Introduction
Chronic rhinosinusitis (CRS) is a heterogenous
disease of the sinuses and upper airways that affects a large
proportion of the population worldwide (1,2). CRS is
classified into CRS with nasal polyps (CRSwNP) and CRS without
nasal polyps (CRSsNP), and is typically characterized by two or
more of the following symptoms: Nasal obstruction, drainage, loss
of sense of smell, headaches and facial pain or pressure (3,4). CRSsNP
is associated with the symptoms of headache and facial pain or
pressure, whereas CRSwNP is more closely associated with the
symptoms of nasal obstruction and loss of sense of smell (5–7). In the
clinic, CRSwNP is frequently resistant to medical therapy, and
therefore, management of patients with CRSwNP is costly.
Furthermore, a proportion of patients with CRSwNP do not benefit
from topical corticosteroids, long-term systemic
glucocorticosteroids and repeated sinus surgery (8–10). Thus,
a better understanding of the reason why CRSwNP is not controlled
by current approaches and the underlying molecular mechanisms is
required.
MicroRNAs (miRNAs/miRs) are a class of small
non-coding RNAs that are 21–24 nucleotides in length and have key
roles in the pathogenesis of numerous diseases. miRNAs either
suppress target transcripts or inhibit translation in order to
post-transcriptionally regulate protein expression (11–13).
Aberrant miRNA expression is a common feature of a diverse range of
diseases (14,15). Previous studies indicate a distinct
expression pattern of inflammatory and re-modeling mediators in
CRSwNP (8). With the development of
high-throughput gene detection techniques and bioinformatics, a new
era of miRNA investigation has rapidly developed for numerous types
of allergic airway disease, including allergic rhinitis (16) and asthma (17,18).
However, the genome-scale miRNA expression pattern of CRSwNP
remains to be fully elucidated.
In the present study, the different miRNA expression
patterns of CRSwNP and CRSsNP were investigated using
next-generation sequencing, and the expression of a candidate miRNA
and its predicted targets was subsequently validated in CRSwNP
using reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis.
Materials and methods
Patients
A total of 100 patients requiring endoscopic sinus
surgery were recruited at The First Affiliated Hospital of Xinjiang
Medical University (Urumqi, China) between October 2016 and
February 2017, including 61 patients with CRSwNP (38 male and 23
female patients; median age, 44 years old; age range, 23–63 years
old) and 39 patients with CRSsNP (27 male and 12 female patients;
median age, 41 years old; age range, 20–65 years old). All subjects
met the criteria for CRS as defined by the European position paper
on rhinosinusitis and NPs 2012 (10). An additional 35 patients (22 male and
13 female patients; median age, 39 years old; age range, 19–57
years old) that required septoplasty and did not have any
inflammatory nasal disease were enrolled as control subjects.
Patients that were pregnant or dependent on systemic steroid
therapy were excluded. During surgery, ethmoidal mucosa or polyp
tissues were collected from the patients with CRS and inferior
turbinate tissues were obtained from the control patients. The
specimens from 5 of the patients with CRSwNP, 4 of the patients
with CRSsNP and 4 of the control patients were used for miRNA
sequencing. Subsequently, the RNA of 26 of the patients was
excluded due to low quality (Fig.
S1). Finally, the RNA of 96 of the patients was used in the
RT-qPCR validation assay.
Total RNA extraction and complementary
(c)DNA synthesis
Total RNA was extracted from each specimen using
TRIzol (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Small RNAs with 18–30 nt fragments were
enriched using denaturing polyacrylamide gel electrophoresis (15%
gel). According to the manufacturer's protocol, the fragments were
ligated to 5 and 3 adaptors and reverse-transcribed into cDNA using
a Mir-X™ miRNA First-Strand Synthesis kit (cat. no. 638313; Takara
Bio Inc.) after purification. The quality of these RNA samples was
assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies,
Inc.). For mRNA quantification, total RNA was subjected to reverse
transcription using a PrimeScript™ 1st Strand cDNA Synthesis Kit
(cat. no. 6110A; Takara Bio Inc.). Only samples meeting the
criteria of optical density ratio at 260/280 nm ≥1.8 and RNA
integrity number ≥8.0 were used for all of the analyses. The RNA
integrity of each sample was also assessed using a denaturing
agarose gel in order to determine the intensity ratio of the 28S
and 18S ribosomal RNA bands.
miRNA sequencing (miRNA-seq)
Following RT of the RNA into cDNA, the cDNA was
subjected to deep sequencing, which was performed by Genesky
Biotechnologies. The small RNA libraries were evaluated using
FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
After removing adaptor and low-quality sequences, the remaining
high-quality sequences were analyzed with the miRBase (19) (http://www.mirbase.org/) and Rfam (20) (http://rfam.xfam.org/) databases.
Differentially expressed gene (DEG)
screening
The R package edgeR (http://www.bioconductor.org/packages/2.4/bioc/html/edgeR.html)
was used to identify DEGs, with the cut-off values of |log2 fold
change (FC)|≥1 and adjusted P-value of <0.05.
2-Dimensional (2D) hierarchical
clustering analysis
A gene expression matrix of the DEGs was prepared
and analyzed with the pheatmap package (http://CRAN.R-project.org/package=pheatmap) in order
to perform unsupervised clustering in R after normalization.
Target prediction and functional
enrichment
The miRWalk database version 2.0 (mirwalk.uni-hd.de/) was used for predicting
miRNA-target interactions (21).
Subsequently, all of the predicted targets were subjected to gene
ontology (GO) (22) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) (23) pathway enrichment analysis in
WebGestalt (http://www.webgestalt.org/), with a criterion of
P<0.01.
qPCR
In order to investigate the expression of miR-4492,
interleukin (IL)-10 and tumor necrosis factor α (TNF-α) in the
specimens, qPCR was performed using SYBR Advantage qPCR Premix
(cat. no. 638322; Takara Bio Inc.) on an ABI 7300 Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH
served as a housekeeping gene for standardization of protein-coding
gene expression, and U6 served as the reference for standardization
of miRNA expression. All assays were performed at least in
triplicate and gene expression was quantified using the
2−∆∆Cq method (24). The
sequences of the primer pairs were as follows: miR-4492 forward,
5′-CTGGGCGCGCGCCAAA-3′; IL-10 forward, 5′-TCAAGGCGCATGTGAACTCC-3′
and reverse, 5′-CCACGGCCTTGCTCTTGTTT-3′; TNF-α forward,
5′-TTCTGCCTGCTGCACTTTGG-3′ and reverse,
5′-TGTCACTCGGGGTTCGAGAAG-3′. The universal miRNA reverse primer was
in the Mir-X miRNA First-Strand Synthesis kit.
