Introduction
Ovarian cancer (OC) is one of the most fatal types
of gynecological malignancy. According to a recent survey, OC
caused ~14,240 deaths in 2017 worldwide (1). However, the mechanisms underlying OC
progression have remained largely elusive. Long non-coding RNAs
(lncRNA) are a novel class of RNA transcripts with a length of
>200 nucleotides that lack any protein-coding capacity. Emerging
studies have demonstrated that certain lncRNAs are dysregulated in
tumor cells and have crucial roles in the progression of cancer,
including prostate cancer, breast cancer, glioma and OC (2). Mechanistically, lncRNAs bind to RNAs,
DNA and proteins to regulate multiple biological functions,
including cell proliferation, apoptosis, epithelial-mesenchymal
transition and cell migration. Several lncRNAs have been reported
to be associated with OC progression. For instance, LINK-A was
reported to promote OC cell migration and invasion by activating
the transforming growth factor-β pathway (3). Furthermore, Shu et al (4) indicated that lncRNA regulator of Akt
signaling associated with hepatocellular carcinoma (HCC) and renal
cell carcinoma promoted OC cell proliferation and invasion by
association with HuR and the microRNA (miRNA/miR)-200 family.
Therefore, exploring the expression pattern of lncRNAs in OC is
useful for the identification of novel biomarkers for OC.
LncRNA zinc finger nuclear transcription factor,
X-box binding 1-type containing 1 antisense RNA 1 (ZFAS1) has been
reported to be upregulated in several types of cancer, including
HCC, colorectal cancer, gastric cancer and glioma. In gastric
cancer, ZFAS1 was demonstrated to be involved in regulating cancer
progression through influencing Wnt/β-catenin signaling and
epigenetically repressing Kruppel-like factor 2 (KLF2) and NKD
inhibitor of WNT signaling pathway 2 (NKD2) (5). In colorectal cancer, ZFAS1 promotes
proliferation and metastasis via regulating zinc finger E-box
binding homeobox 1 (ZEB1) expression and interacting with cyclin D
kinase 1 (6). ZFAS1 was reported to
promote OC cell malignancy by interacting with miR-150-5p (7). These reports suggested ZFAS1 may serve
as a regulator in cancer progression. However, the expression
pattern and functional roles of ZFAS1 in OC progression have
remained to be elucidated.
In the present study, the expression levels of ZFAS1
were compared between OC and normal samples and the association of
ZFAS1 expression with clinicopathological features in OC was
determined by analyzing publicly available datasets. The potential
biological functions of ZFAS1 were analyzed by performing a
Bioinformatics analysis. The present study may provide clues that
validate ZFAS1 as a novel biomarker for OC.
Materials and methods
Dataset analysis
The raw data were obtained from The Cancer Genome
Atlas (TCGA) data from Genomic Data Commons Data Portal (portal.gdc.cancer.gov) and the University of
California Santa Cruz (UCSC) Xena project (http://xena.ucsc.edu). The data were analyzed using
Gene Expression Profiling Interactive Analysis (GEPIA) (8), a newly developed interactive web server
for analyzing RNA sequencing (RNA-seq) expression data. RNA-seq
datasets used by GEPIA are based on the UCSC Xena project, using a
standard processing pipeline. P<0.05 was selected as the cutoff
for identifying significantly differentially expressed genes.
Protein-protein interaction (PPI)
network construction and analysis
In the present study, genes co-expressed with ZFAS1
were determined. The Starbase (9)
and TargetScan (10,11) databases were used to predict the
miRNAs targeting ZFAS1-mRNA pairs. The Pearson correlation
coefficient was used as the threshold for selecting co-expressed
ZFAS1-gene pairs. An absolute value of the Pearson correlation
coefficient of ≥0.3 was used as the threshold. The Search Tool for
the Retrieval of Interacting Genes/Proteins version 10.0 (STRING;
string-db.org) (12,13) was
used for the exploration of potential ZFAS1 protein interactions.
The PPI networks of DEGs generated by STRING were derived from
validated experiments. A combined score of >0.9 was considered
significant. P<0.05 was considered to indicate a statistically
significant difference. The PPI networks were visualized using
Cytoscape (14).
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis
MAS 3.0 (mas.capitalbiotech.com/mas3) may be used to analyze
inter-gene correlations and for gene function annotation. In the
present study, the DAVID tool (https://david.ncifcrf.gov/tools.jsp) was used to
perform GO and KEGG pathway analysis using the genes co-expressed
with ZFAS1. P<0.05 was selected as the threshold of
significance.
Cell culture
The OVCA429, SKOV3, A2780 and COV644 OC cell lines
and normal human ovarian surface epithelial (HOSE) cells were
purchased from the Cell Bank of the Chinese Academy of Sciences.
All cell lines were verified by using short tandem repeat analysis.
The OVCA429, SKOV3, A2780 and COV644 cell lines were cultured in
Dulbecco's modified Eagle's medium (Gibco-BRL; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and antibiotics (Gibco-BRL; Thermo
Fisher Scientific, Inc.). HOSE cells were cultured in medium
composed of a 1:1 mixture of MCDB 105 (Sigma-Aldrich, Merck KGaA)
and M199 (Gibco; Thermo Fisher Scientific, Inc.) medium containing
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.).
Cells were cultured in a humidified atmosphere containing 5%
CO2 at 37°C.
Transfection
Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to perform transfection according to the
manufacturer's instructions. The SKOV3 cells were transfected with
siZFAS1 and siNC at a final concentration of 50 nM. After 48 h, the
cells were used to perform cell counting kit-8 (CCK-8) and cell
cycle assays. The following siRNA sequences were used: siZFAS1,
5′-AGCGGTTTGGTGCGGTGTGAAGCGCGACAT-3′ and siNC,
5′-UUCUCCGAACGUGUCACGUdTdT-3′.
CCK-8 viability assay
The transfected cells (3×103 cells/well)
were seeded in 96-well culture plates and incubated with 10 µl
CCK-8 reagent (Beyotime Institute of Biotechnology) per well at
37°C for 2 h. Viability ability was detected at the selected time
points (0, 1, 2 and 3 days following seeding). The optical density
was determined at a wavelength of 450 nm.
