Introduction
Cartilage injury is difficult to repair since the
cartilage tissue lacks the self-restoration ability (1). Therefore, effective ways to repair a
cartilage defect are a high priority research area (2). Despite some progress, current
therapeutic methods are relatively inefficient and costly (3,4).
Improved formation of chondrocytes differentiated from the
mesenchymal stem cells (MSCs) by genetic regulation is one of many
potentially promising therapeutic options (5–7). Bone
marrow-derived mesenchymal stem cells (BMSCs) are pluripotent stem
cells that can differentiate into osteoblasts, chondrocytes,
fibroblasts and adipocytes (8).
Differentiation of MSCs into chondrocytes is a complex process
strictly regulated by a complex transcriptional network (9). SOX9 is a critical transcription factor
for mesenchymal condensation prior to chondrogenesis (10). A large body of evidence has suggested
that the SOX9 expression is essential for the survival of
chondrocytes in order to progress on to hypertrophy (11,12).
MicroRNA (miRNA or miR) is a class of small
non-coding RNA molecules (21–25 nt) (13). miRNAs play a critical role in many
biological processes by promoting mRNA degradation by interacting
with the 3′untranslated regions (UTRs) of specific mRNAs (14). Various miRNAs have been reported to
differentially express during chondrogenic differentiation
(15–17). Since miRNAs are small endogenous
molecules, they can be used as a tool for genetic engineering
(18,19). miR-30a, miR-195, miR-216b and miR-15a
were selected for the current study. miR-30a expression has been
shown to increase in MSCs during chondrogenic differentiation and
regulate chondrogenic differentiation (20). It was identified that miR-195 was
downregulated in MSC during osteogenic differentiation and
regulated MSC osteogenic differentiation (21). Moreover, it has been reported that
miR-216b directly regulated SOX9 in cell proliferation (22). miR-15a expression decreased in
adipose-derived MSC under normoxia treatment (23).
The aim of the current study was to elucidate the
interaction between miR-30a and SOX9 during the chondrogenic
differentiation of MSCs. Additionally, the authors investigated
whether inhibition of miR-30a has an inhibitory effect on MSC
chondrogenic differentiation via SOX9.
Materials and methods
Ethics statement
The study protocol was approved by the Ethics
Committee of Xiangya Hospital of Central South University (protocol
ID: 201703358). Each bone marrow donor signed an informed consent
before the study.
Cell culture
Human (h)BMSCs from healthy volunteers were
extracted from bone marrow as described in our previous research
(24). In brief, volunteers were
recruited from patients undergoing bone marrow aspiration at
Xiangya Hospital (Changsha, Hunan, China) from January 2015 to
December 2016. Volunteers with immune system diseases and blood
test abnormal were excluded. A total 8 patients (3 male and 5
female) from age 22–45 years old (mean age 36 years) were
recruited. Bone marrow aspiration were performed to acquire the
bone marrow. Ficoll density gradient media (density 1.077
g/cm3; GE Healthcare) method was using to isolate hBMSC.
Buffy coats, which including hBMSCs, were collected after
centrifugation (1,100 × g; 20 min at 37°C). After isolation from
the bone marrow aspirates, hBMSCs were incubated in a complete
hBMSC medium supplemented with 10% fetal bovine serum (FBS;
HUXMA-90011; Cyagen Biosciences, Inc.), 100 U/ml
penicillin/streptomycin and glutamine at a density of
107 cells per 100 mm dishes at 37°C in a 5%
CO2 incubator. The third generation of the cells was
subjected to subsequent experiments. The HEK293 cell line (Thermo
Fisher Scientific, Inc.) was cultured in high-glucose (4,500 mg/l)
DMEM (HyClone; GE Healthcare Life Sciences) supplemented with 10%
FBS and 1% penicillin/streptomycin at 37°C in a 5% CO2
incubator.
Adipogenic, osteogenic and
chondrogenic differentiation of MSCs
For adipogenic and osteogenic differentiation, the
passage 3 hMSCs were harvested with 0.25% trypsin and centrifuged
(168 × g at 37°C for 4 min, then suspended at 2×104
cells/cm2 for subsequent differentiation. For adipogenic
differentiation, hMSCs were cultured in an adipogenic
differentiation medium (GUXMX-90031; Cyagen Biosciences, Inc.; kit
components included mesenchymal stem cell adipogenic
differentiation basal medium, 10% FBS, 1% penicillin-streptomycin,
50 µg/ml glutamine, insulin 2 µl/ml, isobutylmethylxanthine,
dexamethasone 1 µl/ml, dexamethasone 1 µl/ml and rosiglitazone 1
µl/ml) at 37°C and 5% CO2 incubator for 21 days.
Osteogenic differentiation was performed as
described previously (24). Briefly,
hBMSCs were incubated with Human Mesenchymal Stem Cell Growth
Medium (HUXMX-90011; Cyagen Biosciences, Inc., kit components
included human mesenchymal stem cell basal medium, 10% human
mesenchymal stem cell-qualified fetal bovine serum, 1%
penicillin-streptomycin and 2 mmol/l glutamine), after isolation
from bone marrow at a 37°C and 5% CO2 incubator.
For chondrogenic differentiation, 1×106
hMSCs were suspended and centrifuged (300 × g; 4 min; 37°C) to form
a cell mass in a V-shape polypropylene culture tube in a 37°C
incubator in 5% CO2. The cell mass was incubated in a
Mesenchymal Stem Cell Chondrogenic Differentiation Medium
(GUXMX-90041, Cyagen Biosciences, Inc.; kit components included 194
ml basal medium, 100 nM dexamethasone, 50 µg/ml ascorbate, 6.25
µg/ml insulin-transferred selenium-A, 100 µg/ml sodium pyruvate, 50
µg/ml proline and 10 ng/ml TGF-β3) in a V-shape polypropylene
culture tube for 21 days. The induction medium was changed every 2
days.
Staining
Oil red O staining, alizarin red S staining and
toluidine blue staining were performed to assess the
multi-differentiation potential of MSCs. For Oil red O staining,
cell at 80% confluence were fixed with 4% paraformaldehyde solution
for 15 min at room temperature and incubated in 0.18% Oil red O
(Sigma-Aldrich; Merck KGaA) solution for 30 min at room
temperature. For alizarin red S staining, cells were fixed in 4%
paraformaldehyde solution for 15 min and incubated in 3% alizarin
red S (Sigma-Aldrich; Merck KGaA) solution for 30 min at room
temperature. For toluidine blue staining, the cell mass was fixed
in 4% paraformaldehyde solution for 24 h, dehydrated by in an
ethanol gradient (70–96%) at room temperature then paraffin
embedded. Sections (10 µm) were stained with 0.5% toluidine blue
(Sigma-Aldrich; Merck KGaA) solution for 20 min at room
temperature. The stained cells were observed under a phase contrast
microscope (Olympus Corporation).