Statistical analysis
Statistical analysis was performed with SPSS
software version 16.0 (SPSS Inc.). All of the assays were performed
at least in triplicate and values are expressed in bar charts as
the mean ± standard error of the mean. Significant differences
between groups were analyzed using Student's t-test or one-way
analysis of variance followed by Duncan's multiple-comparisons
test. P<0.05 was considered to indicate a statistically
significant difference. Pearson correlation analysis was performed
in order to assess the co-expression of miR-4492 and IL-10 in the
96 specimens.
Results
Identification of differentially
expressed miRNAs in CRSwNP
A total of 6 DEGs (6 downregulated miRNAs) were
identified between CRSwNP and CRSsNP. One DEG (1 downregulated
miRNA) was identified between CRSwNP and the control, and no DEGs
were identified between CRSsNP and the control. The details of the
DEGs are presented in Table I.
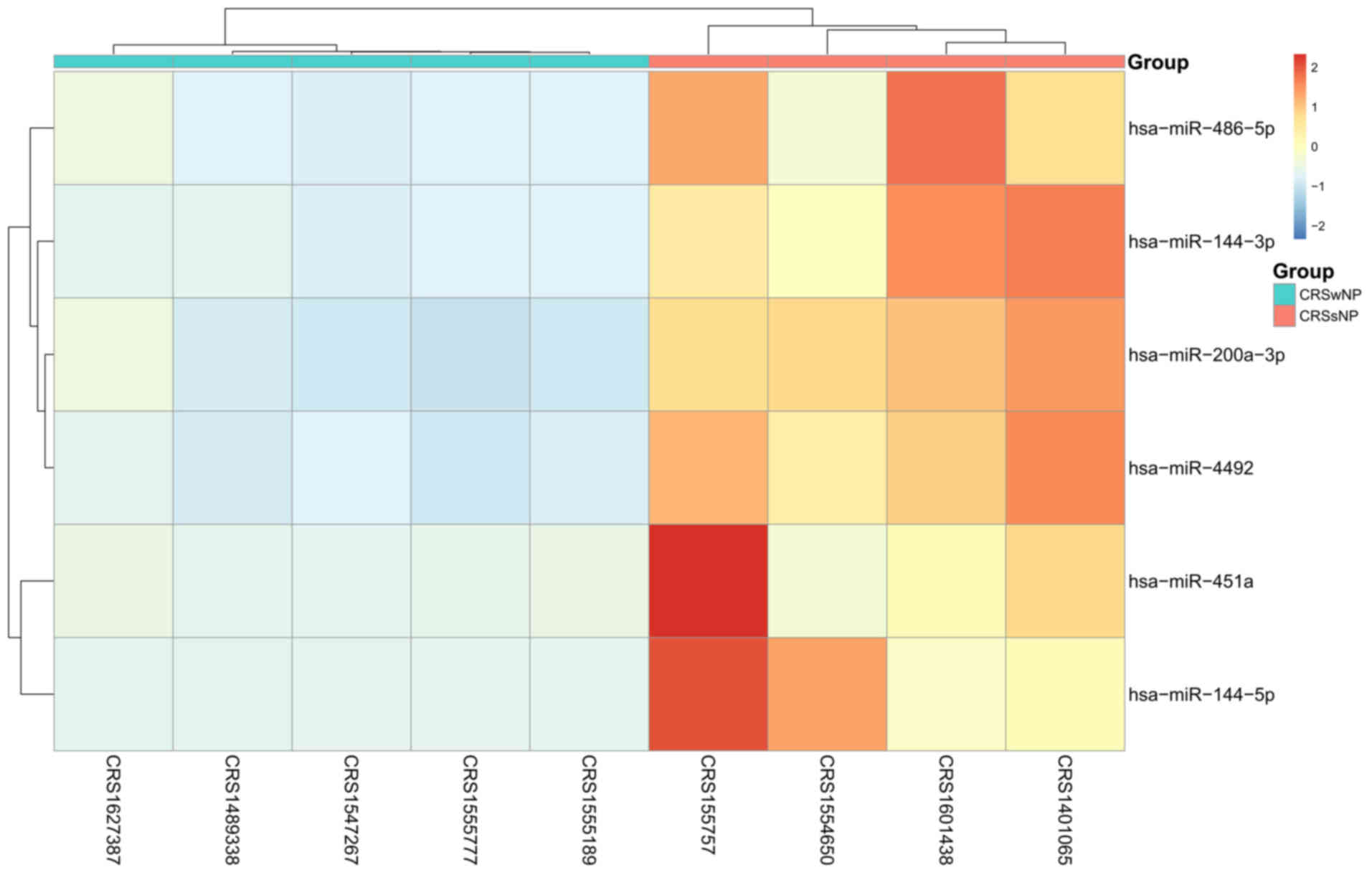
Subsequently, the expression data of six DEGs were subjected to 2D
hierarchical clustering analysis. The distinctive clusters for
CRSwNP and CRSsNP were segregated for all of the CRS specimens
(Fig. 1), revealing differences in
the miRNA expression profiles of the two subgroups of CRS.
 | Table I.Details of aberrantly expressed
miRNAs. |
Table I.
Details of aberrantly expressed
miRNAs.
|
| CRSwNP vs.