Reverse transcription-quantitative
(RT-q)PCR analysis
For RT-qPCR, an Ultrapure RNA kit (CWBIO) was used
to extract RNA. A PrimeScript RT reagent kit (Takara) was used to
perform RT. Reaction parameters were as follows: 42°C for 15 min,
85°C for 5 sec and 4°C for 5 min. AceQ Universal SYBR qPCR Master
Mix (Vazyme) was used to perform Real-time PCR on an ABI 7500
system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Reaction parameters were as follows: Initial denaturation at 95°C
for 5 min; 40 cycles of 95°C for 10 sec and 60°C for 30 sec. The
sequences of the primers were as follows: ZFAS1 forward,
5′-ACGTGCAGACATCTACAACCT-3′ and reverse, 5′-TACTTCCAACACCCGCAT-3′;
β-actin forward, 5′-GAGCTACGAGCTGCCTGACG-3′ and reverse,
5′-CCTAGAAGCATTTGCGGTGG-3′. β-actin was used as a reference. The
2−ΔΔCq method was used to quantify the data (15).
Statistical analysis
Statistical analysis was performed using SPSS 19.0
software (IBM Corp.). Student's t-test or Mann-Whitney U-test was
used to perform statistical comparisons between two groups
depending on the test condition. One-way analysis of variance was
used to perform statistical comparisons among multiple groups
followed by Dunnett's post-hoc test. P<0.05 was considered to
indicate statistical significance.
Results
LncRNA ZFAS1 is upregulated in
advanced- vs. early-stage OC
In order to assess the roles of ZFAS1 in OC, the
expression levels of ZFAS1 in OC and normal samples were first
analyzed using the GEPIA dataset. Of note, the analysis revealed
that ZFAS1 was significantly downregulated in OC compared with that
in normal samples (Fig. 1A). This
result was not consistent with that of a previous study (7).
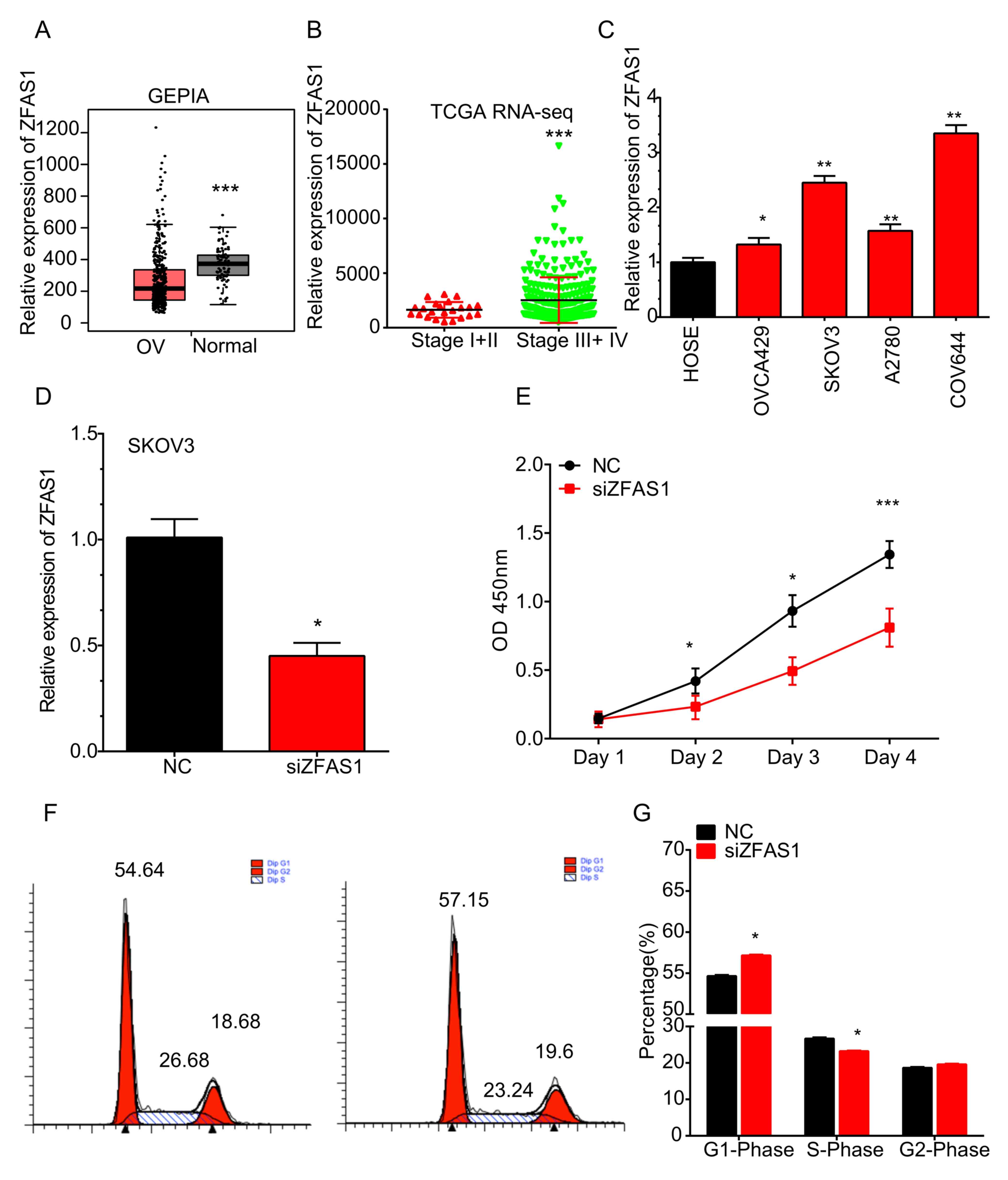 | Figure 1.ZFAS1 is upregulated in OC vs. normal
ovarian cells and in advanced- vs. early-stage OC tissues. (A) In
the GEPIA dataset, ZFAS1 was downregulated in OC compared to normal
samples. ***P<0.05 vs. OC. (B) In TCGA RNA-seq dataset, ZFAS1
was overexpressed at higher-stage (stage III/IV) OC compared with
that in the early-stage OC (stage I/II) samples. ***P<0.001 vs.
stage I+II. (C) ZFAS1 was significantly upregulated in the OVCA429,
SKOV3, A2780 and COV644 OC cell lines compared with that in normal
HOSE cells. *P<0.05 and **P<0.01 vs. HOSE. (D) Silencing
efficiency. (E) Knockdown of ZFAS1 markedly inhibited SKOV3 cell
proliferation. (F) Knockdown of ZFAS1 in SKOV3 cells significantly
increased the percentage of cells in G1 phase, but decreased the
percentage of cells in S phase. (G) Cell cycle data were expressed
as the mean ± standard deviation. *P<0.05 and ***P<0.001 vs.