Flow cytometry analysis
Flow cytometry analysis was performed using an hMSC
characterization kit (HUXMX-09011; Cyagen Biosciences, Inc.) to
detect the MSC surface-specific antigens. In brief, MSCs were
suspended (3×106 cells/ml) and diluted with PBS with
0.1% BSA, mixed 100 µl cell suspension with 2 µl various
fluorescently labeled monoclonal antibodies, including anti-human
CD45, CD14, CD44, CD105, CD90 and CD34, and then incubated at room
temperature for 30 min in the dark. The cells were washed by PBS
with 0.1% BSA twice and then centrifuged (250 × g; 5 min; room
temperature). The supernatant was discarded and cells re-suspended
with PBS with 0.1% BSA and incubated with 2 µl fluorescein
isothiocyanate (FITC)/propidium iodide (PI) goat anti-mouse IgG
antibodies at room temperature for 30 min in the dark. Then samples
were washed with 1 ml PBS with 0.1% BSA twice, centrifuged (250 ×
g; 5 min; 4°C). The supernatant was discarded and cells were
resuspended with 400 µl PBS with 0.1% BSA and analyzed immediately
using a BD FACSCalibur flow cytometer (BD Biosciences). An
unstained sample (1×106) was used as a negative control;
the data were analyzed using BD FACSuite software v1.0 (BD
Biosciences).
Cell transfection
Prior to chondrogenic differentiation, MSCs were
transfected with the miR-30a mimic, miR-30a inhibitor or their
negative controls by using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The miR-30a mimic, miR-30a inhibitor and their negative controls
were purchased from GenePharma Co., Ltd. and the sequences were as
follows: miR-30a mimic, 5′-UGUAAACAUCCUCGACUGGAAG-3′ (sense);
miR-30a inhibitor, 5′-CUUCCAGUCGAGGAUGUUUACA-3′; negative control,
5′-UUCUCCGAACGUGUCACGUTT-3′ (sense). In brief, MSCs were seeded
into six-well plate (1×105/well) and were subjected to
transfection after 24 h. Transfection of 50 nM miR-30a mimic,
miR-30a inhibitor and their negative controls were performed. The
cells were incubated at a 37°C and 5% CO2 incubator for
4 h. In order to exclude the potential influence of transfection
reagent, a MOCK transfection group which transfected without
RNA.
The transfection efficiency was evaluated by the
visualization of fluorescence by Cy3-labeled miRNA transfection. In
brief, Cy3-labeled miRNA (cat. no. AM17120; Thermo Fisher
Scientific, Inc.) was designed for monitoring transfection
efficiency before miRNA mimic or inhibitor transfection
experiments. MSCs were seeded into six-well plates
(1×105/well) and were subjected to transfection after 24
h. A total of 50 nM Cy3-labeled miRNA was transfected with
Lipofectamine 2000 at a 37°C and 5% CO2 incubator. After
4 h of transfection, cells with fluorescence were observed and
counted under a laser-scanning microscope with ×20 magnification
(FV10-ASW 1.7; Olympus Corporation). The transfection efficiency
was 50% (result not shown). A total of 24 h after the transfection,
the cells were subjected to the chondrogenic differentiation assay
with subsequent detection of the results.
Luciferase reporter assay
Pairing between miR-30a and SOX9 mRNA was reported
by the miRNA databases, miRBase (http://www.mirbase.org/) and TargetScan (http://www.targetscan.org/vert_72/). SOX9 3′UTR
and 5-bp base mutated SOX9 3UTR regions were amplified and cloned
into the pGL4.26 luciferase reporter plasmid (Promega Corporation).
HEK293 cells were divided into 6 groups randomly and co-transfected
with a random DNA sequence (5′-UUCUCCGAACGUGUCACGUTT-3′), the
miR-30a mimic or the negative control mimic and the wild-type or
mutant recombinant reporter plasmid at 40% confluence using
Lipofectamine 2000. Luciferase reporter assays were performed 24 h
post-transfection using Dual Luciferase® Reporter assay
system (Promega Corporation) according to the manufacturer's
protocol. Luciferase activities were determined and the ratio of
firefly luciferase to Renilla luciferase activity was
calculated. All transfection experiments were performed in
triplicate.
Western blotting
MSCs were harvested in a lysis buffer (Beyotime
Institute of Biotechnology) containing protease inhibitors and then
centrifuged at 10,000 × g for 20 min at 4°C. Protein concentration
was determined with the BCA Protein Assay reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After
quantification, 20 µg of the sample was loaded into 10% gels,
separated using SDS-PAGE and then transferred to a PVDF membrane
(Bio-Rad Laboratories, Inc.). After blocking with 5% non-fat milk
in Tris-buffered saline and Polysorbate 20 for 1 h at room
temperature, the blots were incubated with the primary antibodies
(all Abcam) including SOX9 (1:1,000; cat. no. ab185966), aggrecan
(1:1,000; cat. no. ab3778), collagen II (1:1,000; cat. no.
ab34712), β-catenin (1:500; cat. no. ab32572) and β-actin (1:5,000;
cat. no. ab8226). After incubating the membrane with the
appropriate secondary antibody (1:10,000; goat anti-rabbit IgG
antibody, cat no. ab150077 and goat anti-mouse IgG antibody, cat
no. ab150117; Abcam) in the blocking buffer for 1 h at room
temperature, protein expression was quantified by Chemiluminescence
Protein Detection Module (Millipore, Inc.). β-actin were used as an
internal reference. Band intensity was determined by Image J (v
1.4.0; National Institutes of Health) program.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the MSCs using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. For reverse transcription of miRNA,
miRNA cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.) was used.
In brief, mature miRNAs were extended by an adaptor sequence of 3′
poly-A tailing and 5′ ligation prior to reverse transcription. Then
a standard reverse transcription reaction (42°C for 15 min, 85°C
for 5 min, then hold at 4°C) was performed. qPCR was performed
using a SYBR Green Realtime PCR master mix (Takara Bio, Inc.) in
PTC-220 Real-Time PCR Machine (Bio-Rad Laboratories, Inc.)
according to the manufacturer's protocol. U6 was used as an
internal reference for miRNA expression analysis. The thermocycling
conditions were as follows: Incubation at 95°C for 10 min, followed
by 40 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C for 30
sec, then incubation at 4°C for 5 min. Specific primers were used
for quantitative PCR as follows: U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; miR-30a forward,
5′-ACACTCCAGCTGGGTGTAAACATCCTCGACTG-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; miR-195 forward,
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGAGCCAAT-3′ and
reverse, 5′-GGGGTAGCAGCAGCACAGAAAT-3′; miR-216b forward,
5′-GCCGCGCTAAAGTGCTTATAGTG-3′ and reverse, 5′-CACCAGGGTCCGAGGT-3′;
and miR-15a forward,
5′-TTGTAATACGACTCACTATAGGGAGAGAGTCGATGTGTTCTTC-3′ and reverse
5′-ACATGGGTTTAGCCATCCAGAAACCCACC-3′. The relative expression levels
of the miRNAs were calculated using the 2−ΔΔCq method
(25).