CRSsNP | CRSwNP vs. NC | CRSsNP vs. NC |
|---|
|
|
|
|
|
|---|
| Gene names | logFC | FDR | logFC | FDR | logFC | FDR |
|---|
| hsa-miR-144-3p |
−3.065465067 |
0.002555901 |
−1.820765547 |
0.821307633 |
0.857082137 |
0.996058565 |
| hsa-miR-144-5p |
−3.160003078 |
0.003750538 |
−1.835835772 |
0.821307633 |
0.856915634 |
0.996058565 |
| hsa-miR-486-5p |
−2.135055827 |
0.013129379 |
−1.258258512 |
0.939213466 |
0.713031482 |
0.996058565 |
|
hsa-miR-200a-3p |
−2.187602496 |
0.013129379 |
−0.284138738 |
0.998572989 |
1.406274811 |
0.996058565 |
| hsa-miR-451a |
−2.09844785 |
0.015772606 |
−1.231243617 |
0.998572989 |
0.720894 |
0.996058565 |
| hsa-miR-4492 |
−1.745117094 |
0.019422342 |
−1.513875624 |
0.012444032 |
0.284184 |
0.996058565 |
Gene enrichment and functional
annotation of the predicted targets of the DEGs
A total of 4,395 predicted target genes of the six
DEGs were obtained from the miRWalk database. Among these genes, a
diverse range of GO terms were significantly enriched in the
following three categories: Biological process (BP), cellular
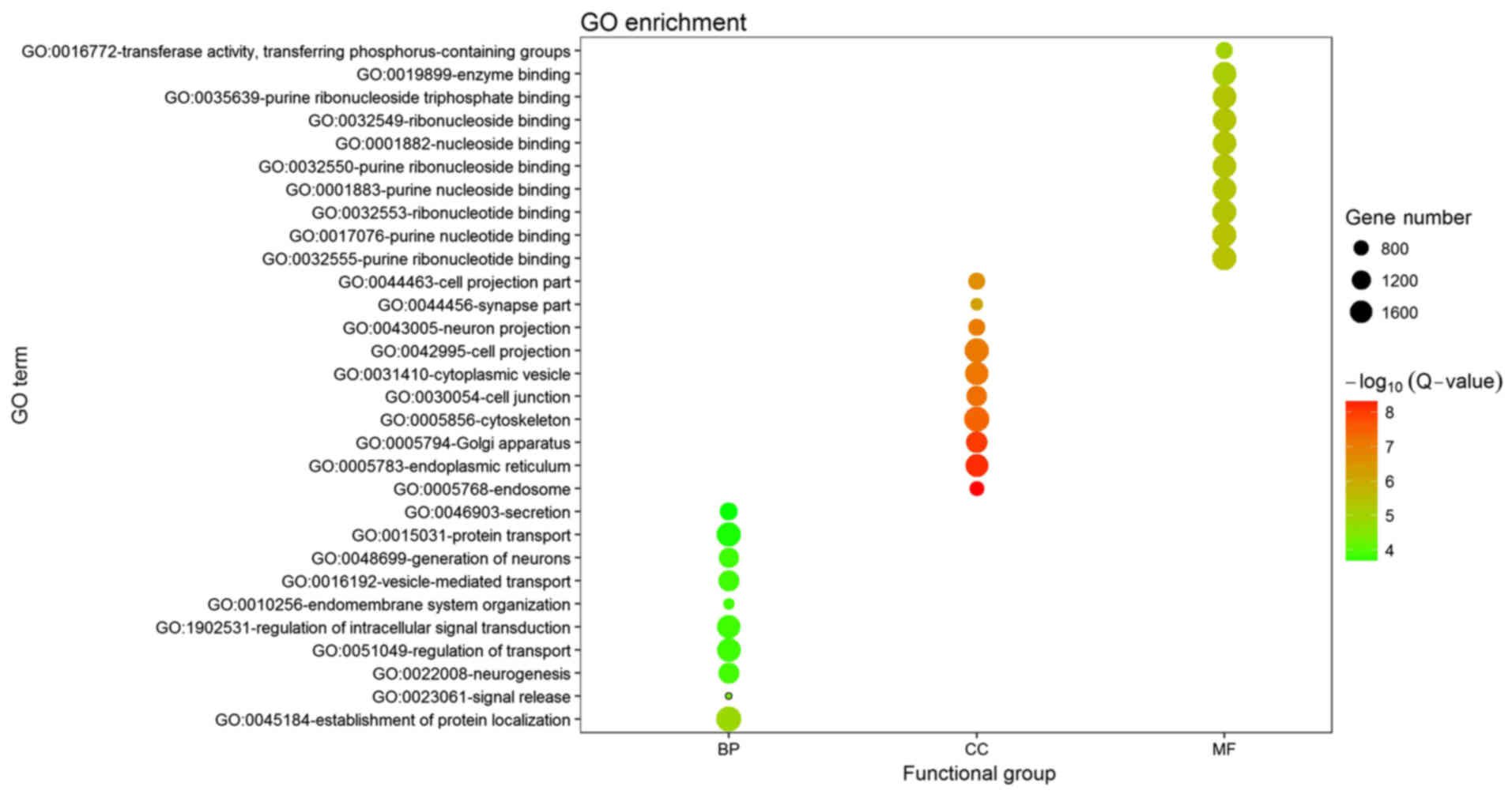
component (CC) and molecular function (MF). The top 5 enriched
terms in each category are provided in Fig. 2, of which establishment of protein
localization, endosome and purine ribonucleotide binding were the
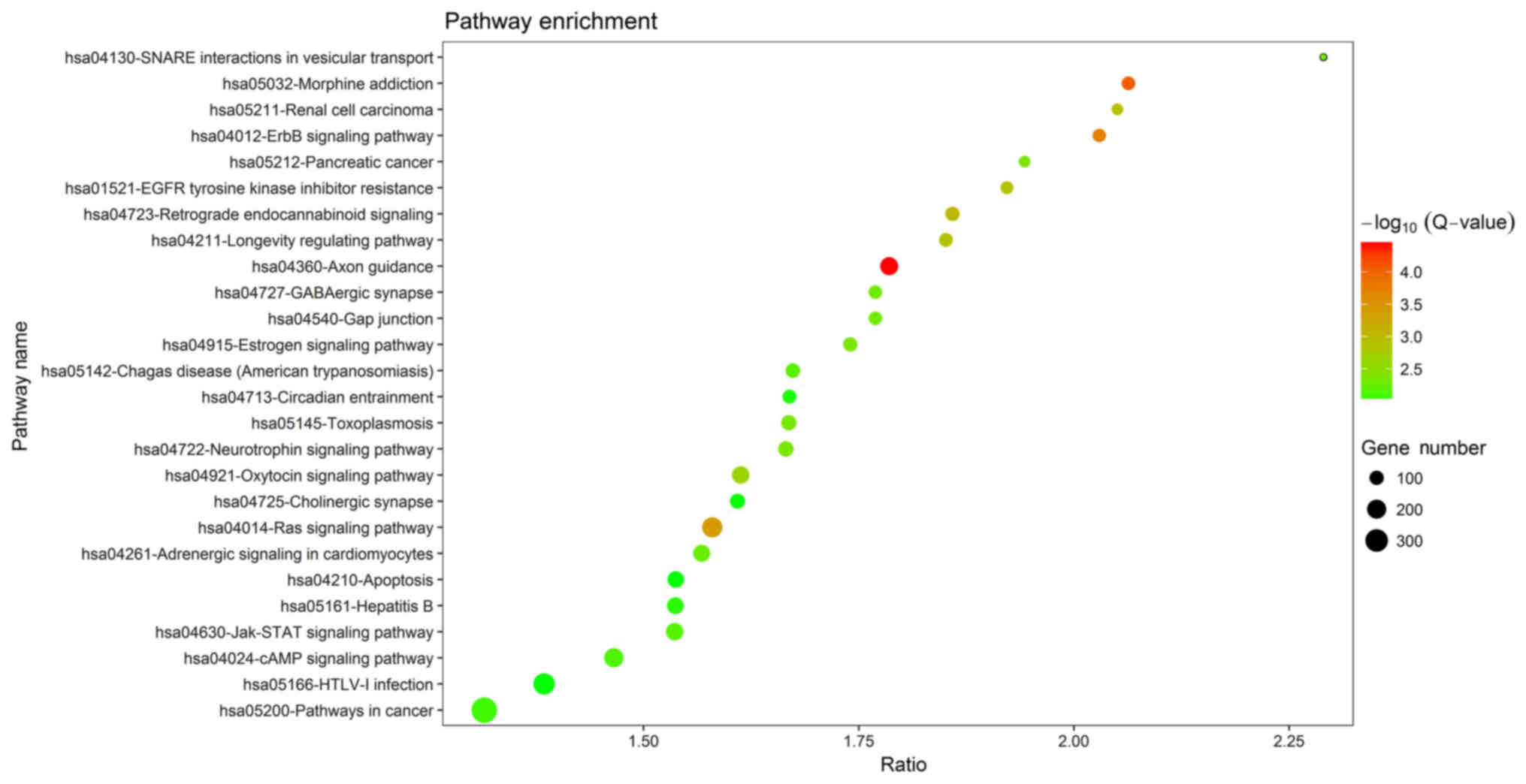
most significant terms in each group, respectively. The KEGG
pathway enrichment analysis identified a total of 26 pathways as
being enriched by the DEGs (Fig. 3),
several of which are associated with the ErbB, Ras, cyclic
adenosine monophosphate and Janus kinase (Jak)/signal transducer
and activator of transcription (STAT) signaling pathways. These
pathways my therefore be involved in the development of CRSwNP.
