Introduction
Colorectal cancer (CRC) is one of the most common
malignancies worldwide, and results in ~600,000 deaths per year
(1). The 5-year overall survival
(OS) rate for patients with early-stage CRC is ~90%; however, in
patients with advanced CRC OS is only ~10% (2). Outcomes of patients with CRC may be
improved by targeting key molecules associated with progression of
CRC (3). Therefore, improving our
understanding of carcinogenesis of CRC may assist in the design of
novel molecular targeting strategies.
MicroRNAs (miRNAs) are single-stranded non-coding
RNAs that can suppress gene expression primarily through binding to
the 3′-untranslated regions (3′-UTRs) of target mRNAs (4). miRNAs are able to interact with
multiple mRNAs, influencing various cellular functions (5). Studies have demonstrated that miR-190b
is abnormally expressed in multiple types of cancer and functions
as an either oncogene or tumor suppressor (6–10). For
instance, miR-190b upregulation modulated human Wilms' tumor cell
growth, invasion, migration and apoptosis by targeting PTEN
(6), and miR-190b expression was
significantly enhanced in hormone-dependent breast cancer (7). Furthermore, miR-190b overexpression
resulted in downregulation of insulin-like growth factor and
results in poorer OS of patients with hepatocellular carcinoma
(8). Contrary to these studies,
miR-190b was demonstrated to exhibit tumor suppressive roles in
osteosarcoma and gastric cancer (9,10).
miR-190b inhibited growth of osteosarcoma cells and induced
apoptosis by regulating B-cell lymphoma-2 (9). In addition, miR-190b was found to be
downregulated in radiotherapy-resistant gastric cancer cells, and
miR-190b overexpression decreased cell viability and enhanced
radio-sensitivity (10). However,
there has been limited study of the role of miR-190b in CRC.
The aim of the present study was to determine the
significance of miR-190b in CRC. miR-190b expression in CRC cell
lines was compared with a normal colon epithelial cell line.
Furthermore, we explored the role of miR-190b in regulating
proliferation of CRC cells, colony formation and invasion, as well
as the mechanism underlying its effects.
Materials and methods
Cell lines and cell culture
Human CRC cell lines HT29, SW480, SW620 and the
normal colon epithelial cell line, FHC, were purchased from
American Type Culture Collection. These cell lines were
authenticated by ATCC using STR profiling and associated methods.
The CRC cells were cultured in RPMI-1640 medium (Invitrogen; Thermo
Fisher Scientific, Inc.), whereas the FHC cells were cultured in
DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with
100 U/ml Penicillin, 50 mg/ml streptomycin, and 10% FBS (all from
Gibco; Thermo Fisher Scientific, Inc.) at 37°C with a humidified
atmosphere containing 5% CO2.
Cell transfection
miR-190b inhibitor (5′-AACCCAAUAUCAAACAUAUCA-3′),
negative control (NC) for miR-190b inhibitor (NC-inhibitor;
5′-UUCUCCGAACGUGUCACGU-3′), small interfering (si)RNA for forkhead
box protein P2 (si-FOXP2; 5′-CGACAGAGACAAUAAGCAACA-3′) and NC-siRNA
(5′-GAAACCAAACGACGACAGUAA-3′) were synthetized by Shanghai GeneChem
Co., Ltd. pcDNA3.1 containing an open reading frame of FOXP2 was
purchased from GenScript. Transfection was performed by mixing
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) with synthesized miRNAs (50 nmol/l), siRNAs (20
nmol/l), or pFOXP2 (4 µg) according to manufacturer's protocols.
Cells were used for subsequent experiments after 48 h of
transfection.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cultured cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. miR-190b expression
levels were detected using an ABI 7500 (Applied Biosystems Thermo
Fisher Scientific, Inc.) using One Step TB Green®
PrimeScript™ RT-PCR Kit (Takara Bio, Inc.) after transcribing the
extracted RNA into complementary DNA (cDNA) using MMLV Reverse
Transcriptase First Strand cDNA Synthesis kit (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 15 min, 85°C for 5s, and 4°C
for 60 min. The sequences of the primers used were as follows:
miR-190b forward, 5′-GGGTGATATGTTTGATAT-3′ and reverse,
5′-CAGTGCGTGTCGTGGAGT-3′; U6 small nuclear RNA forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′;
FOXP2 forward, 5′-AACAACAGCAGGCTCTCCAG-3′ and reverse,
5′-GGCACCTGCAGTGGTCTC-3′; and GAPDH forward,
5′-GGTGGTCTCCTCTGACTTCAACA-3′ and reverse,
5′-GTTGCTGTAGCCAAATTCGTTGT-3′. Expression levels of miR-190b or
FOXP2 were calculated using the 2−∆∆Cq method with U6
snRNA or GAPDH as endogenous controls (11). Experiments were performed in
triplicate. The thermocycling conditions were as follows: Initial
denaturation at 95°C for 30s, followed by 40 cycles of 95°C for 10s
and 56°C for 30s.