Cell proliferation
To assess cell proliferation after the miR-30a mimic
transfection, Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) assay was performed according to the
manufacturer's protocol. Briefly, MSCs (5×103
cells/well) were transfected with the 5 nM miR-30a mimics and
negative controls and plated in 96-well plates for 24 or 48 h at
37°C. Then, 10 µl of the CCK-8 reagent (Dojindo Molecular
Technologies, Inc.) was added to each plate for 4 h at 37°C.
Absorbance at 450 nm was determined and used to calculate cell
viability.
Immunofluorescence analysis
Immunofluorescence analysis was performed to
determine SOX9 translocation in transfected MSCs. After 10 days of
chondrogenic differentiation, MSCs were fixed with 4%
paraformaldehyde (pH 7.4) for 10 min at room temperature and
permeabilized with 0.1% Triton X-100 for 5 min. Then, the slides
were incubated with SOX9 primary antibodies (1:200) for 1 h at room
temperature. Slides were washed and incubated with a fluorescently
labeled secondary antibody (1:200, goat anti-rabbit IgG secondary
antibody, cat. no. A32731, Invitrogen; Thermo Fisher Scientific,
Inc.) for 1 h at room temperature in the dark, and the nuclei were
stained with DAPI (Sigma-Aldrich; Merck KGaA) for 30 min at room
temperature. The images were acquired using a laser-scanning
microscope (FV10-ASW 1.7; Olympus Corporation).
Statistical analysis
The results are presented as mean ± SD and analyzed
using SPSS 13.0 software (SPSS, Inc.). Changes between the groups
were assessed using analysis of variance followed by Bonferroni's
post-hoc test. A value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Characterization of cultured MSCs
To identify the cultured MSCs extracted from the
bone marrow, multi-lineage differentiation capacity and expression
of the characteristic cell surface MSC markers were determined by
flow cytometric analysis after the induction of various types of
differentiation. As shown in Fig. 1,
osteogenic, adipogenic and chondrogenic differentiation were
induced in the cultured MSCs. After 21 days of differentiation, the
results of alizarin red, oil red O or toluidine blue staining were
positive and revealed that cultured MSCs have the multi-lineage
differentiation ability (Fig. 1A).
CD90, CD105 and CD44 are specific cell surface markers of MSCs
while CD34, CD14 and CD45 are the hematopoietic stem cell markers
that were not expressed in the MSCs (26). Flow cytometry analysis showed that
the cultured MSCs expressed CD90, CD105 and CD44, but did not
express CD34, CD14 and CD45 (Fig.
1B).
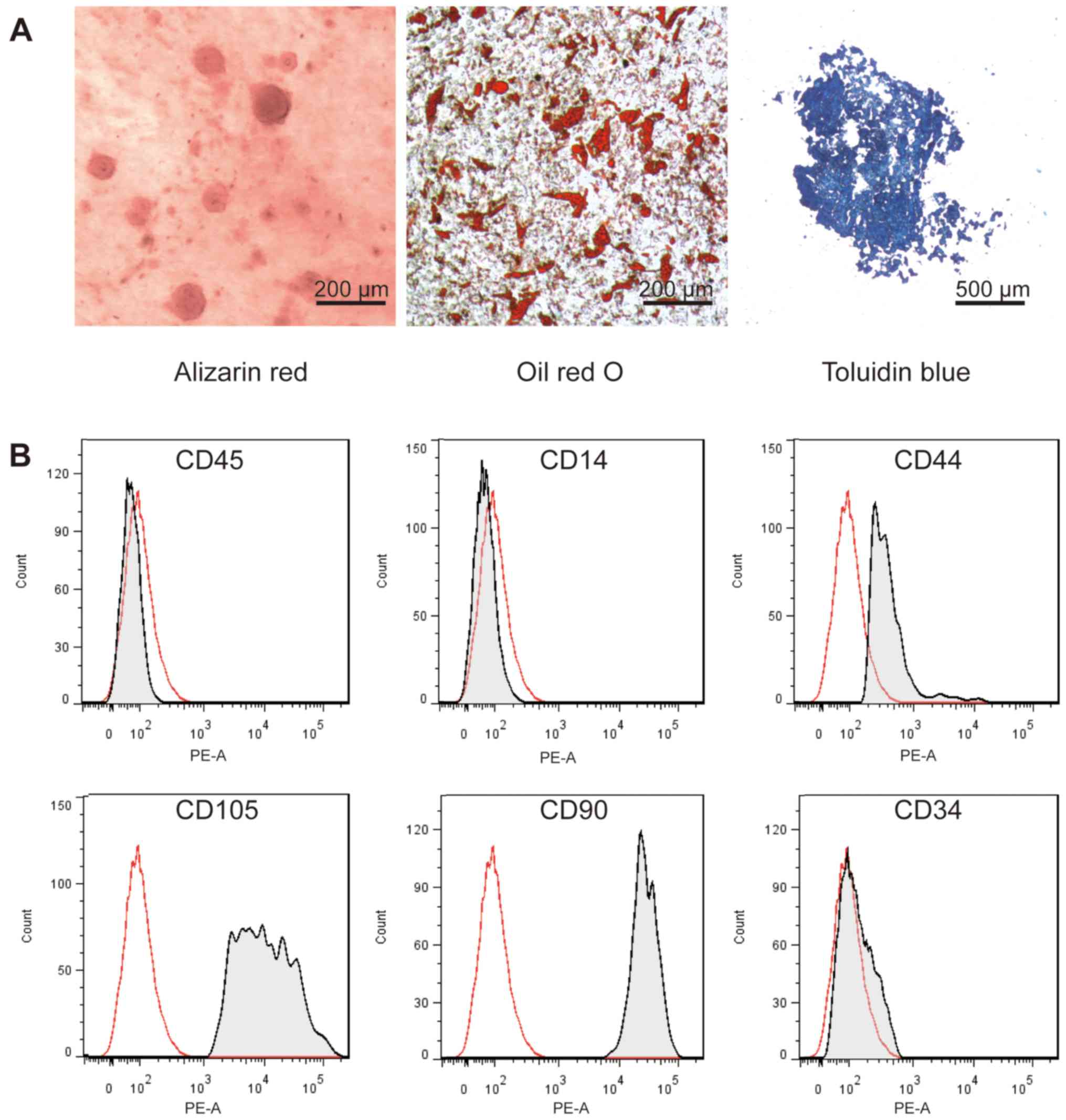 | Figure 1.Characterization of hBMSCs. (A) MSCs
from the human bone marrow were isolated and cultured. Alizarin red
staining, Oil red O staining and toluidine blue staining were
performed to assess the osteogenic, adipogenic and chondrogenic