NC. OC, ovarian cancer; ZFAS1, zinc finger nuclear transcription
factor, X-box binding 1-type containing 1 antisense RNA 1; HOSE,
human ovarian surface epithelium; TCGA, The Cancer Genome Atlas;
NC, negative control; siZFAS1, small interfering RNA targeting
ZFAS1; OD, optical density; RNA-seq, RNA sequencing; GEPIA, Gene
Expression Profiling Interactive Analysis. |
It was then assessed whether ZFAS1 expression is
associated with the stage of OC by analyzing TCGA RNA-seq dataset.
As presented in Fig. 1B, ZFAS1 was
overexpressed at higher stages (stage III/IV) of OC compared with
that in early-stage (stage I/II) OC samples.
The expression of ZFAS1 in OC cell lines was
detected by RT-qPCR. It was revealed that the expression levels of
ZFAS1 were significantly higher in the OVCA429, SKOV3, A2780 and
COV644 cell lines than those in normal HOSE cells (Fig. 1C).
Knockdown of ZFAS1 suppresses SKOV3
cell viability
The effect of ZFAS1 on OC cell viability was
detected using a Cell Counting Kit (CCK)-8 assay following ZFAS1
knockdown. The silencing efficiency was confirmed using RT-qPCR
analysis (Fig. 1D). As presented in
Fig. 1E, knockdown of ZFAS1 markedly
inhibited SKOV3 cell viability at 72 h post-transfection. Cell
cycle analysis demonstrated that knockdown of ZFAS1 in SKOV3 cells
significantly increased the percentage of cells in G1 phase, but
decreased the percentage of cells in S phase (Fig. 1F and G). These results suggested that
knockdown of ZFAS1 suppressed SKOV3 cell viability by inducing cell
cycle arrest.
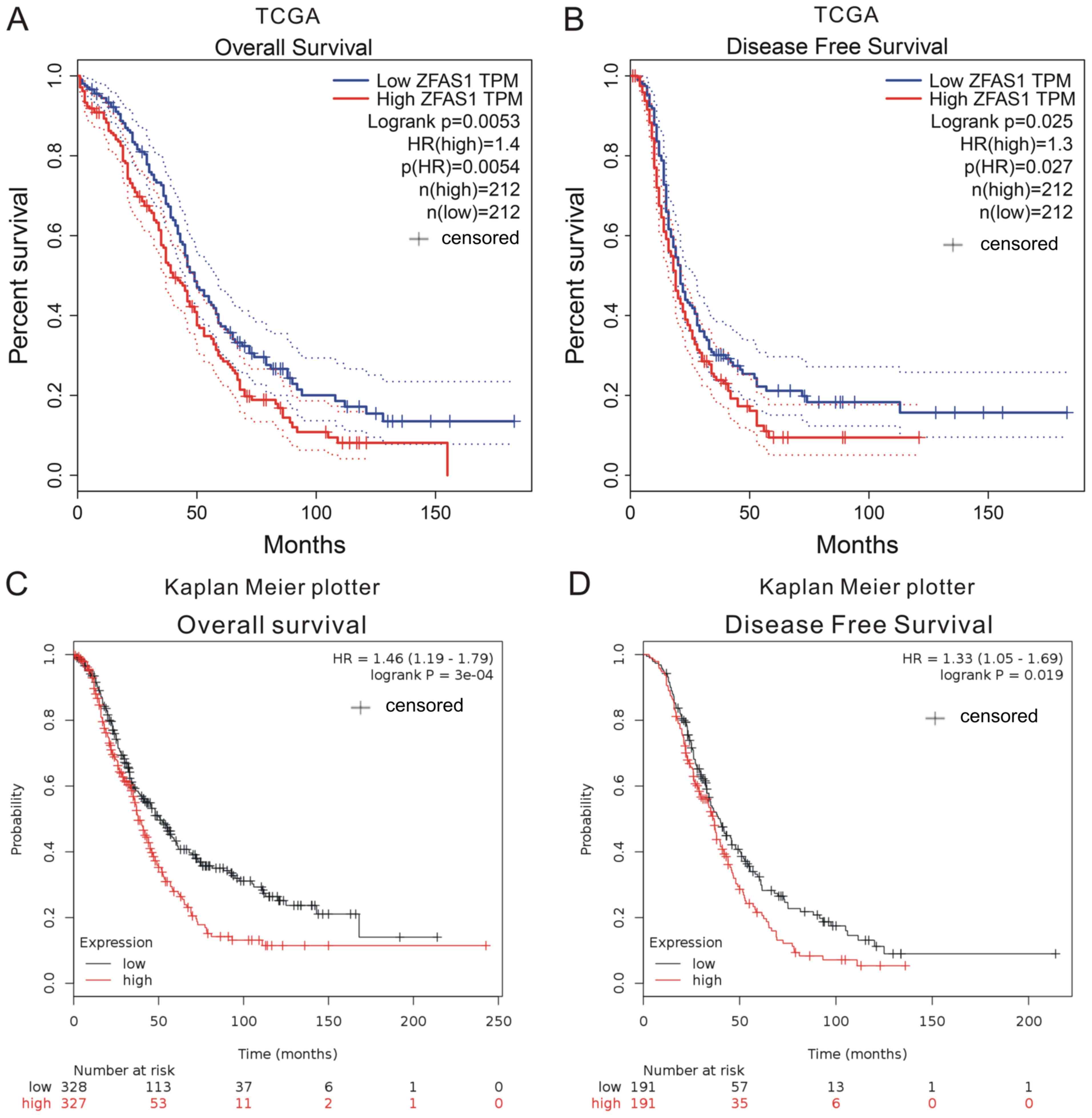
Higher ZFAS1 expression in OC tissues
is associated with poor prognosis
The association between ZFAS1 expression and
prognosis was investigated using TCGA and Kaplan-Meier plotter
datasets. The expression of ZFAS1 in OC tissues was categorized as
high or low according to the median value. By analyzing TCGA
datasets, it was revealed that higher ZFAS1 expression levels were
associated with shorter overall survival time (OS; Fig. 2A) and disease-free survival time
(DFS; Fig. 2B).