Protein extraction and western
blotting
Total protein was isolated from cultured cells using
RIPA lysis buffer supplemented with protease inhibitor (both from
Beyotime Institute of Biotechnology). Protein concentration was
measured using a bicinchoninic acid quantitative detection reagent
kit (Beyotime Institute of Biotechnology). Equal quantities (50 µg)
of protein samples were loaded on to a 10% SDS gel and resolved
using SDS-PAGE, and subsequently transferred to PVDF membranes
(Beyotime Institute of Biotechnology). Membranes were blocked with
non-fat milk at 4°C for 2 h and then incubated with primary
antibodies (both supplied by Abcam) against FOXP2 (1:1,000, cat.
no. ab207587) or GAPDH (1:1,000, cat. no. ab181602) at 4°C
overnight. Membranes were then incubated with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody (1:5,000,
cat. no. ab6721; Abcam) at room temperature for 1 h. Signals were
developed using BeyoECL Star (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. Experiments were
performed in triplicate.
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay was used to measure the cell
proliferative capacity. A total of 3×103 cells/well were
seeded in a 96-well plate and cultured for 0, 24, 48 or 72 h after
seeding. CCK-8 reagent (Beyotime Institute of Biotechnology) was
added to the plate and the cells were incubated for another 2 h.
Absorbance was measured at 450 nm using a Varioskan LUX multimode
reader (Thermo Fisher Scientific, Inc.). Experiments were performed
in triplicate.
Colony formation assay
A total of 300 cells/well were seeded in a 6-well
plate and cultured for 2 weeks at 37°C. Cell colonies were stained
with crystal violet at room temperature for 30 min and the numbers
of colonies in five randomly chosen fields were counted under an
IX73 inverted microscope (Olympus Corporation) at a magnification
of ×200. Experiments were performed in triplicate.
Transwell invasion assay
A total of 1×105 cells in FBS-free medium
were seeded into the upper chamber of Matrigel pre-coated inserts
(Becton, Dickinson and Company). The lower chamber was filled with
RPMI-1640 medium supplemented with 10% FBS. After 48 h of
incubation, noninvasive cells were removed while the invasive cells
were fixed with methanol at room temperature for 30 min and stained
with 0.5% crystal violet at room temperature for 15 min. Invasive
cell numbers were counted under an IX73 inverted microscope at a
magnification of ×200 from five randomly chosen fields to assess
the effects of miR-190b and FOXP2. Experiments were performed in
triplicate.
Apoptosis analysis
Annexin V-fluorescein isothiocyanate/propidium
iodide (PI) Apoptosis Staining kit (Beyotime Institute of
Biotechnology) was used to measure cell apoptosis according to the
manufacturer's instructions. Cells were digested with 0.25% trypsin
and stained with Annexin V and PI for 5 min in the dark at room
temperature. Cell apoptosis was analyzed using a BD LSRFortessa
flow cytometer (Becton, Dickinson and Company) with the equipped
FACS Diva version 6.0 software (Becton, Dickinson and Company).
Experiments were performed in triplicate.