differentiation potential, respectively. (B) The cell membrane
markers of MSCs were determined by flow cytometry. Surface markers
CD45, CD14, CD44, CD105, CD29 and CD34 were detected and analyzed
(n=3). MSCs, mesenchymal stem cells; hBMSCs, bone marrow-derived
mesenchymal stem cells; CD, cluster of differentiation. |
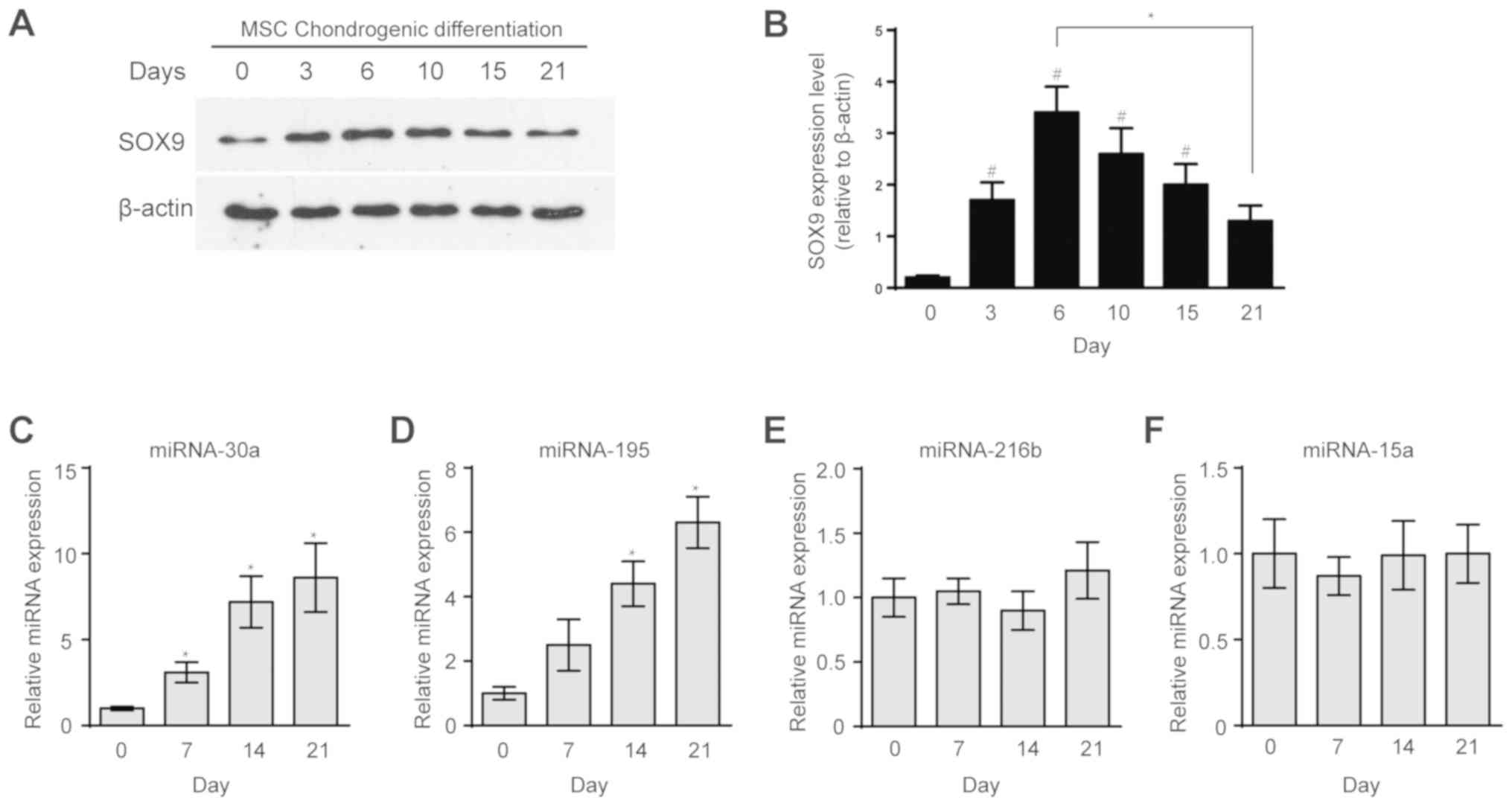
Expression of SOX9 and candidate
miRNAs during MSC chondrogenic differentiation
SOX9 plays the critical role in MSC chondrogenic
differentiation (27–29); therefore, the authors decided to
determine the expression levels of SOX9 at various stages of MSC
chondrogenic differentiation by western blotting. During
chondrogenesis, SOX9 expression was initially increased and then
was decreased when compared to the initial level (Fig. 1A and B). The goal of the present
study was to identify miRNAs that specifically interact with the
SOX9 gene during MSC chondrogenic differentiation. Hence, several
candidate SOX9-associated miRNAs were chosen from the databases and
the literature including miR-30a, miR-195, miR-216b and miR-15a
(20,22,23,30,31).
They were selected from several studies about altered miRNA
expression during chondrogenic differentiation, all the candidate
miRNAs were predicted to interact with SOX9 by the aforementioned
miRNA databases. The expression levels of the candidate miRNAs were
quantified by RT-qPCR. The expression levels of miR-30a and miR-195
consistently increased during MSC chondrogenic differentiation
compared with the untreated group (Fig.
2C-D). In addition, the expression levels of miR-216b and
miR-15a were not changed during chondrogenic differentiation
(Fig. 2E-F).
miR-30a inhibits SOX9 expression in a
cell line and in MSCs
The data indicate that SOX9 mRNA possesses a
potential miR-30a binding site in the 3UTR (Fig. 3A). To confirm this prediction, a
luciferase reporter gene assay was performed using the luciferase
reporter vectors containing the wild-type SOX9 3UTR and the 5-bp
base mutated SOX9 3UTR. The results indicate that the fluorescence
of the wild-type SOX9 3′UTR reporter plasmid was significantly
decreased after the miR-30a mimic transfection, while luciferase
activity was not affected in the case of the mutant reporter
plasmid (Fig. 3B). To further
validate this prediction, the SOX9 expression levels were
determined by western blotting in transfected MSCs. However,
miR-30a transfection efficiency was determined before detection of
interaction relationship between miR-30a and SOX9. After miR-30a
mimic transfection, the increased expression of miR-30a was
determined by RT-qPCR (Fig. 3C). The
expression of SOX9 was significantly decreased in miR-30a
mimic-transfected MSCs compared with the control group (Fig. 3D and E). The results confirmed that
miR-30a can bind to the SOX9 3UTR.