Next, the Kaplan-Meier plotter datasets, which
included 1,287 OC samples with a mean OS of 31 months were
analyzed. In this dataset, OC patients with higher ZFAS1 expression
had a shorter OS and DFS when compared with patients with low
expression (Fig. 2C and D). These
results suggested that ZFAS1 may serve as a novel prognostic marker
in OC patients.
Construction of ZFAS1-associated PPI
networks
In the present study, a PPI network of ZFAS1 was
first generated by calculating the Pearson correlation coefficient
between ZFAS1 and mRNAs in TCGA OC datasets. ZFAS1-mRNA pairs with
an absolute value of the Pearson correlation coefficient of ≥0.3
were selected.
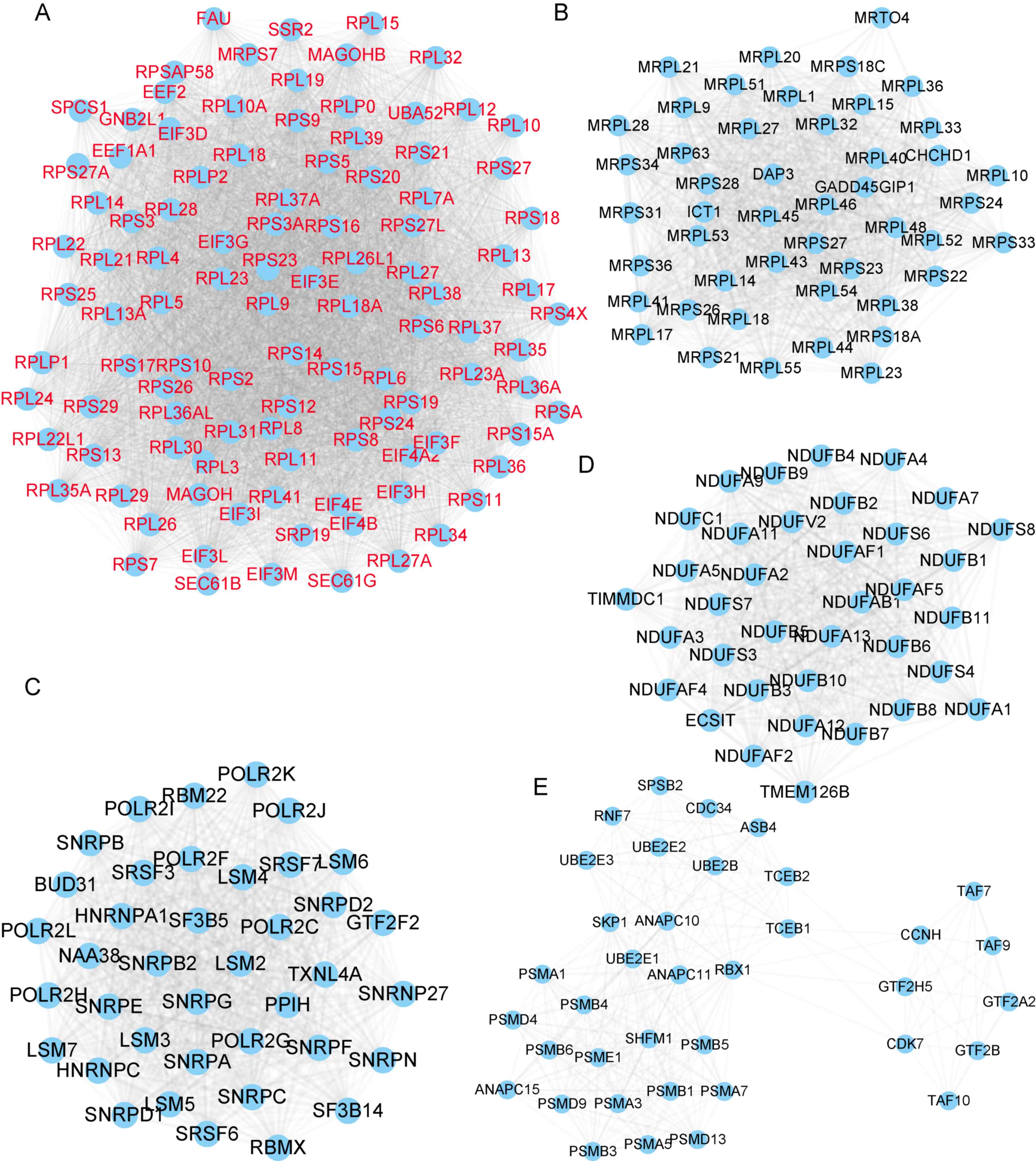
Next, ZFAS1-mediated PPI networks in OC were
constructed by using the STRING database (combined score >0.9).