Dual-luciferase activity reporter
assay
TargetScan version 7.2 (targetscan.org/vert_72/) analysis was performed to
determine the potential targets of miR-190b and a total of 223
genes were identified. Among these genes, FOXP2 was selected for
following studies. Wild-type 3′-UTR of FOXP2 was amplified from the
human CRC cell genome with the primers (Sense,
5′-GGTACCCTTTACAAACAGTTTTGACAG-3′, and antisense,
5′-AAGCTTTGGTGTGAATGATGACTGG-3′) and cloned into pGL3 luciferase
vector (Promega Corporation) and termed pGL3-FOXP2-wt. A
site-direct mutagenesis kit (Takara Bio, Inc.) was used mutate the
FOXP2 3′-UTR using the primers (Fragment 1, forward
5′-CAGCGATCGCGAACTGACTTGTGAAACCTCAGCG-3′, reverse,
5′-CTCGCAGTTACTTCCAGTCCCTCAAAGCC-3′; and fragment 2, forward
5′-GTCTTTGGGTCATGATCAACGAACCGG-3′ and reverse,
5′-TATGTTTAAACTTTATAAATGGGTCAAAAAGAATTAGA-3′) and this was termed
pGL3-FOXP2-mut. Cells were co-transfected with pGL3-FOXP2-mut or
pGL3-FOXP2-wt combined with miR-190b inhibitor or NC-inhibitor
using Lipofectamine® 2000. Luciferase activity was
measured using a Dual-Luciferase reporter system (Promega
Corporation) after 48 h of transfection.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism version 6.0 (GraphPad Software Inc). Data are presented as
the mean ± standard deviation. Differences were analyzed using a
Student's t-test for two groups or an ANOVA with a post hoc Tukey's
test for ≥3 groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
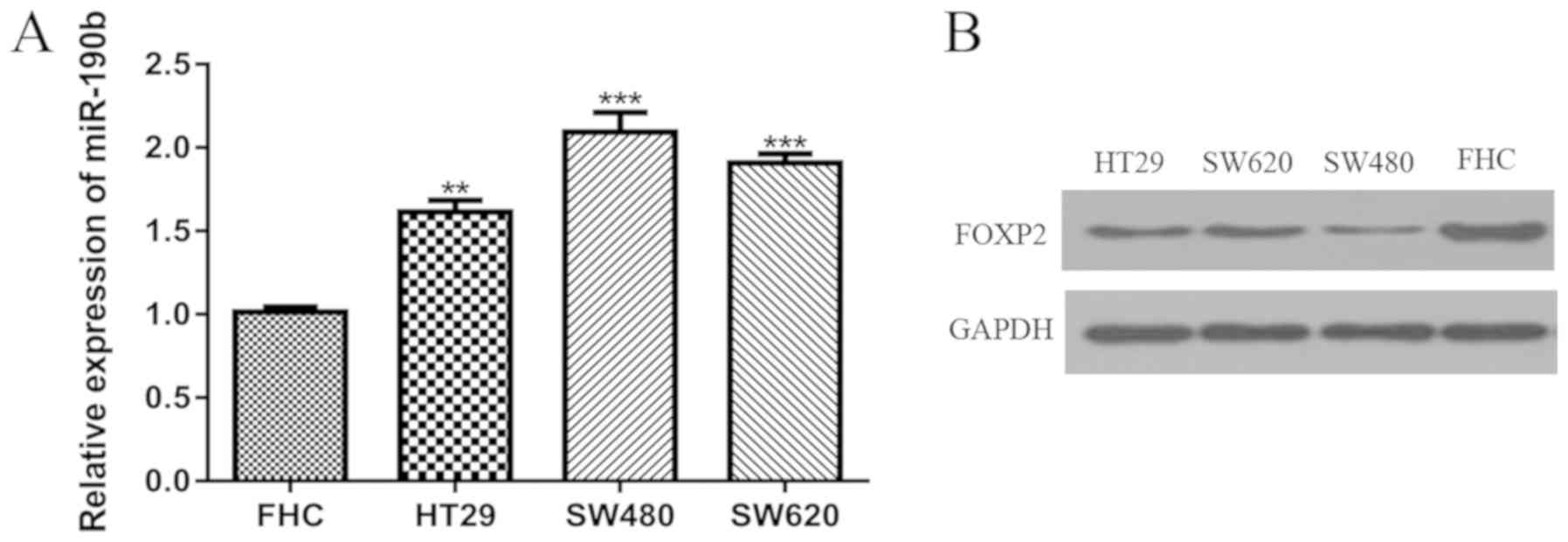
miR-190b expression is upregulated,
whereas FOXP2 expression is downregulated in CRC cell lines
miR-190b expression was significantly higher in the
CRC cell lines compared with the FHC cell line (Fig. 1A). Conversely, FOXP2 expression
appeared to be lower in the CRC cell lines compared the FHC cell
line as determined by western blotting (Fig. 1B). Therefore, the SW480 and SW620
cell lines were selected for the subsequent experiments as they
have the highest and second-highest miR-190b expression level but
lowest and second-lowest FOXP2 expression level.
