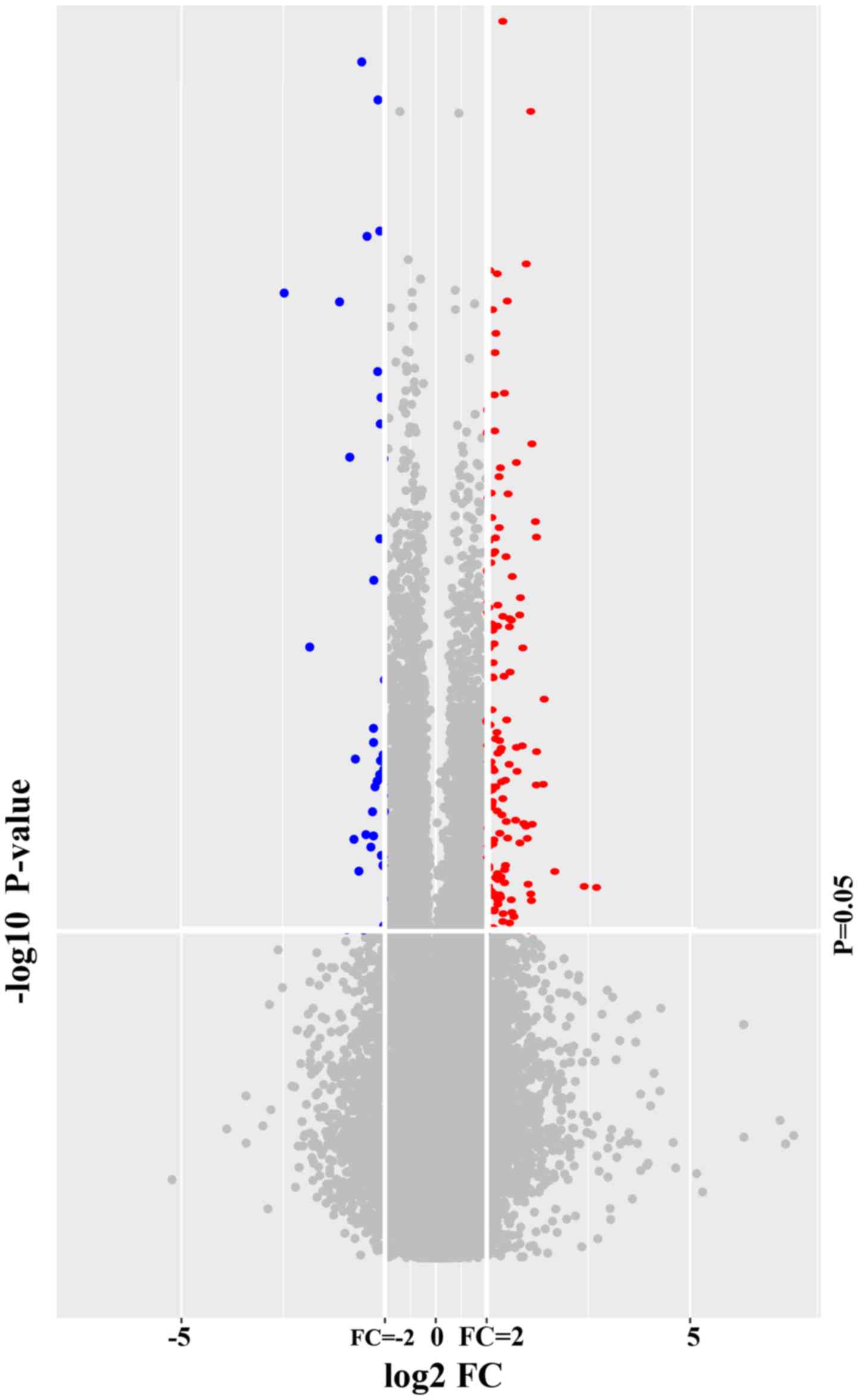
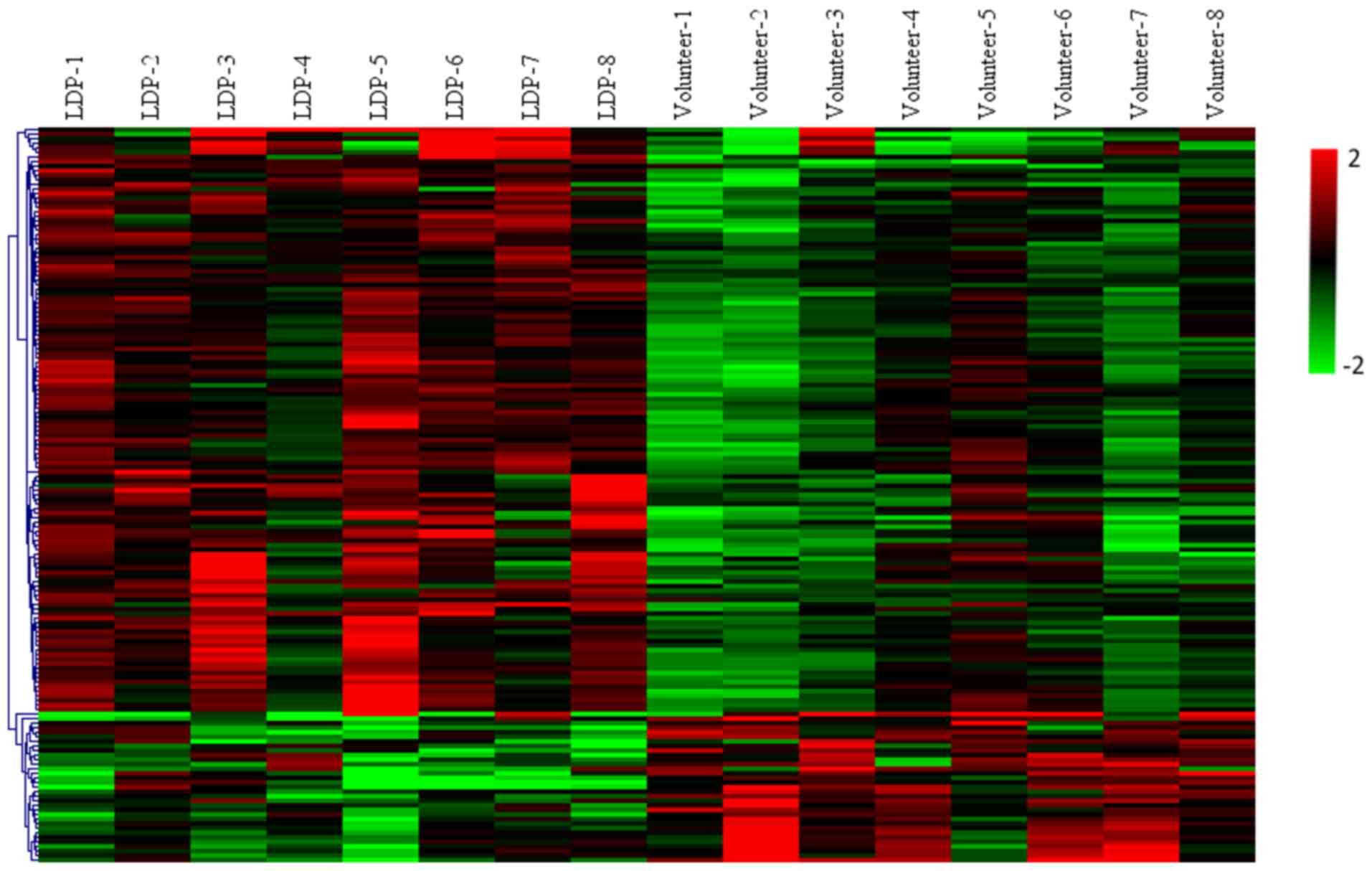
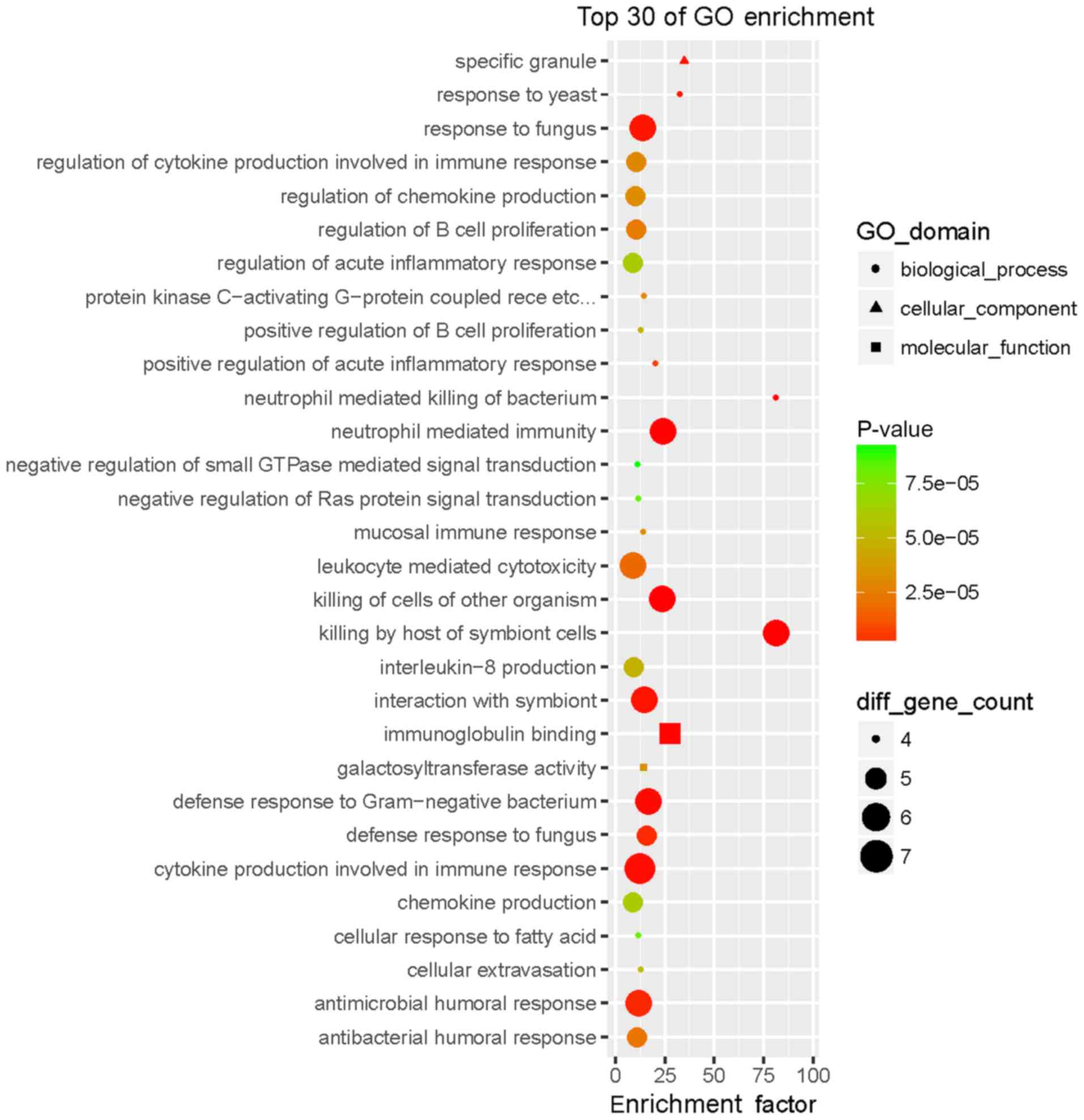
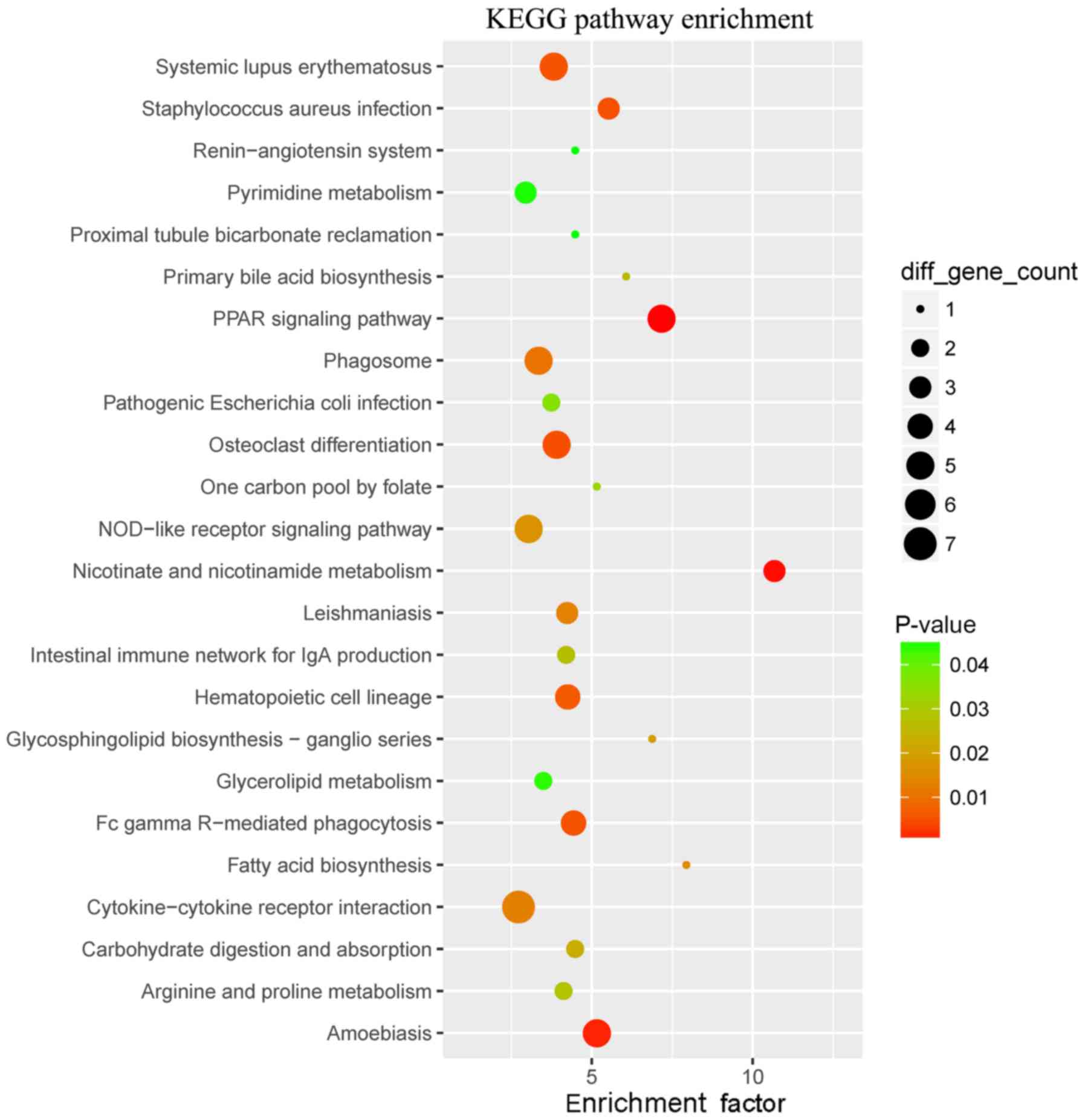
|
1
|
Ahsan K, Najmus-Sakeb, Hossain A, Khan SI
and Awwal MA: Discectomy for primary and recurrent prolapse of
lumbar intervertebral discs. J Orthop Surg (Hong Kong). 20:7–10.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kepler CK, Ponnappan RK, Tannoury CA,
Risbud MV and Anderson DG: The molecular basis of intervertebral
disc degeneration. Spine J. 13:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration, and what causes it? Spine.