Validation of the miR-4492 expression
profile in CRS
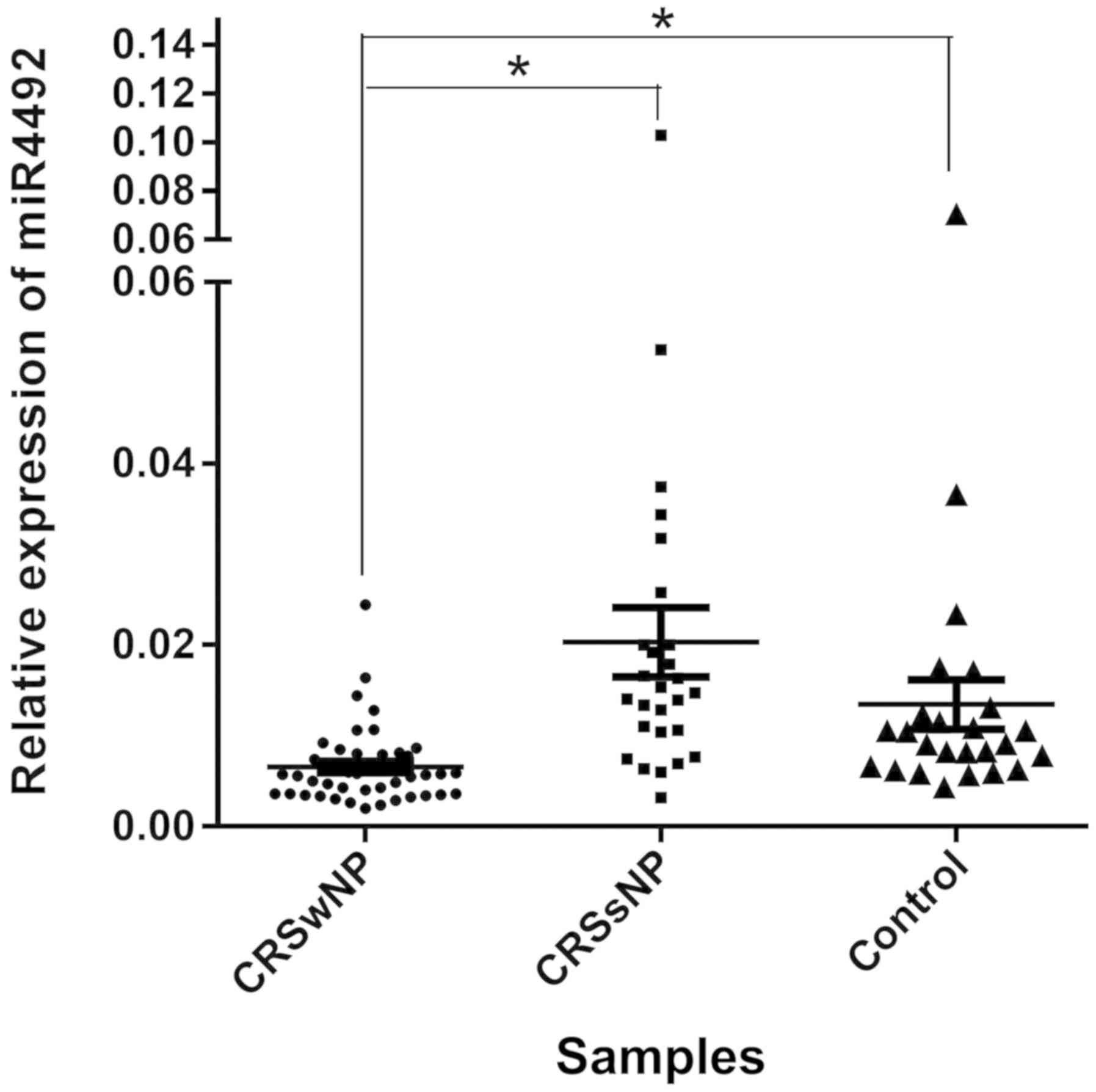
RNA samples from 122 patients were prepared for
RT-qPCR. A total of 26 samples were removed due to a lack of RNA
integrity (Fig. S1). The remaining
96 RNA samples all passed the quality control checks for RNA
integrity and purity. As presented in Fig. 4, the expression levels of miR-4492
were significantly reduced in the CRSwNP samples compared with
those in the CRSsNP samples (P=3.17×10−5). Similarly,
miR-4492 was also markedly downregulated in the CRSwNP group
compared with the control group (P=2.86×10−3). The
difference between the miR-4492 expression levels of the CRSsNP
group and the control group was not significant.