A total of 628 proteins and 12,158 edges were included in the PPI
network of genes positively co-expressed with ZFAS1. Of note,
several hub genes were identified in this network, as presented in
Fig. 3. Module 1 contained 104
proteins and 5,096 edges (Fig. 3A),
Module 2 contained 47 proteins and 1,018 edges (Fig. 3B), Module 3 contained 39 proteins and
732 edges (Fig. 3C), Module 4
contained 36 proteins and 602 edges (Fig. 3D) and Module 5 contained 37 proteins
and 298 edges (Fig. 3E). A total of
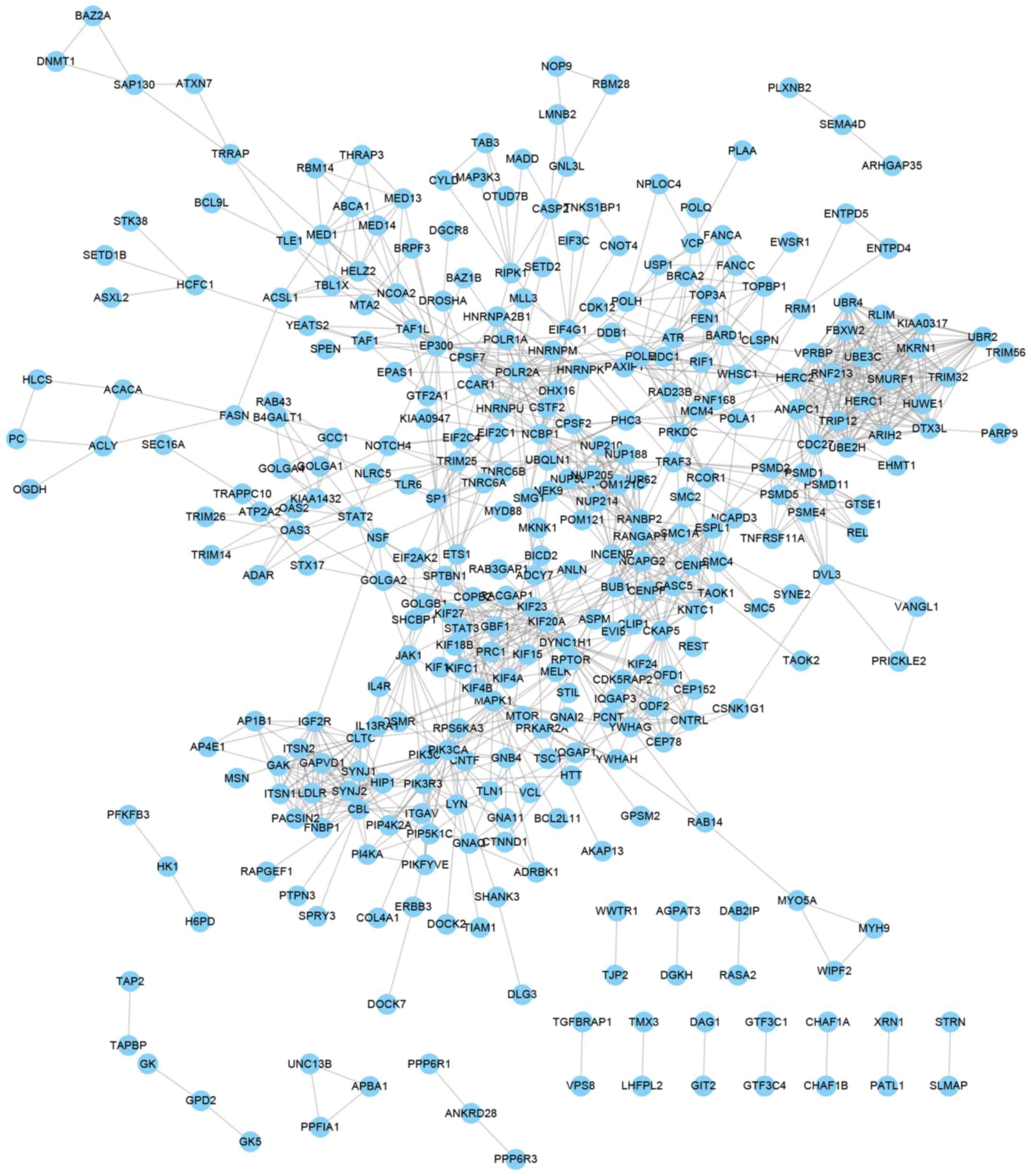
277 proteins and 1,216 edges were included in the PPI network of
genes negatively co-expressed with ZFAS1 (Fig. 4).
Construction of ZFAS1-mediated
competing endogenous (ce)RNA network in OC
Previous studies have indicated that ZFAS1 acts as a
ceRNA in human cancers. In order to explore the potential mechanism
by which ZFAS1 regulates OC progression, a ZFAS1-mediated ceRNA
network was constructed. The Starbase and TargetScan databases were
used to predict the miRNAs targeting ZFAS1-mRNA pairs.
