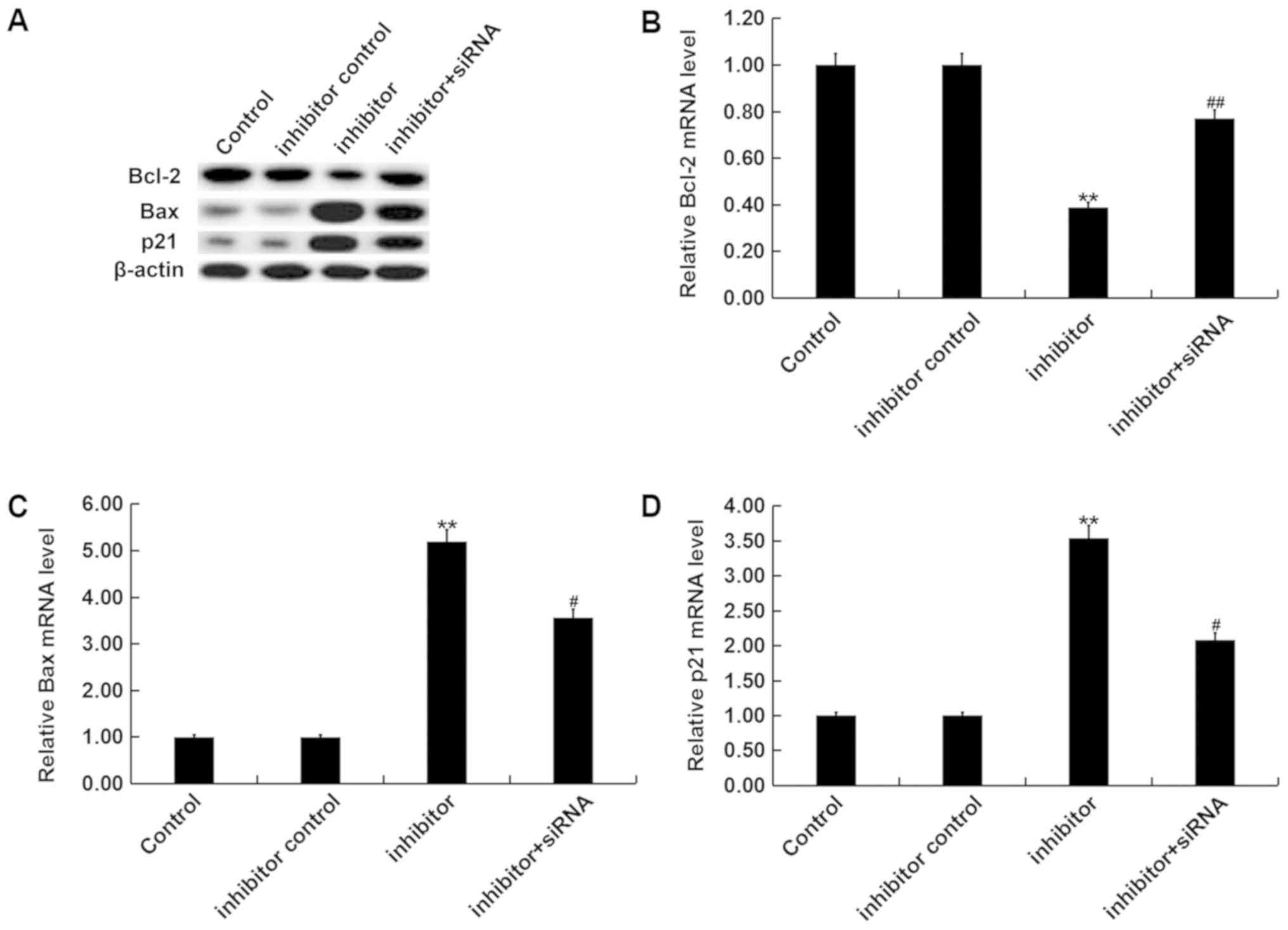
Furthermore, RT-qPCR and western blot analysis revealed that the
miR-3934-5p inhibitor significantly decreased Bcl-2 protein and
mRNA expression, and increased Bax and p21 expression, which was
then reversed via TP53INP1-siRNA (Fig.
6A and D).
Discussion
It has been revealed that miR-3934 may be associated
with cervical and lung cancer (16,17). The
current study demonstrated that miR-3934-5p is highly expressed in
both neuroblastoma tissues and cell lines compared with adjacent
normal tissues and cells. miRNAs serve important roles in many cell
development processes, including cell proliferation, cell
differentiation and apoptosis (27–32).