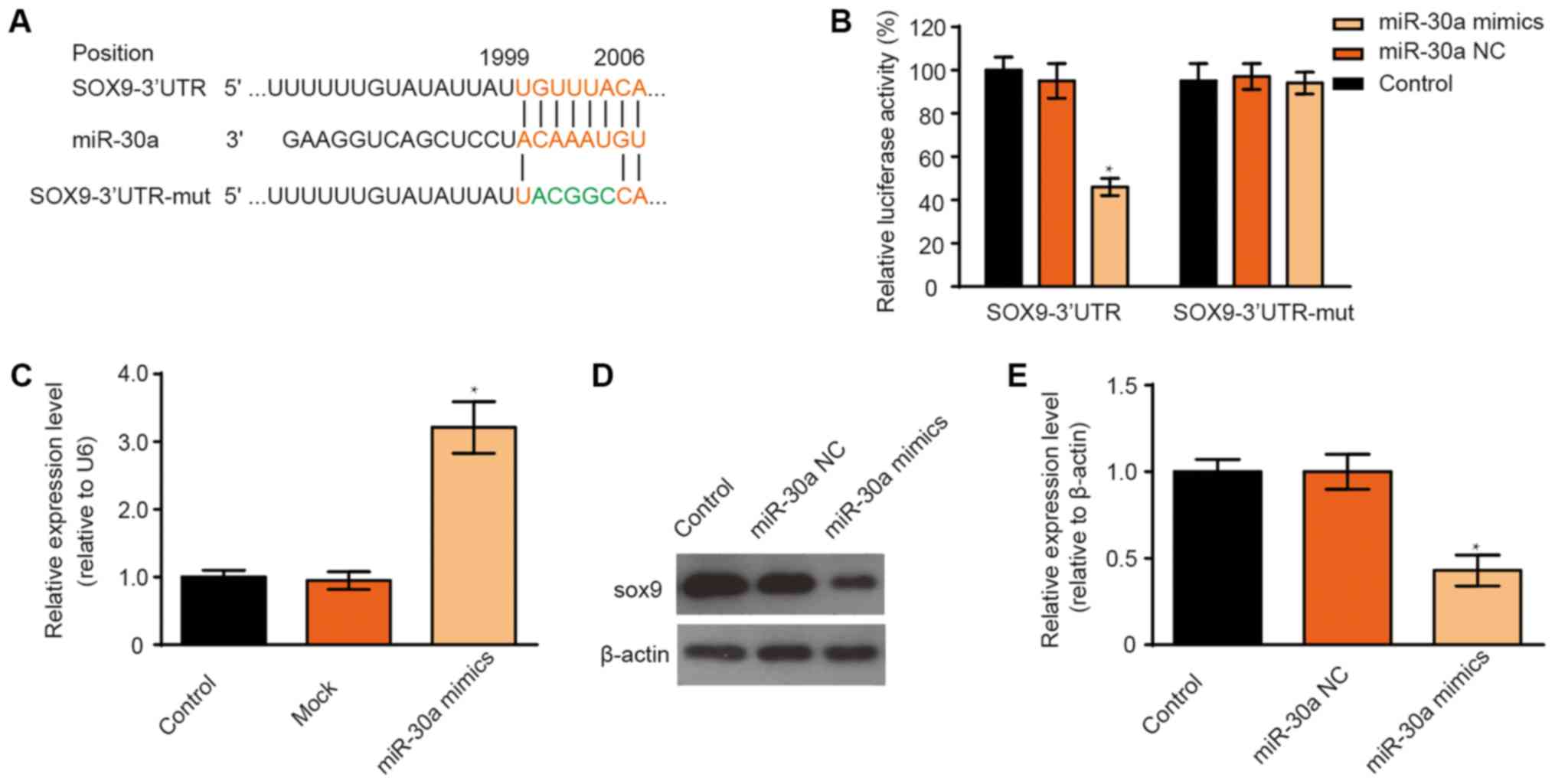 | Figure 3.Effect of miR-30a on SOX9 expression.
(A) Alignment of sequences of the wild-type SOX9, miR-30a and
mutant SOX9 3′UTR. The lines between the sequences shows how
complementary the sequences are. The mutant SOX9 3′UTR contains 5
nucleotide mutations, which are indicated in green. (B) Relative
luciferase activity was assayed after the 293 cells were
co-transfected with a random DNA sequence or the miR-30a mimic or
the negative control mimic and the reporter plasmid with the
wild-type or mutant SOX9 3′UTR. Data are presented as mean ± SD of
three independent experiments. One-way ANOVA, *P<0.05 versus
control. (C) MSCs were transfected with either miR-30a mimic or
transfection reagents, the levels of miR-30a were determined by
reverse transcription-quantitative PCR. (D) Western blotting and
(E) quantification of protein expression. β-actin was used as an
internal reference. Data are presented as mean ± SD of three
independent experiments. One-way ANOVA, *P<0.05 vs. control.
SOX9, transcription factor SOX-9; MSCs, mesenchymal stem cells; miR
or miRNA, microRNA; NC, negative control; UTR, untranslated
region. |
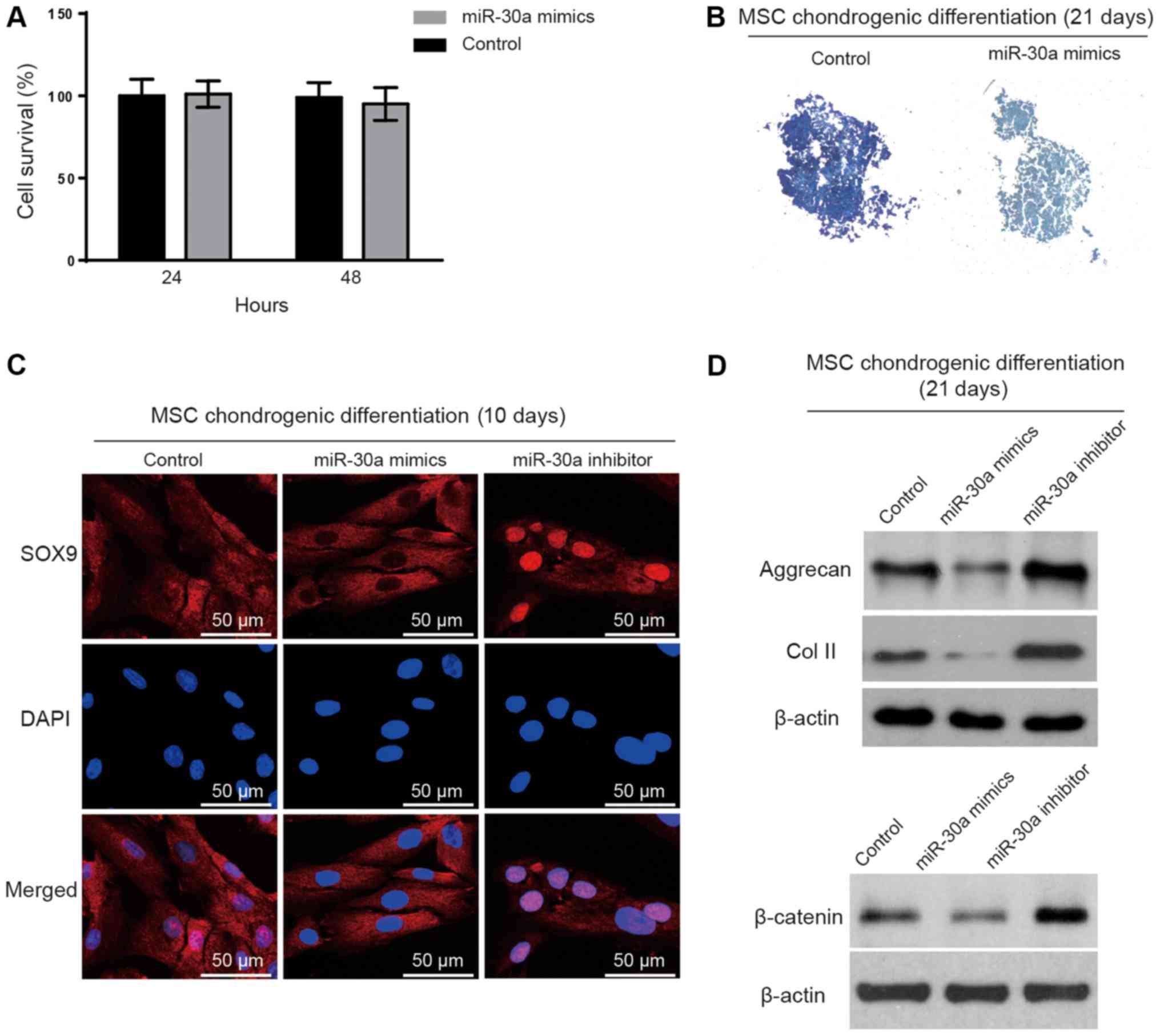
Overexpression of miR-30a suppresses
chondrogenesis
Since miR-30a was upregulated in the early stage of
MSC chondrogenic differentiation, the authors speculated that
miR-30a may affect MSC chondrogenic differentiation via SOX9. To
verify this hypothesis, MSCs were transfected with the miR-30a
mimics, miR-30a inhibitors or negative controls. First, the effect
of the miR-30a mimic transfection on the cell viability was
determined. The results indicate that miR-30a overexpression had no
cytotoxic effect in the MSCs (Fig.