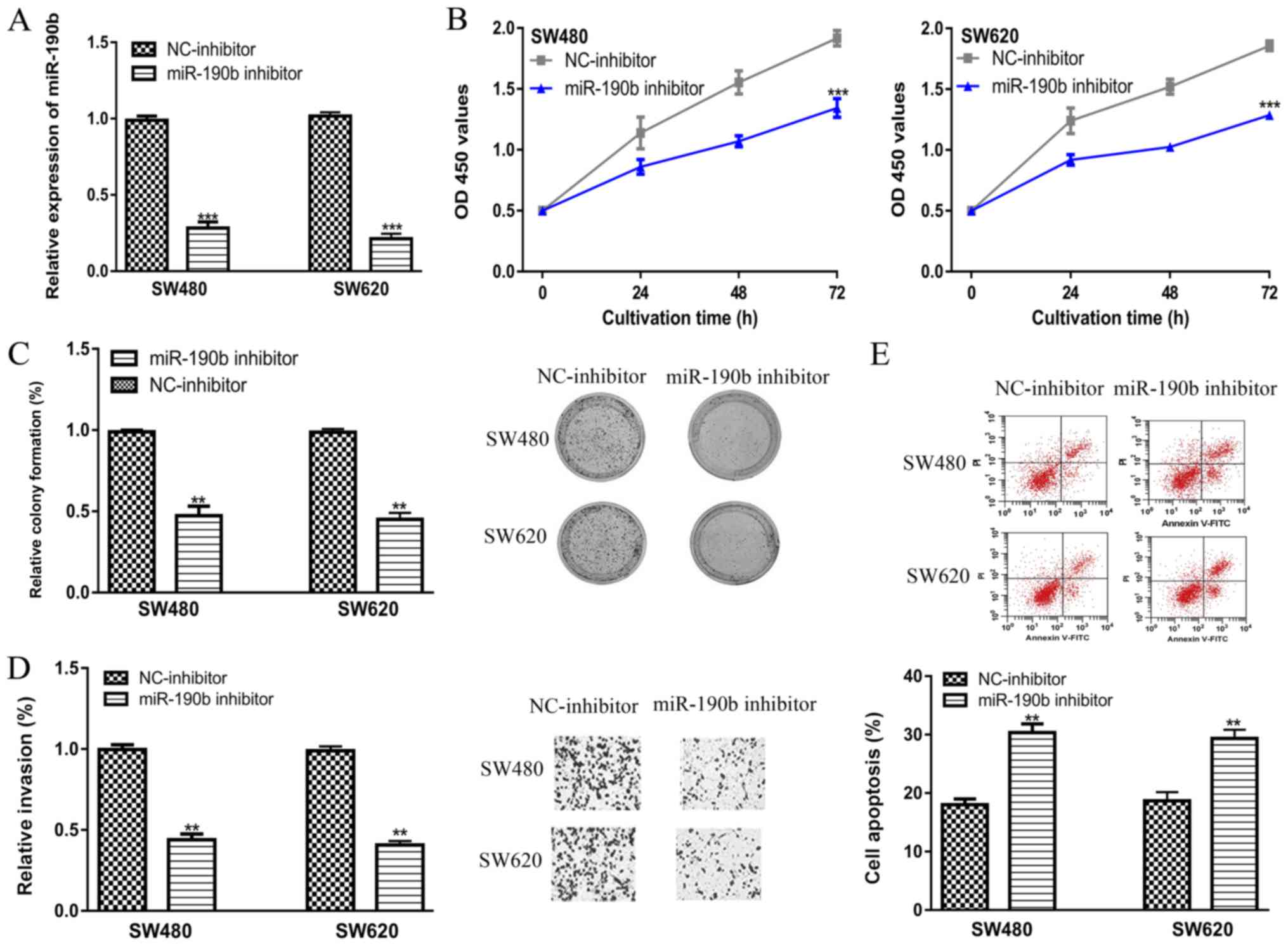
Downregulation of miR-190b reduces
cell proliferation, colony formation and cell invasion, and
increases apoptosis
RT-qPCR analysis showed that transfection of the
miR-190b inhibitor significantly decreased miR-190b levels compared
with the NC-inhibitor (Fig. 2A).