31:2151–2161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Daly C, Ghosh P, Jenkin G, Oehme D and
Goldschlager T: A review of animal models of intervertebral disc
degeneration: Pathophysiology, regeneration, and translation to the
clinic. BioMed Res Int. 2016:59521652016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Beattie PF: Current understanding of
lumbar intervertebral disc degeneration: A review with emphasis
upon etiology, pathophysiology, and lumbar magnetic resonance
imaging findings. J Orthop Sports Phys Ther. 38:329–340. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weber KT, Jacobsen TD, Maidhof R,
Virojanapa J, Overby C, Bloom O, Quraishi S, Levine M and Chahine
NO: Developments in intervertebral disc disease research:
Pathophysiology, mechanobiology, and therapeutics. Curr Rev
Musculoskelet Med. 8:18–31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Colombini A, Lombardi G, Corsi MM and
Banfi G: Pathophysiology of the human intervertebral disc. Int J
Biochem Cell Biol. 40:837–842. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen K, Wu D, Zhu X, Ni H, Wei X, Mao N,
Xie Y, Niu Y and Li M: Gene expression profile analysis of human
intervertebral disc degeneration. Genet Mol Biol. 36:448–454. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo W, Zhang B, Li Y, Duan HQ, Sun C, Xu
YQ and Feng SQ: Gene expression profile identifies potential
biomarkers for human intervertebral disc degeneration. Mol Med Rep.
16:8665–8672. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kazezian Z, Gawri R, Haglund L, Ouellet J,
Mwale F, Tarrant F, O'Gaora P, Pandit A, Alini M and Grad S: Gene
expression profiling identifies interferon signalling molecules and
IGFBP3 in human degenerative annulus fibrosus. Sci Rep.
5:156622015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu C, Fei HD, Sun ZY and Tian JW:
Bioinformatic analysis of the microarray gene expression profile in
degenerative intervertebral disc cells exposed to TNF-α. Eur Rev
Med Pharmacol Sci. 19:3332–3339. 2015.PubMed/NCBI
|
|
12
|
Markova DZ, Kepler CK, Addya S, Murray HB,
Vaccaro AR, Shapiro IM, Anderson DG, Albert TJ and Risbud MV: An
organ culture system to model early degenerative changes of the
intervertebral disc II: Profiling global gene expression changes.
Arthritis Res Ther. 15:R1212013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schubert AK, Smink JJ, Arp M, Ringe J,
Hegewald AA and Sittinger M: Quality assessment of surgical disc
samples discriminates human annulus fibrosus and nucleus pulposus
on tissue and molecular level. Int J Mol Sci. 19:E17612018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Y, Wang S, Liu Y and Wang X:
Microarray analysis of genes and gene functions in disc
degeneration. Exp Ther Med. 7:343–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang YG, Guo X, Sun Z, Jia G, Xu P and
Wang S: Gene expression profiles of disc tissues and peripheral
blood mononuclear cells from patients with degenerative discs. J
Bone Miner Metab. 28:209–219. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsai TT, Lai PL, Liao JC, Fu TS, Niu CC,
Chen LH, Lee MS, Chen WJ, Fang HC, Ho NY, et al: Increased
periostin gene expression in degenerative intervertebral disc
cells. Spine J. 13:289–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thompson JP, Pearce RH, Schechter MT,
Adams ME, Tsang IK and Bishop PB: Preliminary evaluation of a
scheme for grading the gross morphology of the human intervertebral
disc. Spine. 15:411–415. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brinjikji W, Luetmer PH, Comstock B,
Bresnahan BW, Chen LE, Deyo RA, Halabi S, Turner JA, Avins AL,
James K, et al: Systematic literature review of imaging features of
spinal degeneration in asymptomatic populations. AJNR Am J
Neuroradiol. 36:811–816. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luoma K, Riihimäki H, Luukkonen R,
Raininko R, Viikari-Juntura E and Lamminen A: Low back pain in
relation to lumbar disc degeneration. Spine. 25:487–492. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rajan NE, Bloom O, Maidhof R, Stetson N,
Sherry B, Levine M and Chahine NO: Toll-Like Receptor 4 (TLR4)
expression and stimulation in a model of intervertebral disc
inflammation and degeneration. Spine. 38:1343–1351. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nicotra L, Loram LC, Watkins LR and
Hutchinson MR: Toll-like receptors in chronic pain. Exp Neurol.