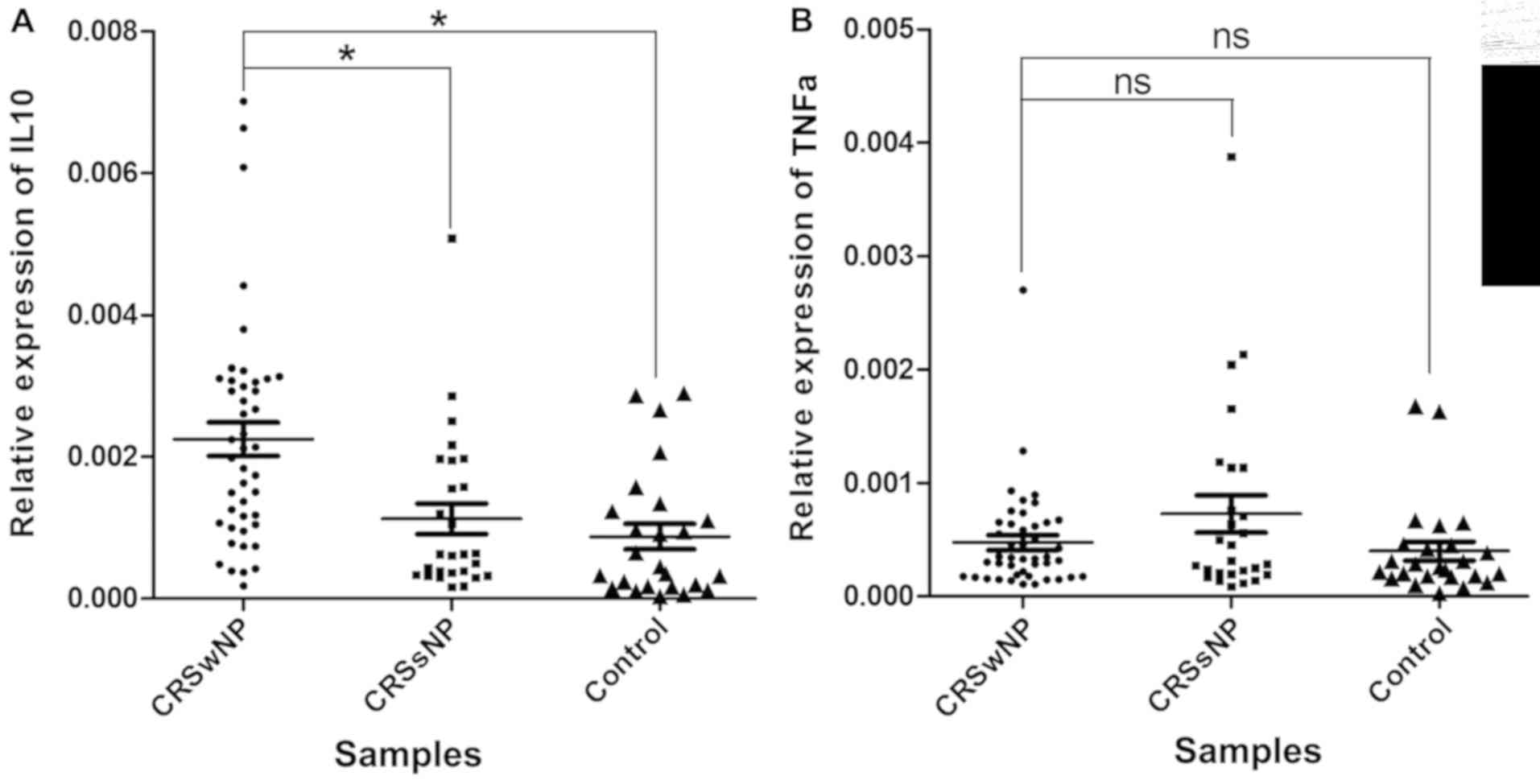
IL-10 and TNF-α expression in CRS
In order to investigate the targets of miR-4492, the
protein-coding target genes were predicted using a Bioinformatics
analysis. From the list of predicted targets of miR-4492, IL-10 and
TNF-α were selected and their expression in the 96 samples was
detected (Fig. 5). While the
difference between the CRSsNP samples and the control samples was
not significant, the expression levels of IL-10 were elevated in
the CRSwNP samples compared with those in the CRSsNP samples
(P=8.76×10−4) and the control samples
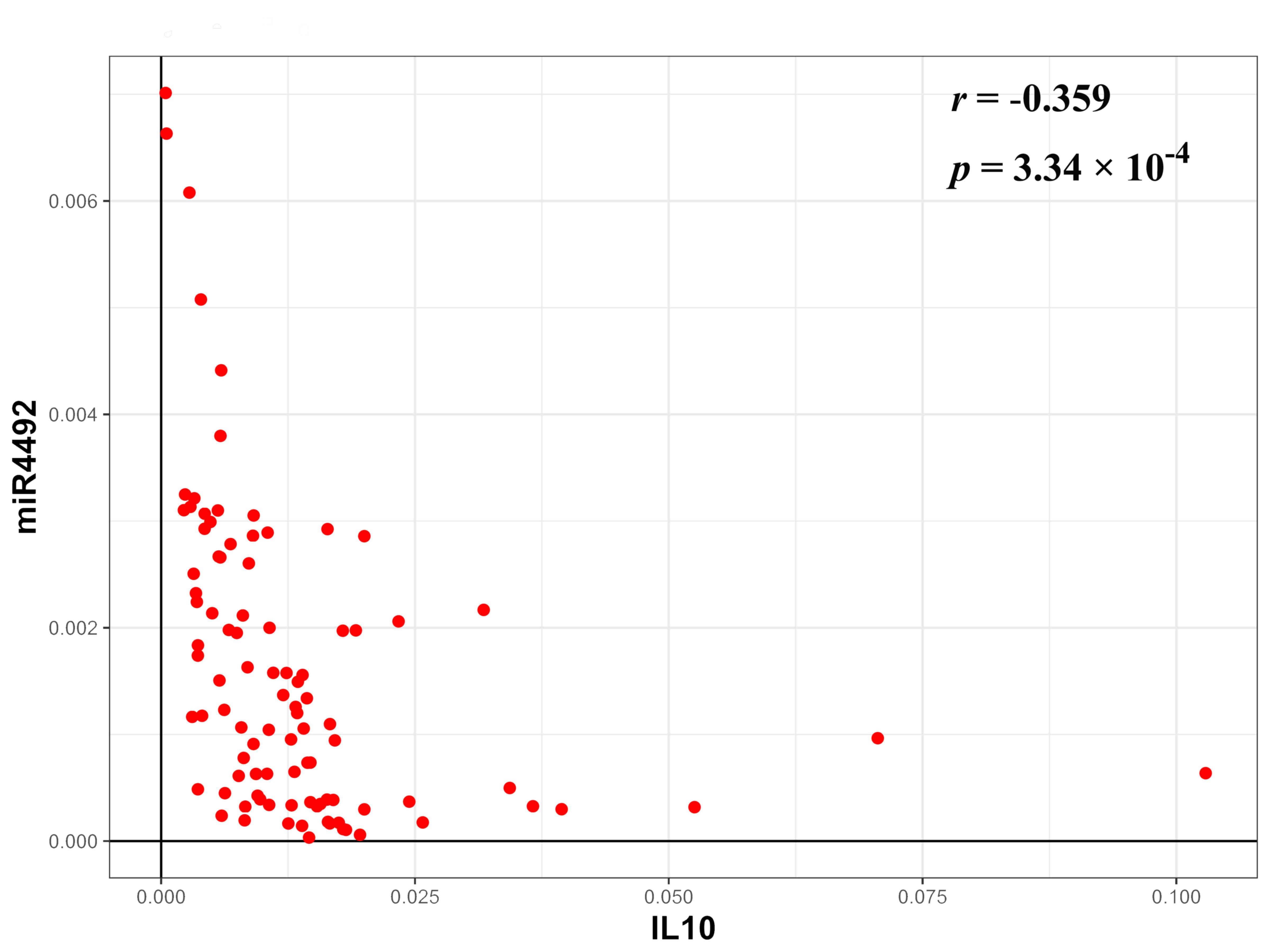
(P=1.74×10−4). In addition, a negative correlation
between the expression of miR-4492 and IL-10 expression was
determined among the 96 specimens (Fig.