4A). Subsequently, the miR-30a mimics were transfected into the
MSCs on day 7 of chondrogenic differentiation. Toluidine blue
staining showed a marked decrease in the synthesis of the
extracellular matrix on day 21 of the chondrogenic differentiation
(Fig. 4B). Additionally, the
translocation of SOX9 was detected by immunofluorescence in the
MSCs transfected with the miR-30a mimic or miR-30a inhibitor on day
7 of chondrogenic differentiation. The nuclear translocation of
SOX9 could be inhibited by the miR-30a mimic transfection. On the
contrary, the miR-30a inhibitor transfection increased the
translocation of SOX9 into the nucleus (Fig. 4C). Furthermore, the levels of the
cartilage marker proteins including aggrecan and collagen II
(32) were evaluated by western
blotting after the transfections. Western blotting results
demonstrated that cartilage markers were enhanced in the miR-30a
inhibitor group compared with the miR-30a mimic group (Fig. 4D). Finally, the protein levels of
β-catenin, the key factor of the Wnt signaling pathway (33), were examined by western blotting to
verify the critical role of SOX9 in the effect of miR-30a on
chondrogenesis. The result showed that β-catenin was inhibited by
miR-30a overexpression (Fig.
4D).
Discussion
Molecular mechanism underlying MSC chondrogenesis
remains poorly understood (34);
thus, the application of mesenchymal cells for cartilage
regeneration and repair is still not feasible (35). In the present study, hBMSCs have been
isolated and induced into chondrogenic differentiation to imitate
the cartilage formation in vitro. The authors of the current
study determined typical MSC phenotype by the expression of
characteristic cell surface markers, including CD45, CD14, CD44,
CD105, CD90 and CD34, and multi-lineage differentiation capacity.
CD34 is expressed in hematopoietic stem cell (36). CD45 and CD14 are lineage commitment
cell antigens (37). In addition,
CD44, CD105 and CD90 were important surface markers express in MSC
(38). Moreover, the SOX9
transcription factor is essential for the mesenchymal cell
aggregation prior to chondrogenesis (39). Hence, the expression levels of
several miRNAs that were reported to interact with the SOX9 3′UTR
were examined. The results indicate that miR-30a and miR-195 were
consistently increased during MSC chondrogenic differentiation.
Additionally, the binding of miR-30a to the SOX9 3′UTR region was
verified. Then, the expression of miR-30a was upregulated and it
was found that MSC chondrogenic differentiation was inhibited. The
results of the current study demonstrate that miR-30a can inhibit
MSC chondrogenic differentiation via SOX9.
Chondrogenic differentiation of the MSCs is an
intricate process that is regulated by multiple factors (40,41). In
this process, SOX9 is the master regulator since high levels of
SOX9 play an essential role in the aggregation of MSCs prior to
cartilage formation. SOX9 expression was initially increased and
then decreased during chondrogenesis. SOX9 activated the expression
of numerous chondrocyte-specific genes including collagen (Col)2a1,
Col9a1, Col11a2 and aggrecan, and the accumulation of the
extracellular matrix (42). This
importance of SOX9 in chondrogenesis suggests that SOX9 can be used
as a regulatory factor. Therefore, the current study focused on
SOX9 to identify its epigenetic regulatory factors.
miRNAs play an important role in stem cell
self-renewal and differentiation as endogenous post-transcriptional
regulators (43). Recently, several
studies suggested that miRNAs may be involved in MSC chondrogenic
differentiation (44–46). During the chondrogenic
differentiation of MSCs, various miRNAs display differential
expression patterns and can regulate the differentiation process by
inhibiting the expression of specific target genes. For example,
miR-140, miR-483, miR-495 and miR-410 regulated chondrogenic
differentiation by modulating HDAC4 (47), SMAD4 (16), SOX9 (17) and Wnt3a (48), respectively. Based on a review of the
published literature and of miRNA databases, the authors of the
current study selected several miRNAs that were reported to
regulate SOX9 and are expressed in various types of stem cells
(21,23). miR-30a, miR-195, miR-216b and miR-15a
were selected for this study. The SOX9 expression pattern during
the chondrogenic differentiation was similar to a parabola
(49). In the current study, the
expression levels of SOX9 decreased in the later stages of the
chondrogenic differentiation while the levels of miR-30a and
miR-195 gradually increase, which is consistent with a previous
study that showed that increased miR-30a and decreased SOX9 were
detected in primary chondrocytes from cartilage taken from patients
with osteoarthritis (20). In the
present study, miR-30a upregulation was identified and it was shown
to suppress SOX9 expression. A luciferase assay also demonstrated
that miR-30a specifically targeted the 3′UTR of SOX9.
Hence, it was hypothesized that these miRNAs inhibit
sustained expression of SOX9 to regulate chondrogenesis. However,
it remained unclear whether they can regulate MSC chondrogenic
differentiation and what are the specific mechanisms of the
process. Therefore, a series of experiments were carried out and
the results indicated that overexpression of miR-30a reduced the
synthesis of extracellular matrix components, indicating that
miR-30a can inhibit MSC chondrogenic differentiation. In the
process, miR-30a inhibited SOX9 protein expression by directly
binding to the SOX9 mRNA 3′UTR region. The authors speculated that
the miR-30a level gradually increases during MSC chondrogenic
differentiation and subsequently inhibits the sustained expression
of SOX9 leading to a decrease in the Sox 9 expression during the
subsequent stage of differentiation. SOX9 translocation from
cytoplasmic to nucleus was detected by immunofluorescence after
miR-30a inhibitor transfection. As SOX9 was a transcription factor
it binds to the promoter of specifically genes in the nucleus
(50). The SOX9 nuclear
translocation means the enhancement of the SOX9 function. In
addition, miR-30a has been reported to serve as a prognostic factor
in urothelial carcinoma of bladder and left ventricular dysfunction
after acute myocardial infarction (51,52). As
the Wnt signaling pathway serves a significantly important role in
cartilage developments and β-catenin is a part of the pathway
(53), its' expression was assessed.