Increasing evidence has also revealed that the aberrant expression
of miRNAs may be associated with different types of cancer, in
which miRNAs may act as tumor suppressors or oncogenes (33–37).
TP53INP1 is a p53-inducible gene that encodes for
two protein isoforms, which modulate p53 biological activities
(38). TP53INP1 over-expression
induces cell cycle arrest in the G1 phase and enhances p53-mediated
apoptosis (39). Many studies have
demonstrated that TP53INP1 is a target gene of many miRNAs
(18–20,25,40–42). To
the best of our knowledge, there are no studies regarding the
association between TP53INP1 and miR-3934-5p in neuroblastoma. In
certain types of cancer, TP53INP1 functions as a tumor suppressor
(21,43). Within the current study, the dual
luciferase reporter assay indicated that miR-3934-5p directly
targets TP53INP1 and that TP53INP1 was downregulated in
neuroblastoma tissues and cell lines. These results indicated that
there was an inverse association between the expression of
miR-3934-5p and TP53INP1 in neuroblastoma. However, in prostate
cancer, TP53INP1 has been reported to act as an oncogene and its
over-expression is associated with castration-resistant prostate
cancer (44). Several miRNAs have
been indicated to negatively regulate TP53INP1 expression
including, miR-155 in pancreatic cancer (45), miR-130b in hepatocellular cancer
(41), miR-125b in type 2
endometrial carcinoma cells (42),
and miR-569 in epithelial cancers (46). In the present study, HUVECs were used
as control cells. However, whether the expression of miR-3934-5p
and TP53INP1 in endothelial cells can be representative of other
normal cells requires further study.
To investigate the effects of miR-3934-5p and
TP53INP1 on neuroblastoma cell proliferation and apoptosis, SK-N-SH
cells were transfected with an inhibitor control, a miR-3934-5p
inhibitor, or miR-3934-5p inhibitor+TP53INP1-siRNA for 48 h. The
results of the CCK-8 assay and flow cytometry revealed that the
miR-3934-5p inhibitor inhibited cell viability and induced cell
apoptosis. In addition, the expression of the anti-apoptotic gene
Bcl-2, the pro-apoptotic gene Bax and the apoptosis regulator p21
were also determined in the current study. The findings of the
current study indicate that the miR-3934-5p inhibitor significantly
decreased Bcl-2 increased Bax and increased p21 expression at both
protein and mRNA levels. Furthermore, all the effects of the
miR-3934-5p inhibitor on neuroblastoma cells were reversed via
TP53INP1-siRNA treatment.
In summary, the data of the current study
demonstrated that miR-3934-5p was downregulated in neuroblastoma
tissues and cell lines. miR-3934-5p downregulation also
significantly inhibited neuroblastoma cell viability and induced
apoptosis by directly regulating the expression of TP53INP1. The
present study has therefore provided a potential novel therapeutic
target for neuroblastoma.
Acknowledgements
The authors would like to acknowledge Dr Yang Jie
and Dr Huang Rongbin (Science and Education Department, Jianou
Municipal Hospital, Jianou, China) who offered valuable suggestions
for the present study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
WY contributed to the study design, data collection,
statistical analysis, data interpretation and manuscript
preparation. FL, CY and MZ contributed to data collection and data
interpretation. DF and XJ contributed to the statistical analysis
and performed the literature search.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Jianou Municipal Hospital (Jianou, China). Written
informed consent was obtained from each patient and their parent or
guardians.
Patient consent for publication
All patients provided consent to publish data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Health Professional Version, .
Neuroblastoma Treatment (PDQ®)-Health Professional
VersionNational Cancer Institute; Bethesda, MD: 2014, https://www.cancer.gov/types/neuroblastoma/hp/neuroblastoma-treatment-pdq
|
|
2
|
Gatta G, Botta L, Rossi S, Aareleid T,
Bielska-Lasota M, Clavel J, Dimitrova N, Jakab Z, Kaatsch P, Lacour
B, et al: Childhood cancer survival in Europe 1999–2007: Results of
EUROCARE-5-a population-based study. Lancet Oncol. 15:35–47. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brodeur GM and Bagatell R: Mechanisms of
neuroblastoma regression. Nat Rev Clin Oncol. 11:704–713. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li P, Gao Y, Ji Z, Zhang X, Xu Q, Li G,
Guo Z, Zheng B and Guo X: Role of urokinase plasminogen activator
and its receptor in metastasis and invasion of neuroblastoma. J
Pediatr Surg. 39:1512–1519. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Matthay KK, Maris JM, Schleiermacher G,
Nakagawara A, Mackall CL, Diller L and Weiss WA: Neuroblastoma. Nat
Rev Dis Primers. 2:160782016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mazar J, Li Y, Rosado A, Phelan P,
Kedarinath K, Parks GD, Alexander KA and Westmoreland TJ: Zika
virus as an oncolytic treatment of human neuroblastoma cells
requires CD24. PLoS One. 13:e02003582018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smith MA, Altekruse SF, Adamson PC, Reaman
GH and Seibel NL: Declining childhood and adolescent cancer
mortality. Cancer. 120:2497–2506. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kwon C, Han Z, Olson EN and Srivastava D:
MicroRNA1 influences cardiac differentiation in Drosophila and
regulates Notch signaling. Proc Natl Acad Sci USA. 102:18986–18991.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ro S, Park C, Young D, Sanders KM and Yan
W: Tissue-dependent paired expression of miRNAs. Nucleic Acids Res.