6). However, no significant correlation of miR-4492 with TNF-α
expression was observed.
Discussion
CRS is an inflammatory upper airway disease with a
high prevalence worldwide (1).
Although CRSwNP is not the most common form of CRS, there is a
higher proportion of patients with CRSwNP in tertiary care
populations that have failed under medical management (25). A considerable number of studies have
confirmed that miRNAs are involved in a diverse range of biological
functions, including cell proliferation, differentiation,
apoptosis, stress responses and signal transduction, and abnormal
miRNA expression is considered to be a common feature of various
diseases (26,27). The key role of miRNAs in inflammation
is demonstrated by studies on the association of miRNA
dysregulation with excessive inflammation and aberrant immune
responses, including pulmonary fibrosis and asthma (17,18).
More recently, several studies have identified that marked changes
in miRNA expression, which is tightly regulated, serve essential
roles in the sustained and exaggerated inflammation of CRS
(26–30). In the present study, next-generation
sequencing was performed in order to detect key miRNAs in CRSwNP.
Compared to the CRSsNP group, six differentially expressed miRNAs,
namely Homo sapiens (hsa)-miR-200a-3p, hsa-miR-144-5p,
hsa-miR-144-3p, hsa-miR-486-5p, hsa-miR-4492 and hsa-miR-451a, were
identified in the CRSwNP samples. Downregulation of miR-200 family
members as part of the epithelial to mesenchymal transition has
been previously implicated in cancer development and progression
(31). Furthermore, previous studies
have confirmed the involvement of miR-200 family members in
inflammatory and immune responses (32). Downregulation of exosome miR-200 has
been detected in the bronchoalveolar lavage fluid of patients with
asthma (33). A recent study
indicated that miR-144 is an important regulator of innate immune
responses, and its major and minor product are involved in
colorectal polyposis, which involves inflammatory polyps at a
different anatomic site (34).
Panganiban et al (35)
detected the expression of 420 plasma miRNAs in healthy individuals
and patients with allergic rhinitis (AR) and asthma using RT-qPCR.
They revealed that miR-144 was commonly dysregulated in patients
with AR and asthma, suggesting that miR-144 is a promising plasma
biomarker in these diseases. Furthermore, a previous study
suggested that miR-541a, another miRNA that was identified as
aberrant in the present study, is also an asthma-associated miRNA
(36). It is widely accepted that
CRS and asthma are closely associated in numerous aspects and they
have similar pathogenic mechanisms and therapeutic principles.
Based on the above, it may be possible to speculate on a concordant
function of miR-200a, miR-144 and miR-451a in these two diseases.
miR-485 has also been demonstrated to be reduced in CRSwNP, as
detected using a microarray-based approach (29). The differential expression of
miR-4492 has been identified in a number of conditions, including
responses to viral infection (37,38),
necrosis (39), Hirschsprung's
disease (40) and breast cancer
(41). However, the exact functions
and mechanisms require further investigation.
The mechanisms underlying the inflammation are
different between the two types of CRS. CRSsNP involves more T
helper type 1 cell (Th1) polarization and collagen deposition,
whereas CRSwNP is characterized by more Th2 polarization and tissue
eosinophilia (42,43). It has been reported that miRNA is
involved in the regulation of innate immune cells that are
recruited to infiltrate the inflamed tissue, as well as the
polarization of Th2 cells (44).
Although relatively few studies have assessed the miRNAs involved
in CRS, a recent study indicated that miRNAs are associated with
the inflammatory pattern of this disease (45). Previous studies have revealed that
miR150-5p, miR-125b and miR-124 promote the development of CRS via
regulating the immune response to inflammation (27,30,46). Xia
et al (26) demonstrated that
7 miRNAs were differentially expressed in CRSwNP by using RT-qPCR,
and the expression was correlated with the occurrence of NPs.
Although these miRNAs were not among the DEGs identified in the
present study, the disparity may be attributed to a number of
reasons. First, different detecting platforms may partly account
for the difference, as the RNA-seq data do not have complete genome
coverage. In addition, in the present study, a relatively small
number of samples were used for RNA-seq. Furthermore, the
distinctive genetic backgrounds of the individuals and the tissue
heterogeneity characteristic of this disease may have also affected
the results. Therefore, a validation analysis of these DEGs was
necessary in the present study. Of note, miR-4492, was the only
differentially expressed miRNA identified between the CRSwNP
samples and the control samples, and was downregulated in the
polyps. Hence, the expression of miR-4492 was validated in the 96
specimens with acceptable RNA quality using RT-qPCR. The RT-qPCR
analysis confirmed the downregulation of this miR in NP. Therefore,
despite the lack of previous studies that support the involvement
of this miRNA in NPs, the present results suggest that miR-4492 may
have important roles in NPs according to the results of the present
study.
Deregulation of a number of cytokines, including
IL-1, IL-2, IL-10 and TNF-α, has been detected in a large number of
diseases, including cancer, autoimmune diseases and inflammatory
diseases. Increasing evidence suggests that cytokines are key
mediators of the inflammation and immune response associated with
NP, among which TNF-α and IL-10 are two predicted targets of
miR-4492. Therefore, these two molecules were selected as
candidates for validation in the present study. TNF-α is one of the
most important pro-inflammatory cytokines produced by macrophages
and T lymphocytes. Previous studies of Parkinson's disease
demonstrate the importance of upregulated TNF-α in immune-mediated
initiation of inflammation (47,48).
Although differential expression of TNF-α has been reported in CRS
with and without NP in previous studies, differences in the
expression of TNF-α were not detected in the present study using
RT-qPCR. A possible explanation for this discrepancy may be the
heterogeneity of individuals from different regions. IL-10 is a
pleiotropic cytokine with important immunoregulatory functions
(49). Dysregulation of IL-10 is
involved in the immunopathologic response to infection and the
development of numerous autoimmune diseases (50). In the present study, elevated levels
of IL-10 were detected in the CRSwNP samples, and IL-10 expression
was negatively correlated with the expression of miR-4492 in CRS.
Consistent with this, a previous study using clinical specimens
demonstrated that increased expression levels of IL-10 have a
pivotal role in the pathogenesis of CRSwNP (51). In a previous study by our group,
dysregulation of miR-4492, miR-200-3p, TNF-α and IL-10 was observed
in paraffin-embedded CRSwNP samples using RNA-seq and RT-qPCR
(52), whereas differences in the
expression of TNF-α were not detected in the present study.