β-catenin expression was decreased after miR-30a mimic
transfection, which means the activation of Wnt signaling pathway
is influenced by miR-30a levels. β-catenin accumulation in the
cytoplasm and nuclear translocation of β-catenin are key procedures
in the activation of canonical Wnt pathway (53).
The present study had some limitations. Firstly, the
results obtained in the current study are not likely to imitate the
effects in vivo, although MSCs were employed. Thus the
results need to be validated with animal studies. Secondly, miR-30a
was selected from a limited candidate group and other potential
miRNAs may exist, which need to be explored in more studies.
Thirdly, it is more convincing to add the control images of various
MSC differentiation stages to elucidate the SOX9 expression during
MSC differentiation, which is lacking in the present study. Lastly,
the cytoplasmic and nuclear SOX9 levels were not verified by
western blotting.
In conclusion, the current study showed that miR-30a
has a negative regulatory effect on MSC chondrogenic
differentiation by targeting SOX9. Interfering with endogenous
expression of miR-30a and attenuating its inhibitory effect on SOX9
may be possible. Therefore, miR-30a and its mechanism of action may
provide for a novel strategy for the treatment of cartilage-related
diseases.
Acknowledgements
Not applicable.
Funding
This work was supported by the Chinese National
Natural Science Foundation (grant nos. 81472145 and 81772298) and
the Central South University Graduate Innovation Fund (grant no.
2017zzts212).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HZ and MT designed the experiments. YW and GY
performed the experiments. HY and ZZ contributed to data analysis.
YW wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of Xiangya Hospital of Central South University (Protocol
ID: 201703358). Each bone marrow donor signed informed consent
before the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Loeser RF, Collins JA and Diekman BO:
Ageing and the pathogenesis of osteoarthritis. Nat Rev Rheumatol.
12:412–420. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Makris EA, Gomoll AH, Malizos KN, Hu JC
and Athanasiou KA: Repair and tissue engineering techniques for
articular cartilage. Nat Rev Rheumatol. 11:21–34. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Roos EM and Arden NK: Strategies for the
prevention of knee osteoarthritis. Nat Rev Rheumatol. 12:92–101.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhattacharjee M, Coburn J, Centola M,
Murab S, Barbero A, Kaplan DL, Martin I and Ghosh S: Tissue
engineering strategies to study cartilage development, degeneration
and regeneration. Adv Drug Deliv Rev. 84:107–122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Evans CH and Huard J: Gene therapy
approaches to regenerating the musculoskeletal system. Nat Rev
Rheumatol. 11:234–242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Craft AM, Rockel JS, Nartiss Y, Kandel RA,
Alman BA and Keller GM: Generation of articular chondrocytes from
human pluripotent stem cells. Nat Biotechnol. 33:638–645. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xia H, Liang C, Luo P, Huang J, He J, Wang
Z, Cao X, Peng C and Wu S: Pericellular collagen I coating for
enhanced homing and chondrogenic differentiation of mesenchymal
stem cells in direct intra-articular injection. Stem Cell Res Ther.
9:1742018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grayson WL, Bunnell BA, Martin E, Frazier
T, Hung BP and Gimble JM: Stromal cells and stem cells in clinical
bone regeneration. Nat Rev Endocrinol. 11:140–150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Visweswaran M, Pohl S, Arfuso F, Newsholme
P, Dilley R, Pervaiz S and Dharmarajan A: Multi-lineage
differentiation of mesenchymal stem cells-To Wnt, or not Wnt. Int J
Biochem Cell Biol. 68:139–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang X, Huang X, Jiang T, Zheng L, Zhao J
and Zhang X: The role of Sox9 in collagen hydrogel-mediated
chondrogenic differentiation of adult mesenchymal stem cells
(MSCs). Biomater Sci. 6:1556–1568. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Loebel C, Czekanska EM, Bruderer M,
Salzmann G, Alini M and Stoddart MJ: In vitro osteogenic potential
of human mesenchymal stem cells is predicted by Runx2/Sox9 ratio.
Tissue Eng Part A. 21:115–123. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ono N, Ono W, Nagasawa T and Kronenberg
HM: A subset of chondrogenic cells provides early mesenchymal
progenitors in growing bones. Nat Cell Biol. 16:1157–1167. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shivdasani RA: MicroRNAs: Regulators of
gene expression and cell differentiation. Blood. 108:3646–3653.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Treiber T, Treiber N and Meister G:
Regulation of microRNA biogenesis and its crosstalk with other
cellular pathways. Nat Rev Mol Cell Biol. 20:5–20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wa Q, He P, Huang S, Zuo J, Li X, Zhu J,
Hong S, Lv G, Cai D, Xu D, et al: miR-30b regulates chondrogenic
differentiation of mouse embryo-derived stem cells by targeting
SOX9. Exp Ther Med. 14:6131–6137. 2017.PubMed/NCBI
|
|
16
|
Anderson BA and McAlinden A: miR-483
targets SMAD4 to suppress chondrogenic differentiation of human
mesenchymal stem cells. J Orthop Res. 35:2369–2377. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee S, Yoon DS, Paik S, Lee KM, Jang Y and
Lee JW: microRNA-495 inhibits chondrogenic differentiation in human
mesenchymal stem cells by targeting Sox9. Stem Cells Dev.
23:1798–1808. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moutinho C and Esteller M: MicroRNAs and
epigenetics. Adv Cancer Res. 135:189–220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Meurs JB, Boer CG, Lopez-Delgado L and
Riancho JA: Role of epigenomics in bone and cartilage disease. J
Bone Miner Res. 34:215–230. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang T, Xie J, Li H, Li D, Liu P and Hu
Y: MicroRNA-30a promotes extracellular matrix degradation in
articular cartilage via downregulation of Sox9. Cell Prolif.
49:207–218. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Almeida MI, Silva AM, Vasconcelos DM,
Almeida CR, Caires H, Pinto MT, Calin GA, Santos SG and Barbosa MA:
miR-195 in human primary mesenchymal stromal/stem cells regulates
proliferation, osteogenesis and paracrine effect on angiogenesis.
Oncotarget. 7:7–22. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu S, Dong H, Dai H, Liu D and Wang Z:
MicroRNA-216b regulated proliferation and invasion of non-small
cell lung cancer by targeting SOX9. Oncol Lett. 15:10077–10083.
2018.PubMed/NCBI
|
|
23
|
Liu XJ, Bai XG, Teng YL, Song L, Lu N and
Yang RQ: miRNA-15a-5p regulates VEGFA in endometrial mesenchymal
stem cells and contributes to the pathogenesis of endometriosis.
Eur Rev Med Pharmacol Sci. 20:3319–3326. 2016.PubMed/NCBI
|
|
24
|
Wang YJ, Zhang HQ, Han HL, Zou YY, Gao QL
and Yang GT: Taxifolin enhances osteogenic differentiation of human
bone marrow mesenchymal stem cells partially via NF-κB pathway.