CCK-8 assay revealed that the knockdown of miR-190b was able to
inhibit cell proliferation (Fig.
2B). Colony formation assay confirmed the results of CCK-8
assay, which showed the introduction of miR-190b inhibitor has a
long-term impact on cell proliferation (Fig. 2C). Transwell invasion showed cell
invasion ability was inhibited by miR-190b inhibitor (Fig. 2D). Furthermore, flow cytometry showed
cell apoptosis was enhanced by miR-190b inhibitor transfection,
which further validated the results of CCK-8 assay and clony
formation assay (Fig. 2E).
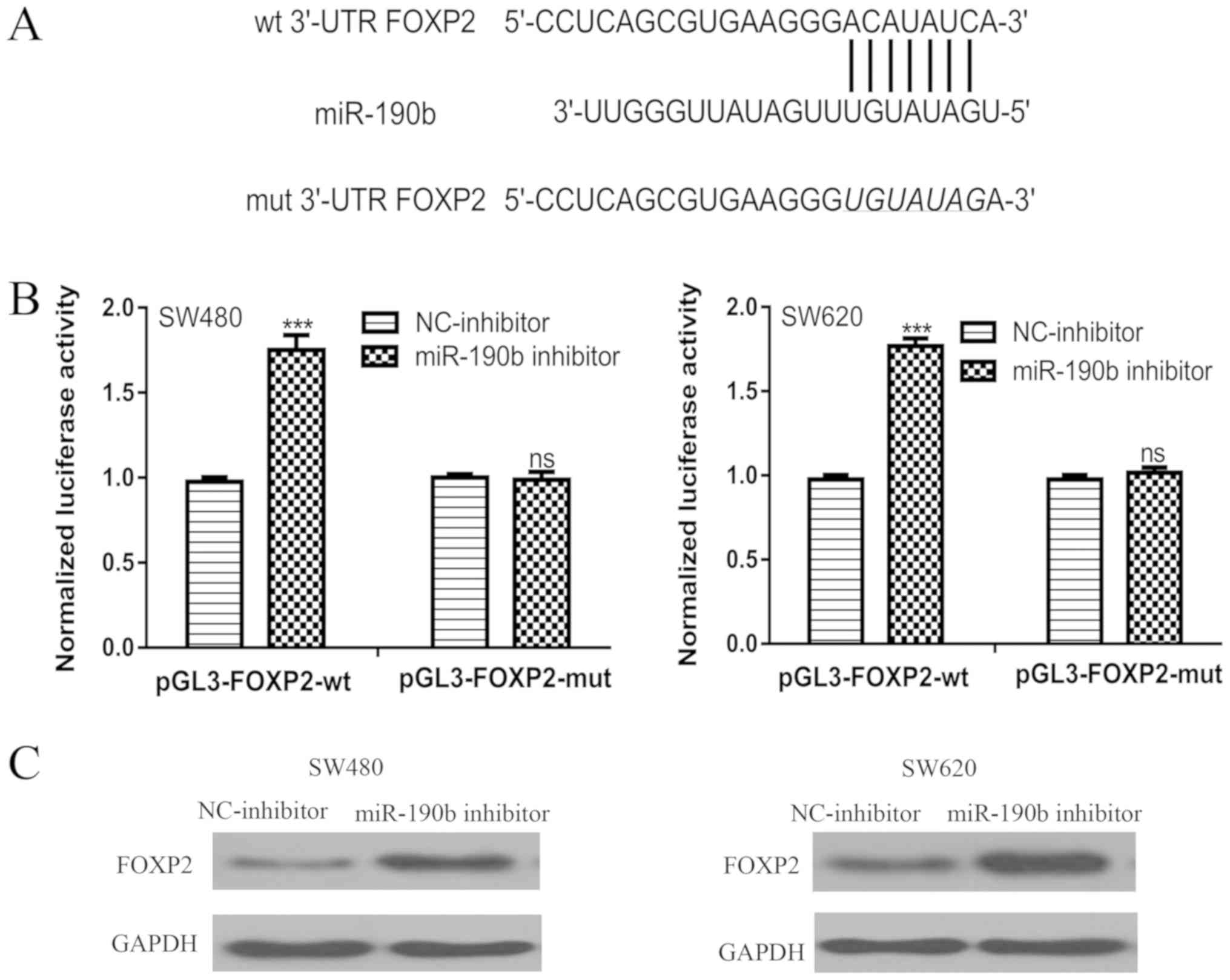
FOXP2 is a direct target of
miR-190b
TargetScan was used to identify candidate targets of
miR-190b. A conserved binding site within the 3′-UTR of FOXP2 for
miR-190b was identified (Fig. 3A). A
luciferase activity reporter assay showed that transfection of the
miR-190b inhibitor increased the luciferase activity of
pGL3-FOXP2-wt, but not pGL3-FOXP2-mut (Fig. 3B). Western blotting showed that FOXP2
protein expression levels were enhanced by transfection of the
miR-190b inhibitor in CRC cell lines (Fig. 3C).