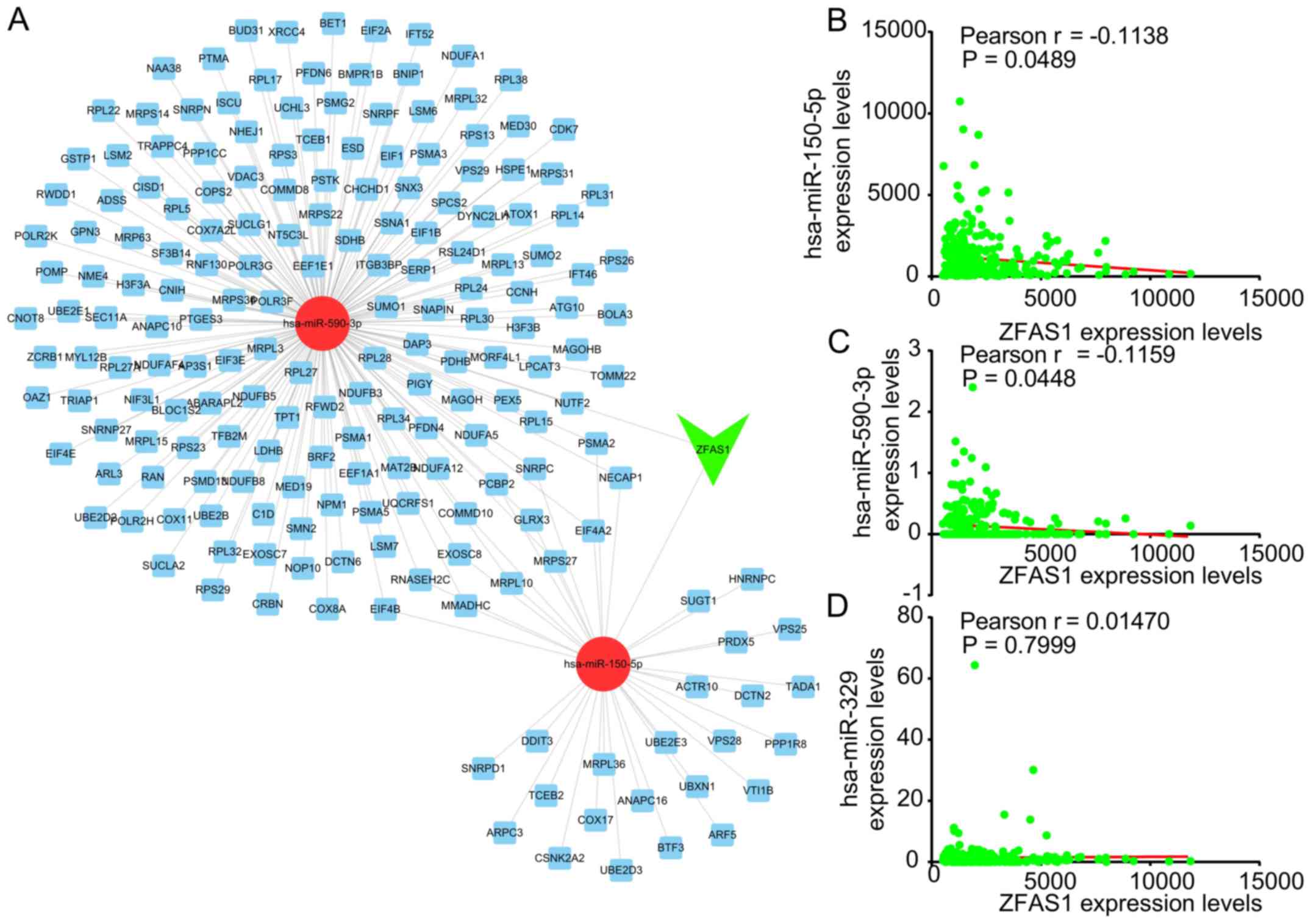
As presented in Fig.
5A, this ceRNA network included 2 miRNAs [Homo sapiens
(hsa)-miR-150-5p and hsa-miR-590-3p] and 195 mRNAs. hsa-miR-150-5p
has been previously reported to be a target of ZFAS1 in OC
(7). The correlation between ZFAS1
and the candidate miRNAs was then analyzed. It was revealed that
ZFAS1 was negatively correlated to the expression of hsa-miR-150-5p
and hsa-miR-590-3p in OC (Fig. 5B and
C). However, the expression of ZFAS1 was not significantly
correlated to miR-329 expression in OC (Fig. 5D), which was previously reported to
be sponged by ZFAS1 in bladder cancer (16).
Enrichment analysis of ZFAS1 in
OC
Enrichment analysis of genes co-expressed with ZFAS1
was then performed by using the DAVID tool. GO analysis indicated
that genes positively co-expressed with ZFAS1 are associated with
translation, translational initiation, ribosomal RNA processing,
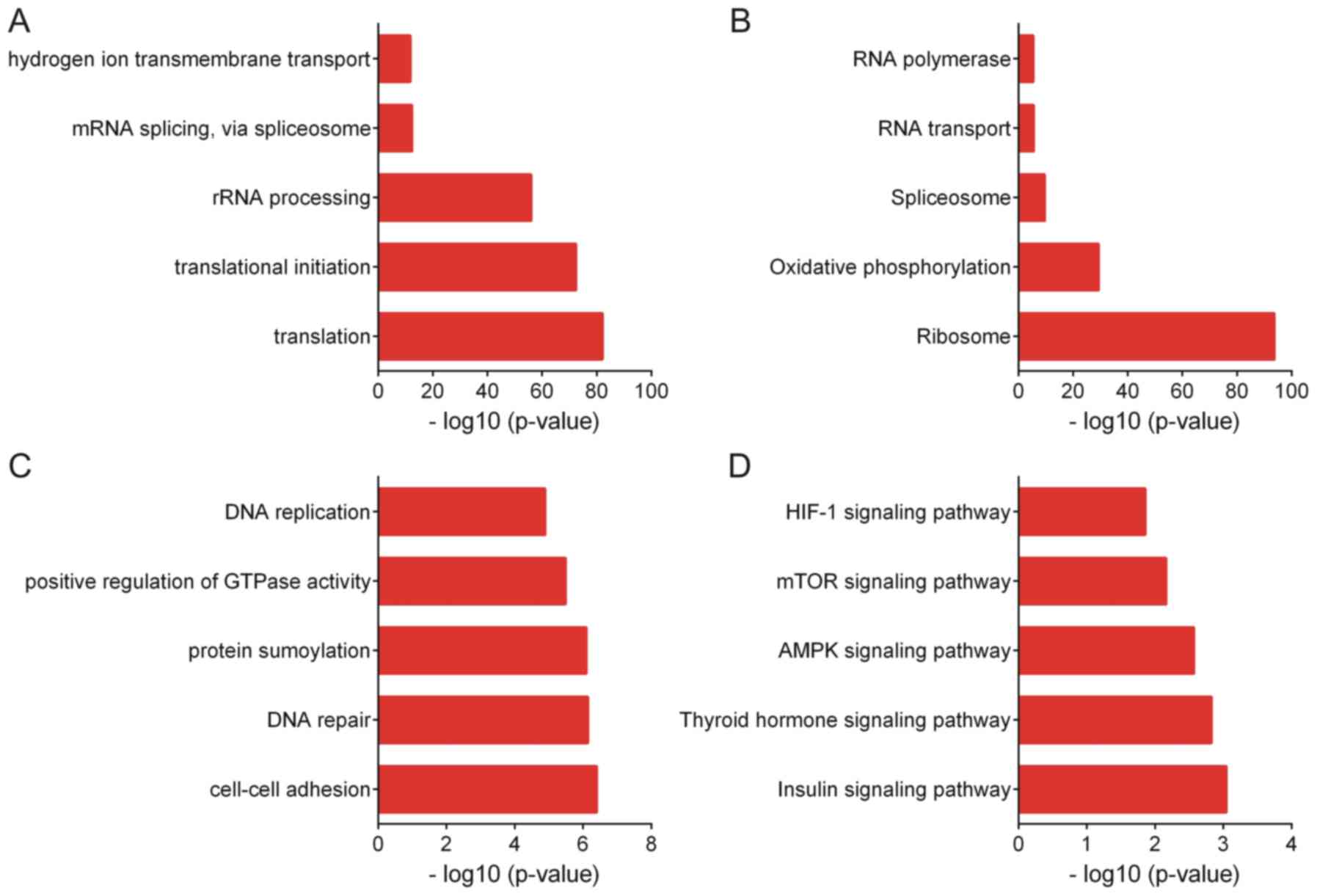
mRNA splicing and hydrogen ion transmembrane transport (Fig. 6A). KEGG analysis suggested that genes
positively co-expressed with ZFAS1 are associated with the pathways
of ribosome, oxidative phosphorylation, spliceosome, RNA transport
and RNA polymerase (Fig. 6B).