234:316–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shamji MF, Setton LA, Jarvis W, So S, Chen
J, Jing L, Bullock R, Isaacs RE, Brown C and Richardson WJ:
Proinflammatory cytokine expression profile in degenerated and
herniated human intervertebral disc tissues. Arthritis Rheum.
62:1974–1982. 2010.PubMed/NCBI
|
|
25
|
Vo NV, Hartman RA, Yurube T, Jacobs LJ,
Sowa GA and Kang JD: Expression and regulation of
metalloproteinases and their inhibitors in intervertebral disc
aging and degeneration. Spine J13. 331–341. 2013. View Article : Google Scholar
|
|
26
|
von Bahr S, Movin T, Papadogiannakis N,
Pikuleva I, Rönnow P, Diczfalusy U and Björkhem I: Mechanism of
accumulation of cholesterol and cholestanol in tendons and the role
of sterol 27-hydroxylase (CYP27A1). Arterioscler Thromb Vasc Biol.
22:1129–1135. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Escher G, Krozowski Z, Croft KD and
Sviridov D: Expression of sterol 27-hydroxylase (CYP27A1) enhances
cholesterol efflux. J Biol Chem. 278:11015–11019. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Zhao Y, Wang M, Si M, Li J, Hou
Y, Jia J and Nie L: Serum lipid levels are positively correlated
with lumbar disc herniation - a retrospective study of 790 Chinese
patients. Lipids Health Dis. 15:802016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kauppila LI: Atherosclerosis and disc
degeneration/low-back pain--a systematic review. Eur J Vasc
Endovasc Surg. 37:661–670. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bank RA, Bayliss MT, Lafeber FP, Maroudas
A and Tekoppele JM: Ageing and zonal variation in
post-translational modification of collagen in normal human
articular cartilage. The age-related increase in non-enzymatic
glycation affects biomechanical properties of cartilage. Biochem J.
330:345–351. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ali R, Le Maitre CL, Richardson SM,
Hoyland JA and Freemont AJ: Connective tissue growth factor
expression in human intervertebral disc: Implications for
angiogenesis in intervertebral disc degeneration. Biotech
Histochem. 83:239–245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nasto LA, Robinson AR, Ngo K, Clauson CL,
Dong Q, St Croix C, Sowa G, Pola E, Robbins PD, Kang J, et al:
Mitochondrial-derived reactive oxygen species (ROS) play a causal
role in aging-related intervertebral disc degeneration. J Orthop
Res. 31:1150–1157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Flekac M, Skrha J, Hilgertova J, Lacinova
Z and Jarolimkova M: Gene polymorphisms of superoxide dismutases
and catalase in diabetes mellitus. BMC Med Genet. 9:302008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Landis GN and Tower J: Superoxide
dismutase evolution and life span regulation. Mech Ageing Dev.
126:365–379. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nel A, Xia T, Mädler L and Li N: Toxic
potential of materials at the nanolevel. Science. 311:622–627.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tian Y, Yuan W, Fujita N, Wang J, Wang H,
Shapiro IM and Risbud MV: Inflammatory cytokines associated with
degenerative disc disease control aggrecanase-1 (ADAMTS-4)
expression in nucleus pulposus cells through MAPK and NF-κB. Am J
Pathol. 182:2310–2321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang H, Tian Y, Wang J, Phillips KL, Binch
AL, Dunn S, Cross A, Chiverton N, Zheng Z, Shapiro IM, et al:
Inflammatory cytokines induce NOTCH signaling in nucleus pulposus
cells: Implications in intervertebral disc degeneration. J Biol
Chem. 288:16761–16774. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Molinos M, Almeida CR, Caldeira J, Cunha
C, Gonçalves RM and Barbosa MA: Inflammation in intervertebral disc
degeneration and regeneration. J R Soc Interface. 12:201504292015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Boos N, Weissbach S, Rohrbach H, Weiler C,
Spratt KF and Nerlich AG: Classification of age-related changes in
lumbar intervertebral discs: 2002 Volvo Award in basic science.
Spine. 27:2631–2644. 2002. View Article : Google Scholar : PubMed/NCBI
|