FOXP2 is involved in miR-190b mediated
cell behaviors
Transfection of si-FOXP2 transfection decreased both
the mRNA and protein expression levels of FOXP2 in CRC cells
(Fig. 4A and B). The effect of
miR-190b inhibitor on FOXP2 expression was abolished by si-FOXP2
(Fig. 4A). FOXP2 downregulation
enhanced CRC cell proliferation, colony formation and cell
invasion, but decreased apoptosis compared with the NC-siRNA
(Fig. 4C-F). Overexpression of FOXP2
decreased CRC cell proliferation, colony formation and cell
invasion, but increased apoptosis compared with transfection with
the pcDNA3.1 empty vector (Fig.
4C-F). In addition, rescue experiments revealed that the
inhibitory effects of miR-190b inhibitor on CRC cell behaviors
could be partially reversed by si-FOXP2 (Fig. 4C-F).
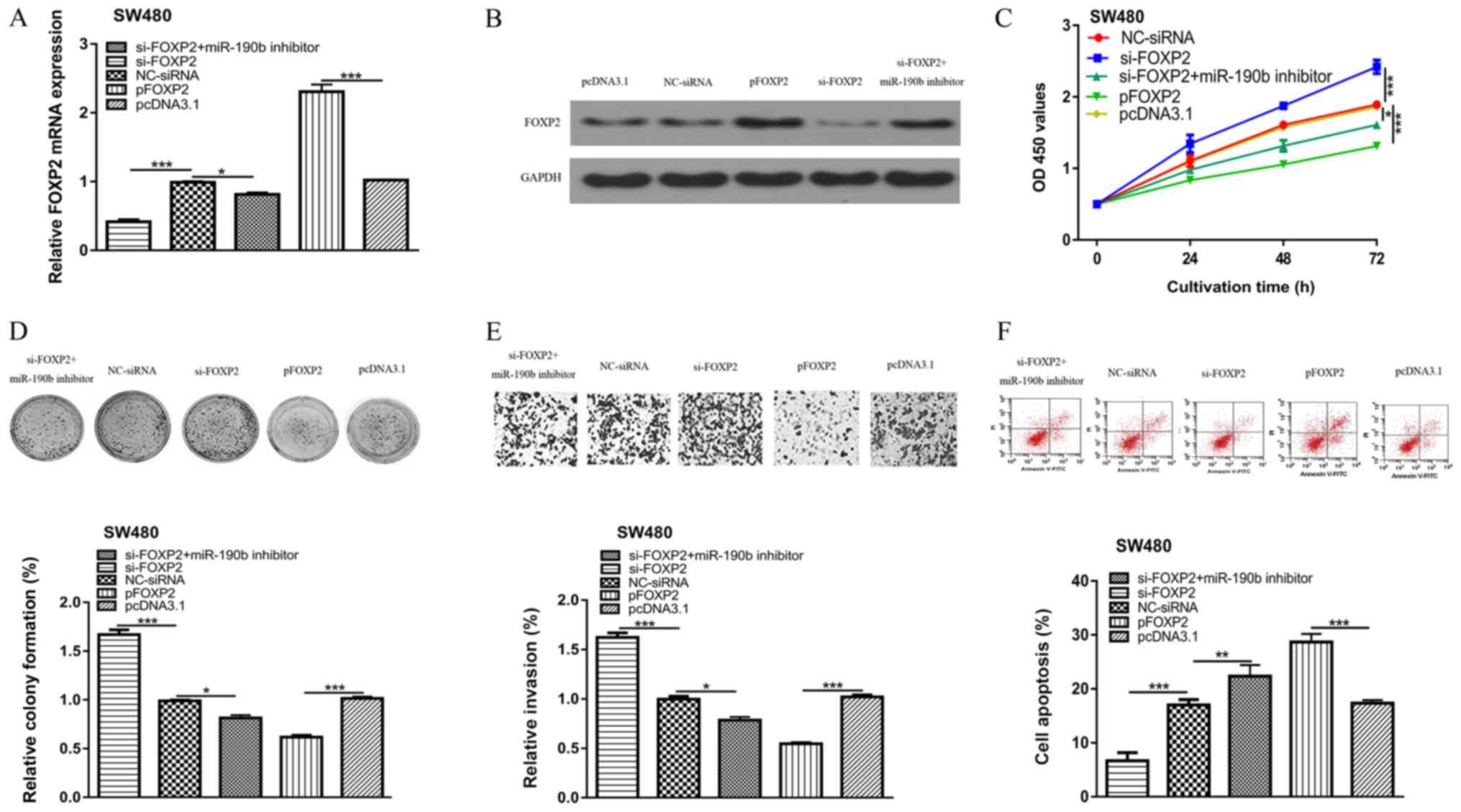 | Figure 4.FOXP2 is involved in miR-190b-mediated
alterations to CRC cell behavior. FOXP2 (A) mRNA and (B) protein
expression levels were reduced in cells transfected with si-FOXP2,
and increased in cells transfected with the FOXP2 overexpression
vector. (C) Cell proliferation, (D) colony formation, (E) cell
invasion (×200) and (F) cell apoptosis in colorectal cancer cell
transfected with si-FOXP2, NC-siRNA, si-FOXP2 and miR-190b
inhibitor, pcDNA3.1, or pFOXP2. *P<0.05, **P<0.01,
***P<0.001. miR-190b, microRNA-190b; FOXP2, forkhead box protein
P2; si, small interfering; NC, negative control. |
Discussion
Advancements in the understanding of CRC
tumorigenesis have identified biomarkers to improve early diagnosis
and molecular targeted therapies (3,12).
Numerous studies have highlighted the importance of
cancer-associated miRNAs in the initiation and progression of CRC
(13,14). Li et al (13) demonstrated that miR-452 was
upregulated in CRC and its upregulation activated the Wnt/β-catenin
signaling pathway, which promoted cancer progression in
vitro and in vivo (13).
Another study by Liu et al (14) revealed that miR-7702 acted as a tumor
suppressor in CRC by targeting transcriptional adaptor 1 (14).
In the present study, miR-190b expression was found
significantly upregulated in the assessed CRC cell lines compared
with the normal colon epithelial cell line, FHC. Functional assays