GO analysis revealed that genes negatively
co-expressed with ZFAS1 were associated with cell-cell adhesion,
DNA repair, protein sumoylation, positive regulation of GTPase
activity and DNA replication (Fig.
6C). KEGG analysis indicated that genes negatively co-expressed
with ZFAS1 were associated with the insulin, thyroid hormone, AMP
kinase, mTOR and hypoxia-inducible factor-1 signaling pathways
(Fig. 6D).
Discussion
OC is one of the most fatal types of gynecological
malignancy. Of note, the prognosis of OC has remained poor with the
5-year survival rate of advanced-stage OC being <30%. LncRNAs
act as novel regulators in cancer progression and were observed to
be differentially expressed in various human cancer types,
including OC. Certain lncRNAs have been revealed to have crucial
roles in OC by regulating cell viability, metastasis and resistance
to chemotherapeutics. For instance, lncRNA small NF90-associated
RNA was reported to promote OC viability by upregulating growth
factor receptor-bound protein 2-associated binding protein
(17), lncRNA prostate cancer gene
expression marker 1-induced OC tumorigenesis through the Ras
homolog family member pathway (18)
and metastasis-associated lung adenocarcinoma transcript 1
regulated OC cell viability, migration and apoptosis through the
phosphoinositide 3-kinase/AKT pathway (19). Of note, the expression pattern and
functional roles of the vast majority of lncRNAs in OC have
remained elusive.
ZFAS1 has been reported to be a potential biomarker
in HCC, colorectal cancer, gastric cancer and glioma. For instance,
Wang and Xing (20) reported that
upregulation of ZFAS1 in colorectal cancer tissues predicted poor
prognosis. Furthermore, a meta-analysis by Song et al
(21) indicated that high ZFAS1
expression in solid tumors was associated with a shorter OS and
recurrence-free survival. However, the expression pattern of ZFAS1
in OC has remained largely elusive. In the present study, the
expression levels of ZFAS1 in OC and normal samples was first
analyzed using the GEPIA dataset. It was revealed that ZFAS1 was
downregulated in OC compared to normal samples. This result was not
consistent with that of a previous study by Xia et al
(7), reporting that ZFAS1 was
upregulated in OC (7). Thus, the
present study hypothesized that ZFAS1 expression may be associated
with the progression of OC. The results of TCGA analysis suggested
that ZFAS1 was overexpressed in advanced-stage OC compared with
that in early-stage OC samples. Furthermore, by detecting ZFAS1
expression levels in OC cell lines, it was revealed that ZFAS1 was
overexpressed in OC cell lines. GEPIA was based on TCGA and
Genotype-Tissue Expression project (GTEx) data. The TCGA and GTEx
data were produced by two different groups, which may be a possible
reason why the results of the GEPIA analysis were not consistent
with those of Xia et al (7).
In order to validate whether ZFAS1 may serve as a prognostic marker
for OC, the association between ZFAS1 expression in OC tissues and
survival time was then analyzed, revealing that OC patients with
higher ZFAS1 expression had a shorter OS and DFS. Taken together,
these results suggested that ZFAS1 may serve as a novel biomarker
for OC.
LncRNAs exert their roles in cancer cells by
interacting with other RNA molecules, proteins and DNA. ZFAS1 was
reported to promote the occurrence of nasopharyngeal carcinoma by
activating the Wnt/β-catenin pathway (22). Knockdown of ZFAS1 repressed cell
viability via inducing KLF2 and NKD2 expression and inhibited cell
migration and invasion via reducing ZEB1 and ZEB2 expression in
gastric cancer (5). ZFAS1 was also
identified to be a ceRNA by sponging miR-329 in bladder cancer
(9), miR-940 in prostate cancer
(23), miR-486 in osteosarcoma
(24) and miR-484 in colorectal
cancer (25). In the present study,
a loss-of-function assay was performed to examine the effect of
ZFAS1 on OC viability. The results suggested that knockdown of
ZFAS1 significantly suppressed OC cell viability and induced cell
cycle arrest. These in vitro results were consistent with
previous study and showed that ZFAS1 played as an oncogene in OC.
In addition, a Bioinformatics analysis was performed to reveal the
potential mechanisms underlying the role of ZFAS1 in OC
progression. The proteins co-expressed with ZFAS1 were determined
and PPI networks were constructed from them. Furthermore, a
Bioinformatics analysis was performed to determine the GO terms and
KEGG pathways enriched by genes co-expressed with ZFAS1. GO
analysis indicated that the genes positively co-expressed with
ZFAS1 are associated with translation, mRNA splicing, cell-cell
adhesion, DNA repair, protein sumoylation, positive regulation of
GTPase activity and DNA replication. In addition, a ZFAS1-mediated
ceRNA network was constructed in OC, which included 2 miRNAs
(hsa-miR-150-5p and hsa-miR-590-3p) and 195 mRNAs. Of note, Xia
et al (7) also reported that
ZFAS1 interacted with miR-150-5p to promote Sp1 expression. The
present analyses were consistent with the study by Xia et al
(7) in terms of ZFAS1 being
upregulated in OC and miR-150-5p being a target of ZFAS1. The
present study provided novel information contributing to the
understanding of the functions of ZFAS1. For instance, it indicated
that miR-590-3p was also a potential target of ZFAS1, which may
regulate >150 ZFAS1 co-expressing genes in OC.