35:5944–5953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mallory AC and Vaucheret H: MicroRNAs:
Something important between the genes. Curr Opin Plant Biol.
7:120–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carrington JC and Ambros V: Role of
microRNAs in plant and animal development. Science. 301:336–338.
2001. View Article : Google Scholar
|
|
16
|
Chen JY, Yao DS, He CJ, et al:
Differential expression of microRNA in cervical squamous cell
carcinoma. J Pract Med. 30:83–87. 2014.
|
|
17
|
Sathipati SY and Ho SY: Identifying the
miRNA signature associated with survival time in patients with lung
adenocarcinoma using miRNA expression profiles. Sci Rep.
7:2017.
|
|
18
|
Chen Q, Zhou Y, Richards AM and Wang P:
Up-regulation of miRNA-221 inhibits hypoxia/reoxygenation-induced
autophagy through the DDIT4/mTORC1 and Tp53inp1/p62 pathways.
Biochem Biophys Res Commun. 474:168–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu CG, Yang MF, Fan JX and Wang W: MiR-30a
and miR-205 are downregulated in hypoxia and modulate
radiosensitivity of prostate cancer cells by inhibiting autophagy
via TP53INP1. Eur Rev Med Pharmacol Sci. 20:1501–1508.
2016.PubMed/NCBI
|
|
20
|
Wang W, Liu J and Wu Q: MiR-205 suppresses
autophagy and enhances radiosensitivity of prostate cancer cells by
targeting TP53INP1. Eur Rev Med Pharmacol Sci. 20:92–100.
2016.PubMed/NCBI
|
|
21
|
Seillier M, Peuget S, Gayet O, Gauthier C,
NGuessan P, Monte M, Carrier A, Iovanna JL and Dusetti NJ:
TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family
proteins through the LC3-interacting region (LIR) and promotes
autophagy-dependent cell death. Cell Death Differ. 19:1525–1535.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Saadi H, Seillier M and Carrier A: The
stress protein TP53INP1 plays a tumor suppressive role by
regulating metabolic homeostasis. Biochimie. 118:44–50. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xue X, Wang X, Zhao Y, Hu R and Qin L:
Exosomal miR-93 promotes proliferation and invasion in
hepatocellular carcinoma by directly inhibiting
TIMP2/TP53INP1/CDKN1A. Biochem Biophys Res Commun. 502:515–521.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Sun H, Zhang D, Fan D, Zhang Y,
Dong X, Liu S, Yang Z, Ni C, Li Y, et al: TP53INP1 inhibits
hypoxia-induced vasculogenic mimicry formation via the ROS/snail
signalling axis in breast cancer. J Cell Mol Med. 22:3475–3488.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cai Q, Zeng S, Dai X, Wu J and Ma W:
miR-504 promotes tumour growth and metastasis in human osteosarcoma
by targeting TP53INP1. Oncol Rep. 38:2993–3000. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu X, Li Z, Shen J, Wu WK, Liang J, Weng X
and Qiu G: MicroRNA-10b promotes nucleus pulposus cell
proliferation through RhoC-Akt pathway by targeting HOXD10 in
intervetebral disc degeneration. PLoS One. 8:e830802013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Z, Yu X, Shen J, Wu WK and Chan MT:
MicroRNA expression and its clinical implications in Ewings
sarcoma. Cell Prolif. 48:1–6. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu X and Li Z: MicroRNAs regulate vascular
smooth muscle cell functions in atherosclerosis (review). Int J Mol
Med. 34:923–933. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li M, Yu M, Liu C, Zhu H, He X, Peng S and
Hua J: miR-34c works downstream of p53 leading to dairy goat male
germline stem-cell (mGSCs) apoptosis. Cell Prolif. 46:223–231.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie
T, Zhang J, Peng C, Lin Y and Chen J: MicroRNA-25 regulates
chemoresistance-associat-ed autophagy in breast cancer cells, a
process modulated by the natural autophagy inducer
isoliquiritigenin. Oncotarget. 5:7013–7026. 2014.PubMed/NCBI
|
|
32
|
Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei
Y, Li Y, Yang Q, Liu J, Wei JJ, et al: miR-145 inhibits tumor
growth and metastasis by targeting metadherin in high-grade serous
ovarian carcinoma. Oncotarget. 5:10816–10829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y,
Liu C, Song W, Wang F, Zhang J, et al: DNA methylation
downregulated mir-10b acts as a tumor suppressor in gastric cancer.