showed that downregulation of miR-190b reduced proliferation,
colony formation and invasion of CRC cells, whilst increasing
apoptosis in vitro. These results suggest that miR-190b
functions as an oncogenic miRNA during progression of CRC, which is
in consistent with its role in Wilms' tumor, breast cancer, and
hepatocellular carcinoma (6–8). It is well documented that miRNAs
function as tumor suppressors or oncogenes by regulating the
expression of their specific target genes (3,8–10,13,14). For
example, genes including PTEN, IGF1, Bcl-2 have been identified as
targets for miR-190b and involved in the tumor suppressive or
oncogenic roles of miR-190b in cancers (6,8–10). To identify the potential target of
miR-190b, TargetScan bioinformatical analysis tool was employed,
and we found FOXP2 was a putative target for miR-190b as it has
previously been demonstrated to be abnormally expressed in a number
of different types of cancer (15–17).
FOXP2 downregulation promotes migration and invasion of breast
cancer by influencing the tumor growth factor β/SMAD signaling
pathway, indicating the tumor suppressive role of FOXP2 (15). FOXP2 was also found to be regulated
by miRNAs including miR-196b and miR-23a in different types of
cancer (16,17). Here, FOXP2 was validated as a direct
target of miR-190b through a luciferase activity reporter assay and
western blotting. Knockdown of FOXP2 expression resulted in the
stimulation effects on the malignancy behaviors associated with
progression of CRC, whereas the reverse effects were observed when
FOXP2 was overexpressed. These results indicated that FOXP2
functions as a tumor suppressor gene in CRC progression, which is
in consistent with its roles reported in other cancer types
(15–17). Additionally, rescue experiments
showed that FOXP2 could reverse the inhibitory effects of miR-190b
on CRC cells. miR-190b has opposing roles, dependent on the type of
cancer (6–10), whereas FOXP2 functions as a tumor
suppressor irrespective of the cancer (15–17). The
results of the present study demonstrated that a miR-190b/FOXP2
axis contributed to the determination of cancer cell behavior. A
limitation of the present study was the lack of in vivo
studies to validate the conclusions of the in vitro studies.