Biochem Biophys Res Commun. 490:36–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Uder C, Bruckner S, Winkler S, Tautenhahn
HM and Christ B: Mammalian MSC from selected species: Features and
applications. Cytometry A. 93:32–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao C, Jiang W, Zhou N, Liao J, Yang M,
Hu N, Liang X, Xu W, Chen H, Liu W, et al: Sox9 augments
BMP2-induced chondrogenic differentiation by downregulating Smad7
in mesenchymal stem cells (MSCs). Genes Dis. 4:229–239. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liao J, Hu N, Zhou N, Lin L, Zhao C, Yi S,
Fan T, Bao W, Liang X, Chen H, et al: Sox9 potentiates BMP2-induced
chondrogenic differentiation and inhibits BMP2-induced osteogenic
differentiation. PLoS One. 9:e890252014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Almalki SG and Agrawal DK: Key
transcription factors in the differentiation of mesenchymal stem
cells. Differentiation. 92:41–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang M, Lin H, Fu H, Wang B, Han G and
Fan M: MicroRNA-195-5p regulates osteogenic differentiation of
periodontal ligament cells under mechanical loading. J Cell
Physiol. 232:3762–3774. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eguchi T, Watanabe K, Hara ES, Ono M,
Kuboki T and Calderwood SK: OstemiR: A novel panel of microRNA
biomarkers in osteoblastic and osteocytic differentiation from
mesencymal stem cells. PLoS One. 8:e587962013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luo Y, Sinkeviciute D, He Y, Karsdal M,
Henrotin Y, Mobasheri A, Önnerfjord P and Bay-Jensen A: The minor
collagens in articular cartilage. Protein Cell. 8:560–572. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duan P and Bonewald LF: The role of the
wnt/β-catenin signaling pathway in formation and maintenance of
bone and teeth. Int J Biochem Cell Biol. 77:23–29. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deng A, Zhang H, Hu M, Liu S, Wang Y, Gao
Q and Guo C: The inhibitory roles of Ihh downregulation on
chondrocyte growth and differentiation. Exp Ther Med. 15:789–794.
2018.PubMed/NCBI
|
|
35
|
Deng ZH, Li YS, Gao X, Lei GH and Huard J:
Bone morphogenetic proteins for articular cartilage regeneration.
Osteoarthritis Cartilage. 26:1153–1161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Viswanathan C, Kulkarni R, Bopardikar A
and Ramdasi S: Significance of CD34 negative hematopoietic stem
cells and CD34 positive mesenchymal stem cells-A valuable dimension
to the current understanding. Curr Stem Cell Res Ther. 12:476–483.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Szade K, Zuba-Surma E, Rutkowski AJ,
Jozkowicz A and Dulak J: CD45-CD14+CD34+ murine bone marrow
low-adherent mesenchymal primitive cells preserve multilineage
differentiation potential in long-term in vitro culture. Mol Cells.
31:497–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Baptista LS, do Amaral RJ, Carias RB,
Aniceto M, Claudio-da-Silva C and Borojevic R: An alternative
method for the isolation of mesenchymal stromal cells derived from
lipoaspirate samples. Cytotherapy. 11:706–715. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Healy C, Uwanogho D and Sharpe PT:
Regulation and role of Sox9 in cartilage formation. Dev Dyn.
215:69–78. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Somoza RA, Welter JF, Correa D and Caplan
AI: Chondrogenic differentiation of mesenchymal stem cells:
Challenges and unfulfilled expectations. Tissue Eng Part B Rev.
20:596–608. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Barry F and Murphy M: Mesenchymal stem
cells in joint disease and repair. Nat Rev Rheumatol. 9:584–594.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jo A, Denduluri S, Zhang B, Wang Z, Yin L,
Yan Z, Kang R, Shi LL, Mok J, Lee MJ and Haydon RC: The versatile
functions of Sox9 in development, stem cells, and human diseases.
Genes Dis. 1:149–161. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shim J and Nam JW: The expression and
functional roles of microRNAs in stem cell differentiation. BMB
Rep. 49:3–10. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Martin EC, Qureshi AT, Llamas CB, Burow
ME, King AG, Lee OC, Dasa V, Freitas MA, Forsberg JA, Elster EA, et
al: Mirna biogenesis pathway is differentially regulated during
adipose derived stromal/stem cell differentiation. Adipocyte.
7:96–105. 2018.PubMed/NCBI
|
|
45
|
Zeng ZL, Lin XL, Tan LL, Liu YM, Qu K and
Wang Z: MicroRNAs: Important regulators of induced pluripotent stem
cell generation and differentiation. Stem Cell Rev. 14:71–81. 2018.
View Article : Google Scholar
|
|
46
|
Ran X, Xiao CH, Xiang GM and Ran XZ:
Regulation of embryonic stem cell self-renewal and differentiation
by MicroRNAs. Cell Reprogramm. 19:150–158. 2017. View Article : Google Scholar
|
|
47
|
Papaioannou G, Mirzamohammadi F, Lisse TS,
Nishimori S, Wein MN and Kobayashi T: MicroRNA-140 provides
robustness to the regulation of hypertrophic chondrocyte
differentiation by the PTHrP-HDAC4 pathway. J Bone Miner Res.
30:1044–1052. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Y, Huang X and Yuan Y: MicroRNA-410
promotes chondrogenic differentiation of human bone marrow
mesenchymal stem cells through down-regulating Wnt3a. Am J Transl
Res. 9:136–145. 2017.PubMed/NCBI
|
|
49
|
Suchorska WM, Augustyniak E, Richter M and
Trzeciak T: Gene expression profile in human induced pluripotent
stem cells: Chondrogenic differentiation in vitro, part A.
Mol Med Rep. 15:2387–2401. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Oh CD, Yasuda H, Zhao W, Henry SP, Zhang
Z, Xue M, de Crombrugghe B and Chen D: SOX9 directly regulates
CTGF/CCN2 transcription in growth plate chondrocytes and in nucleus
pulposus cells of intervertebral disc. Sci Rep. 6:299162016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Maciejak A, Kostarska-Srokosz E, Gierlak
W, Dluzniewski M, Kuch M, Marchel M, Opolski G, Kiliszek M, Matlak
K, Dobrzycki S, et al: Circulating miR-30a-5p as a prognostic
biomarker of left ventricular dysfunction after acute myocardial
infarction. Sci Rep. 8:98832018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang C, Ma X, Du J, Yao Z, Shi T, Ai Q,
Chen X, Zhang Z, Zhang X and Yao X: MicroRNA-30a as a prognostic
factor in urothelial carcinoma of bladder inhibits cellular
malignancy by antagonising Notch1. BJU Int. 118:578–589. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Perugorria MJ, Olaizola P, Labiano I,
Esparza-Baquer A, Marzioni M, Marin JJG, Bujanda L and Banales JM:
Wnt-β-catenin signalling in liver development, health and disease.
Nat Rev Gastroenterol Hepatol. 16:121–136. 2019. View Article : Google Scholar : PubMed/NCBI
|