Differences in sample type and sample size may at least partly
account for this discrepancy. In addition, Luo et al
(53) indicated that miR-19a has a
crucial role in nasal polyposis through the suppression of IL-10 in
peripheral dendritic cells. Furthermore, in KEGG pathway analysis,
IL-10 was enriched in the Jak/STAT signaling pathway. Combined with
the results of the present study, it is indicated that hsa-miR-4492
may serve roles in nasal polyposis via the hsa-miR-4492/IL-10
interaction in the Jak/STAT signaling pathway (52). However, the present study has a
number of limitations, including the relatively small number of
samples for miRNA sequencing and lack of a functional validation
assay. Therefore, further studies are required in order to
elucidate the interactional mechanism of this pathway in
CRSwNP.
In conclusion, the present study demonstrated that
miR-4492 is downregulated and IL-10 is upregulated in NPs, and
suggests that the involvement of the hsa-miR-4492/IL-10-axis in the
Jak/STAT signaling pathway may be a key mechanism in CRSwNP.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the
National Natural Science Foundation of China (grant no. 81460094)
and the Natural Science Foundation of Xinjiang Province (grant no.
2015211C076).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LL and JYo conceived and designed the study. LL, JF,
DZ, YW, JYa and RH performed the data analysis. LL wrote the
manuscript. LL and JYo edited the manuscript. All authors reviewed
the manuscript and approved the final version.
Ethics approval and informed consent
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Xinjiang Medical
University (Urumqi, China; permit no. 20160114-05) and it was
performed with written informed consent from the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeConde AS and Soler ZM: Chronic
rhinosinusitis: Epidemiology and burden of disease. Am J Rhinol
Allergy. 30:134–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hoffmans R, Schermer T, van der Linde K,
Bor H, van Boven K, van Weel C and Fokkens W: Rhinosinusitis in
morbidity registrations in dutch general practice: A retro-spective
case-control study. BMC Fam Pract. 16:1202015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meltzer EO, Hamilos DL, Hadley JA, Lanza
DC, Marple BF, Nicklas RA, Bachert C, Baraniuk J, Baroody FM,
Benninger MS, et al: Rhinosinusitis: Establishing definitions for
clinical research and patient care. J Allergy Clin Immunol. 114((6
Suppl)): S155–S212. 2004. View Article : Google Scholar
|
|
4
|
Benninger MS, Ferguson BJ, Hadley JA,
Hamilos DL, Jacobs M, Kennedy DW, Lanza DC, Marple BF, Osguthorpe
JD, Stankiewicz JA, et al: Adult chronic rhinosinusitis:
Definitions, diagnosis, epidemiology, and pathophysiology.
Otolaryngol Head Neck Surg. 129((3 Suppl)): S1–S32. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Banerji A, Piccirillo JF, Thawley SE,
Levitt RG, Schechtman KB, Kramper MA and Hamilos DL: Chronic
rhinosinusitis patients with polyps or polypoid mucosa have a
greater burden of illness. Am J Rhinol. 21:19–26. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koskinen A, Numminen J, Markkola A,
Karjalainen J, Karstila T, Seppälä M, Julkunen A, Lemmetyinen R,
Pekkanen J, Rautiainen M, et al: Diagnostic accuracy of symptoms,
endoscopy, and imaging signs of chronic rhinosinusitis without
nasal polyps compared to allergic rhinitis. Am J Rhinol Allergy.
32:121–131. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dietz de Loos DA, Hopkins C and Fokkens
WJ: Symptoms in chronic rhinosinusitis with and without nasal
polyps. Laryngoscope. 123:57–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Avdeeva K and Fokkens W: Precision
medicine in chronic rhinosinusitis with nasal polyps. Curr Allergy
Asthma Rep. 18:252018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fokkens WJ, Lund VJ, Mullol J, Bachert C,
Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, et
al: European position paper on rhinosinusitis and nasal polyps
2012. Rhinol. Suppl 23:3 p preceding table of contents. 1–298.
2012. View Article : Google Scholar
|
|
10
|
Fokkens WJ, Lund VJ, Mullol J, Bachert C,
Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, et
al: EPOS 2012: European position paper on rhinosinusitis and nasal
polyps 2012. A summary for otorhinolaryngologists. Rhinology.
50:1–12. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vishnoi A and Rani S: MiRNA biogenesis and
regulation of diseases: An overview. Methods Mol Biol. 1509:1–10.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ni WJ and Leng XM: miRNA-dependent
activation of mRNA translation. Microrna. 5:83–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Beilharz TH, Humphreys DT and Preiss T:
miRNA effects on mRNA closed-loop formation during translation
initiation. Prog Mol Subcell Biol. 50:99–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fleshner M and Crane CR: Exosomes, DAMPs
and miRNA: Features of stress physiology and immune homeostasis.
Trends Immunol. 38:768–776. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Foley NH and O'Neill LA: miR-107: A
toll-like receptor-regulated miRNA dysregulated in obesity and type
II diabetes. J Leukoc Biol. 92:521–527. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou M, Li W, Xie Z, Ai J, Sun B and Tan G:
Effects of anticholinergic agent on miRNA profiles and
transcriptomes in a murine model of allergic rhinitis. Mol Med Rep.
16:6558–6569. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Solberg OD, Ostrin EJ, Love MI, Peng JC,
Bhakta NR, Hou L, Nguyen C, Solon M, Nguyen C, Barczak AJ, et al:
Airway epithelial miRNA expression is altered in asthma. Am J
Respir Crit Care Med. 186:965–974. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu F, Qin HB, Xu B, Zhou H and Zhao DY:
Profiling of miRNAs in pediatric asthma: Upregulation of miRNA-221
and miRNA-485-3p. Mol Med Rep. 6:1178–1182. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kalvari I, Argasinska J, Quinones-Olvera
N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD and Petrov AI:
Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA
families. Nucleic Acids Res. 46:D335–D342. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dweep H, Gretz N and Sticht C: miRWalk
database for miRNA-target interactions. Methods Mol Biol.
1182:289–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
The Gene Ontology Consortium, . Expansion
of the gene ontology knowledgebase and resources. Nucleic Acids
Res. 45:D331–D338. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lal D, Scianna JM and Stankiewicz JA:
Efficacy of targeted medical therapy in chronic rhinosinusitis, and
predictors of failure. Am J Rhinol Allergy. 23:396–400. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia G, Bao L, Gao W, Liu S, Ji K and Li J:
Differentially expressed miRNA in inflammatory mucosa of chronic
rhinosinusitis. J Nanosci Nanotechnol. 15:2132–2139. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma Z, Shen Y, Zeng Q, Liu J, Yang L, Fu R
and Hu G: MiR-150-5p regulates EGR2 to promote the development of
chronic rhinosinusitis via the DC-Th axis. Int Immunopharmacol.
54:188–197. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li FM and Zheng JF: Application and
progress of miRNA in chronic rhinosinusitis. Lin Chuang Er Bi Yan
Hou Tou Jing Wai Ke Za Zhi. 30:1656–1658. 2016.(In Chinese).