Animal models should be utilized to validate whether targeting the
expression of miR-190b is an effective strategy for cancer
treatment.
In summary, the present study demonstrated that
miR-190b promoted proliferation, colony formation and cell invasion
of CRC cells by negatively regulating the expression of FOXP2. The
results demonstrated that miR-190b functioned as an oncogene in
CRC. To the best of our knowledge, this is the first study to
report the significance of miR-190b in CRC, and may thus lay a
foundation to establish miR-190b as a therapeutic target for CRC in
the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ, CL, QC, XL and CZ contributed to study design.
QZ, CL, QC, XL, QW and CZ performed experiments and data analysis.
QZ and CZ were major contributors in writing the manuscript. All
author have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yiu AJ and Yiu CY: Biomarkers in
colorectal cancer. Anticancer Res. 36:1093–1102. 2016.PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
An NN, Shawn J, Peng JP, Wu MD and Huang
LG: Up-regulation of miR-190b promoted growth, invasion, migration
and inhibited apoptosis of Wilms' tumor cells by repressing the
PTEN expression. Eur Rev Med Pharmacol Sci. 22:961–969.
2018.PubMed/NCBI
|
|
7
|
Cizeron-Clairac G, Lallemand F, Vacher S,
Lidereau R, Bieche I and Callens C: MiR-190b, the highest
up-regulated miRNA in ERα-positive compared to ERα-negative breast
tumors, a new biomarker in breast cancers? BMC Cancer. 15:4992015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hung TM, Ho CM, Liu YC, Lee JL, Liao YR,
Wu YM, Ho MC, Chen CH, Lai HS and Lee PH: Up-regulation of
microRNA-190b plays a role for decreased IGF-1 that induces insulin
resistance in human hepatocellular carcinoma. PLoS One.
9:e894462014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kang M, Xia P, Hou T, Qi Z, Liao S and
Yang X: MicroRNA-190b inhibits tumor cell proliferation and induces
apoptosis by regulating Bcl-2 in U2OS osteosarcoma cells.
Pharmazie. 72:279–282. 2017.PubMed/NCBI
|
|
10
|
Wang C and Qiao C: MicroRNA-190b confers
radio-sensitivity through negative regulation of Bcl-2 in gastric
cancer cells. Biotechnol Lett. 39:485–490. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee JH, Hwang I, Kang YN, Choi IJ and Kim
DK: Genetic characteristics of mitochondrial DNA was associated
with colorectal carcinogenesis and its prognosis. PLoS One.
10:e01186122015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li T, Jian X, He H, Lai Q, Li X, Deng D,
Liu T, Zhu J, Jiao H, Ye Y, et al: MiR-452 promotes an aggressive
colorectal cancer phenotype by regulating a Wnt/β-catenin positive
feedback loop. J Exp Clin Cancer Res. 37:2382018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu H, Li D, Fang H and Ning J:
Species-specific function of microRNA-7702 in human colorectal
cancer cells via targeting TADA1. Am J Transl Res. 10:2579–2589.
2018.PubMed/NCBI
|
|
15
|
Chen MT, Sun HF, Li LD, Zhao Y, Yang LP,
Gao SP and Jin W: Downregulation of FOXP2 promotes breast cancer
migration and invasion through TGFβ/SMAD signaling pathway. Oncol
Lett. 15:8582–8588. 2018.PubMed/NCBI
|
|
16
|
Yu Z, Lin X, Tian M and Chang W:
microRNA-196b promotes cell migration and invasion by targeting
FOXP2 in hepatocellular carcinoma. Oncol Rep. 39:731–738.
2018.PubMed/NCBI
|
|
17
|
Diao H, Ye Z and Qin R: miR-23a acts as an
oncogene in pancreatic carcinoma by targeting FOXP2. J Investig
Med. 66:676–683. 2018. View Article : Google Scholar : PubMed/NCBI
|