In conclusion, the present study indicated that
ZFAS1 was overexpressed in OC compared with normal cells. By
analyzing GEPIA dataset, the present study found that ZFAS1 was
downregulated in OC compared with normal samples; however, it was
highly expressed in advanced stage OC compared to early stage OC
samples. Higher ZFAS1 expression was associated with shorter OS and
DFS in OC patients. Knockdown of ZFAS1 suppressed SKOV3 cell
viability and cell cycle progression. Bioinformatics analysis
indicated that ZFAS1 is associated with translation, mRNA splicing,
cell-cell adhesion, DNA repair, protein sumoylation, positive
regulation of GTPase activity and DNA replication. The present
study suggests that ZFAS1 may serve as a biomarker for OC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets analyzed during the current study are
available in the Genomic Data Commons Data Portal repository
[https://portal.gdc.cancer.gov/exploration?filters=%7B%22op%22%3A%22and%22%2C%22content%22%3A%5B%7B%22op%22%3A%22in%22%2C%22content%22%3A%7B%22field%22%3A%22cases.primary_site%22%2C%22value%22%3A%5B%22Ovary%22%5D%7D%7D%5D%7D].
Authors' contributions
SH and MFX conceived and designed the study. SH, DZL
and MFX developed the methodology, analysed and interpreted the
data, and wrote, reviewed and/or revised the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 671:7–30. 2017. View Article : Google Scholar
|
|
2
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma J and Xue M: LINK-A lncRNA promotes
migration and invasion of ovarian carcinoma cells by activating
TGF-β pathway. Biosci Rep. 38(pii): BSR201809362018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shu C, Yan D, Mo Y, Gu J, Shah N and He J:
Long noncoding RNA lncARSR promotes epithelial ovarian cancer cell
proliferation and invasion by association with HuR and miR-200
family. Am J Cancer Res. 86:981–992. 2018.
|
|
5
|
Nie F, Yu X, Huang M, Wang Y, Xie M, Ma H,
Wang Z, De W and Sun M: Long noncoding RNA ZFAS1 promotes gastric
cancer cells proliferation by epigenetically repressing KLF2 and
NKD2 expression. Oncotarget. 824:38227–38238. 2017.
|
|
6
|
Fang C, Zan J, Yue B, Liu C, He C and Yan
D: Long non-coding ribonucleic acid zinc finger antisense 1
promotes the progression of colonic cancer by modulating ZEB1
expression. J Gastroenterol Hepatol. 326:1204–1211. 2017.
View Article : Google Scholar
|
|
7
|
Xia B, Hou Y, Chen H, Yang S, Liu T, Lin M
and Lou G: Long non-coding RNA ZFAS1 interacts with miR-150-5p to
regulate Sp1 expression and ovarian cancer cell malignancy.
Oncotarget. 8:19534–19546. 2017.PubMed/NCBI
|
|
8
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:e050052015. View Article : Google Scholar
|
|
11
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Franceschini A, Lin J, von Mering C and
Jensen LJ: SVD-phy: Improved prediction of protein functional
associations through singular value decomposition of phylogenetic
profiles. Bioinformatics. 32:1085–1087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 254:402–408. 2001.
View Article : Google Scholar
|
|
16
|
Wang JS, Liu QH, Cheng XH, Zhang WY and
Jin YC: The long noncoding RNA ZFAS1 facilitates bladder cancer
tumorigenesis by sponging miR-329. Bio Pharm. 103:174–181. 2018.
View Article : Google Scholar
|
|
17
|
Huang Y, Hu Y, Jin Z and Shen Z: LncRNA
snaR upregulates GRB2-associated binding protein 2 and promotes
proliferation of ovarian carcinoma cells. Biochem Biophys Res
Commun. 503:2028–2032. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen S, Wang LL, Sun KX, Liu Y, Guan X,
Zong ZH and Zhao Y: LncRNA PCGEM1 induces ovarian carcinoma
tumorigenesis and progression through RhoA pathway. Cell Physiol
Biochem. 474:1578–1588. 2018. View Article : Google Scholar
|
|
19
|
Jin Y, Feng SJ, Qiu S, Shao N and Zheng
JH: LncRNA MALAT1 promotes proliferation and metastasis in
epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med
Pharmacol Sci. 2114:3176–3184. 2017.
|
|
20
|
Wang W and Xing C: Upregulation of long
noncoding RNA ZFAS1 predicts poor prognosis and prompts invasion
and metastasis in colorectal cancer. Pathol Res Pract.
2128:690–695. 2016. View Article : Google Scholar
|
|
21
|
Song W, Tian C, Zhang RJ, Zou SB and Wang
K: Meta-analysis of the prognostic value of lncRNA ZFAS1 in
patients with solid tumors. Oncotarget. 852:90301–90307. 2017.
|
|
22
|
Chen X, Li J, Li CL and Lu X: Long
non-coding RAN ZFAS1 promotes nasopharyngeal carcinoma through
activation of Wnt/β-catenin pathway. Eur Rev Med Pharmacol Sci.
2211:3423–3429. 2018.
|
|
23
|
Chen X, Yang C, Xie S and Cheung E: Long
non-coding RNA GAS5 and ZFAS1 are prognostic markers involved in
translation targeted by miR-940 in prostate cancer. Oncotarget.
91:1048–1062. 2018.
|
|
24
|
Li N, Sun ZH, Fang M, Xin JY and Wan CY:
Long non-coding RNA ZFAS1 sponges miR-486 to promote osteosarcoma
cells progression and metastasis in vitro and vivo. Oncotarget.
861:104160–104170. 2017.
|
|
25
|
Xie S, Ge Q, Wang X, Sun X and Kang Y:
Long non-coding RNA ZFAS1 sponges miR-484 to promote cell
proliferation and invasion in colorectal cancer. Cell Cycle.
172:154–161. 2018. View Article : Google Scholar
|