PubMed/NCBI
|
|
29
|
Ma ZX, Tan X, Shen Y, Ke X, Yang YC, He
XB, Wang ZH, Dai YB, Hong SL and Hu GH: MicroRNA expression profile
of mature dendritic cell in chronic rhinosinusitis. Inflamm Res.
64:885–893. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang XH, Zhang YN, Li HB, Hu CY, Wang N,
Cao PP, Liao B, Lu X, Cui YH and Liu Z: Overexpression of miR-125b,
a novel regulator of innate immunity, in eosinophilic chronic
rhinosinusitis with nasal polyps. Am J Respir Crit Care Med.
185:140–151. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Izumchenko E, Chang X, Michailidi C,
Kagohara L, Ravi R, Paz K, Brait M, Hoque MO, Ling S, Bedi A and
Sidransky D: The TGFβ-miR200-MIG6 pathway orchestrates the
EMT-associated kinase switch that induces resistance to EGFR
inhibitors. Cancer Res. 74:3995–4005. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zidar N, Boštjančič E, Jerala M, Kojc N,
Drobne D, Štabuc B and Glavač D: Down-regulation of microRNAs of
the miR-200 family and up-regulation of Snail and Slug in
inflammatory bowel diseases-hallmark of epithelial-mesenchymal
transition. J Cell Mol Med. 20:1813–1820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Levänen B, Bhakta NR, Torregrosa Paredes
P, Barbeau R, Hiltbrunner S, Pollack JL, Sköld CM, Svartengren M,
Grunewald J, Gabrielsson S, et al: Altered microRNA profiles in
bronchoalveolar lavage fluid exosomes in asthmatic patients. J
Allergy Clin Immunol. 131:894–903. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu CW, Cao X, Berger CK, Foote PH, Mahoney
DW, Simonson JA, Anderson BW, Yab TC, Taylor WR, Boardman LA, et
al: Novel approach to fecal occult blood testing by assay of
erythrocyte-specific microRNA markers. Dig Dis Sci. 62:1985–1994.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Panganiban RP, Wang Y, Howrylak J,
Chinchilli VM, Craig TJ, August A and Ishmael FT: Circulating
microRNAs as biomarkers in patients with allergic rhinitis and
asthma. J Allergy Clin Immunol. 137:1423–1432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Z, Zhang XH, Callejas-Diaz B and
Mullol J: MicroRNA in united airway diseases. Int J Mol Sci.
17:E7162016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xun M, Ma CF, Du QL, Ji YH and Xu JR:
Differential expression of miRNAs in enterovirus 71-infected cells.
Virol J. 12:562015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Egaña-Gorroño L, Guardo AC, Bargalló ME,
Planet E, Vilaplana E, Escribà T, Pérez I, Gatell JM, García F,
Arnedo M, et al: MicroRNA profile in CD8+ T-Lymphocytes from
HIV-infected individuals: Relationship with antiviral immune
response and disease progression. PLoS One. 11:e01552452016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ahn SH, Ahn JH, Ryu DR, Lee J, Cho MS and
Choi YH: Effect of Necrosis on the miRNA-mRNA regulatory network in
CRT-MG human astroglioma cells. Cancer Res Treat. 50:382–397. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gao ZG, Chen QJ, Shao M, Qian YZ, Zhang
LF, Zhang YB and Xiong QX: Preliminary identification of key
miRNAs, signaling pathways, and genes associated with
hirschsprung's disease by analysis of tissue microRNA expression
profiles. World J Pediatr. 13:489–495. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Boo L, Ho WY, Ali NM, Yeap SK, Ky H, Chan
KG, Yin WF, Satharasinghe DA, Liew WC, Tan SW, et al: MiRNA
transcriptome profiling of spheroid-enriched cells with cancer stem
cell properties in human breast MCF-7 cell line. Int J Biol Sci.
12:427–445. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wei CH, Phan L, Feltz J, Maiti R, Hefferon
T and Lu Z: tmVar 2.0: Integrating genomic variant information from
literature with dbSNP and clinvar for precision medicine.
Bioinformatics. 34:80–87. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheng KJ, Xu YY, Zhou ML, Zhou SH and Wang
SQ: Role of local allergic inflammation and staphylococcus aureus
enterotoxins in Chinese patients with chronic rhinosinusitis with
nasal polyps. J Laryngol otol. 131:707–713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mattes J, Collison A, Plank M, Phipps S
and Foster PS: Antagonism of microRNA-126 suppresses the effector
function of TH2 cells and the development of allergic airways
disease. Proc Natl Acad Sci USA. 106:18704–18709. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang XH, Zhang YN and Liu Z: MicroRNA in
chronic rhinosinusitis and allergic rhinitis. Curr Allergy Asthma
Rep. 14:4152014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu CC, Xia M, Zhang YJ, Jin P, Zhao L,
Zhang J, Li T, Zhou XM, Tu YY, Kong F, et al: Micro124-mediated AHR
expression regulates the inflammatory response of chronic
rhinosinusitis (CRS) with nasal polyps. Biochem Biophys Res Commun.
500:141–151. 2018. View Article : Google Scholar
|
|
47
|
Nishimura M, Mizuta I, Mizuta E, Yamasaki
S, Ohta M, Kaji R and Kuno S: Tumor necrosis factor gene
polymorphisms in patients with sporadic Parkinson's disease.
Neurosci Lett. 311:1–4. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lindenau JD, Altmann V, Schumacher-Schuh
AF, Rieder CR and Hutz MH: Tumor necrosis factor alpha
polymorphisms are associated with Parkinson's disease age at onset.
Neurosci Lett. 658:133–136. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mocellin S, Marincola F, Rossi CR, Nitti D
and Lise M: The multifaceted relationship between IL-10 and
adaptive immunity: Putting together the pieces of a puzzle.
Cytokine Growth Factor Rev. 15:61–76. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Iyer SS and Cheng G: Role of interleukin
10 transcriptional regulation in inflammation and autoimmune
disease. Crit Rev Immunol. 32:23–63. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu J, Han R, Kim DW, Mo JH, Jin Y, Rha KS
and Kim YM: Role of interleukin-10 on nasal polypogenesis in
patients with chronic rhinosinusitis with nasal polyps. PLoS One.
11:e01610132016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li LG, Ma T and YR C: MiR-200A-3P and
miR4492 involved in regulation of chronic rhinosinusitis with nasal
polyps. Int J Genet. 41:108–114. 2018.
|
|
53
|
Luo XQ, Shao JB, Xie RD, Zeng L, Li XX,
Qiu SQ, Geng XR, Yang LT, Li LJ, Liu DB, et al: Micro RNA-19a
interferes with IL-10 expression in peripheral dendritic cells of
patients with nasal polyposis. Oncotarget. 8:48915–48921.
2017.PubMed/NCBI
|