Gastric Cancer. 18:43–54. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Z, Yu X, Wang Y, Shen J, Wu WK, Liang J
and Feng F: By downregulating TIAM1 expression, microRNA-329
suppresses gastric cancer invasion and growth. Oncotarget.
6:17559–17569. 2015.PubMed/NCBI
|
|
35
|
Li Z, Yu X, Shen J and Jiang Y: MicroRNA
dysregulation in uveal melanoma: A new player enters the game.
Oncotarget. 6:4562–4568. 2015.PubMed/NCBI
|
|
36
|
Li J, You T and Jing J: MiR-125b inhibits
cell biological progression of Ewings sarcoma by suppressing the
PI3K/Akt signalling pathway. Cell Prolif. 47:152–160. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schirmer U, Doberstein K, Rupp AK, Bretz
NP, Wuttig D, Kiefel H, Breunig C, Fiegl H, Müller-Holzner E,
Zeillinger R, et al: Role of miR-34a as a suppressor of L1CAM in
endometrial carcinoma. Oncotarget. 5:462–472. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang PH, Motoo Y, Garcia S, Iovanna JL,
Pébusque MJ and Sawabu N: Down-expression of tumor protein
p53-induced nuclear protein 1 in human gastric cancer. World J
Gastroenterol. 12:691–696. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tomasini R, Samir AA, Carrier A, Isnardon
D, Cecchinelli B, Soddu S, Malissen B, Dagorn JC, Iovanna JL and
Dusetti NJ: TP53INP1s and homeodomain-interacting protein kinase-2
(HIPK2) are partners in regulating p53 activity. J Biol Chem.
278:37722–37729. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liao D, Li T, Ye C, Zeng L, Li H, Pu X,
Ding C, He Z and Huang GL: miR-221 inhibits autophagy and targets
TP53INP1 in colorectal cancer cells. Exp Ther Med. 15:1712–1717.
2018.PubMed/NCBI
|
|
41
|
Ma S, Tang KH, Chan YP, Lee TK, Kwan PS,
Castilho A, Ng I, Man K, Wong N, To KF, et al: miR-130b Promotes
CD133(+) liver tumor-initiating cell growth and self-renewal via
tumor protein 53-induced nuclear protein 1. Cell Stem Cell.
7:694–707. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiang F, Liu T, He Y, Yan Q, Chen X, Wang
H and Wan X: MiR-125b promotes proliferation and migration of type
II endometrial carcinoma cells through targeting TP53INP1 tumor
suppressor in vitro and in vivo. BMC Cancer. 11:4252011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Seux M, Peuget S, Montero MP, Siret C,
Rigot V, Clerc P, Gigoux V, Pellegrino E, Pouyet L, NGuessan P, et
al: TP53INP1 decreases pancreatic cancer cell migration by
regulating SPARC expression. Oncogene. 30:3049–3061. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Giusiano S, Baylot V, Andrieu C, Fazli L,
Gleave M, Iovanna JL, Taranger-Charpin C, Garcia S and Rocchi P:
TP53INP1 as new therapeutic target in castration-resistant prostate
cancer. Prostate. 72:1286–1294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gironella M, Seux M, Xie MJ, Cano C,
Tomasini R, Gommeaux J, Garcia S, Nowak J, Yeung ML, Jeang KT, et
al: Tumor protein 53-induced nuclear protein 1 expression is
repressed by miR-155, and its restoration inhibits pancreatic tumor
development. Proc Natl Acad Sci USA. 104:16170–16175. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chaluvally-Raghavan P, Zhang F, Pradeep S,
Hamilton MP, Zhao X, Rupaimoole R, Moss T, Lu Y, Yu S, Pecot CV, et
al: Copy number gain of hsa-miR-569 at 3q26.2 leads to loss of
TP53INP1 and aggressiveness of epithelial cancers. Cancer Cell.
26:863–879. 2014. View Article : Google Scholar : PubMed/NCBI
|