Introduction
Endometriosis, a benign disease with malignant
properties, is defined by the presence of active endometrial cells
outside the uterus, including in the pelvis, abdominal cavity,
intestines, rectovaginal septum, abdominal wall and myometrium
(also known as adenomyosis) (1,2).
Patients afflicted with endometriosis are associated with higher
occurrences of anxiety and depression (3). In addition, women with endometriosis
are more likely to experience non-menstrual pelvic pain (36.7% vs.
14.3%), dyspareunia (29.5% vs. 13.4%) and infertility (11.6% vs.
3.4%) compared with women without endometriosis (4). The current gold standard treatments for
endometriosis are surgical resection and hormone suppression;
however, none of these therapies are ideal due to their various
side effects and high recurrence rates (5). In order to improve the treatment
strategies, it is important to study the underlying mechanisms
involved in disease development and progression. In addition,
identifying reliable molecular markers can aid in the diagnosis and
treatment of endometriosis.
The upregulation and downregulation of genes
associated with disease susceptibility serves an important role in
the progression of endometriosis (6). With the development of next-generation
sequencing, gene expression microarrays have been widely performed
to identify the differentially expressed genes (DEGs) that may be
involved in the development and progression of endometriosis
(5,7). However, due to the different sample
size, technology detection platforms and inconsistent data
processing methods across different studies, the DEGs identified in
previous studies are inconsistent or even contradictory. Thus,
there are certain limitations in using a single gene expression
profile. Integrated bioinformatics analysis has emerged as a
promising tool for exploring the molecular markers and signaling
pathways involved in diseases, and has previously been applied to
study ovarian cancer (8), breast
cancer (9) and non-small cell lung
cancer (10).
In the present study, four microarray expression
datasets, namely GSE11691 (11),
GSE23339 (12), GSE25628 (13) and GSE78851 (14) were downloaded from the Gene
Expression Omnibus (GEO) database of the National Center for
Biotechnology Information. A total of 58 samples, including 27
cases of endometriosis and 31 normal samples, were included in the
present study. Firstly, the gene expression profiles were
normalized, and the DEGs were then identified using the limma
package function of R software. Subsequently, Gene Ontology (GO)
enrichment analysis of DEGs was performed on Database for
Annotation, Visualization and Integrated Discovery (DAVID), while
the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were
analyzed via the KOBAS online analysis database. Finally, a
protein-protein interaction (PPI) network was constructed using the
STRING online database. Cytoscape software was applied for further
visualization. The current study identified key signaling pathways
and potential candidate genes involved in the development of
endometriosis, which may facilitate a better understanding of the
underlying molecular mechanisms and provide effective targets for
the diagnosis and treatment of this disease.
Materials and methods
Gene expression data
The keyword ‘endometriosis’ was used to search the
GEO database (http://www.ncbi.nlm.nih.gov/geo), and the gene
expression profiles of GSE11691 (11), GSE23339 (12), GSE25628 (13) and GSE78851 (14) were downloaded. The dataset GSE11691,
based on the platform GPL96 [HG-U133A] Affymetrix Human Genome
U133A Array (Affymetrix; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), included 9 endometriosis tissues and 9 normal endometrial
tissue samples. The platform for GSE23339 was GPL6102 (Illumina
Human-6 v2.0 expression beadchip; Illumina, Inc., San Diego, CA,
USA), including 10 endometrioma samples and 9 control endometrium
specimens. GSE25628 included 7 samples of ectopic endometrioma and
6 samples of normal endometrial tissue, and its platform was GPL571
[HG-U133A_2] Affymetrix Human Genome U133A 2.0 Array (Affymetrix;
Thermo Fisher Scientific, Inc.). The platform for GSE78851,
consisting of 3 tissues from patients with adenomyosis and 5 normal
tissues, was GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST
Array [transcript (gene) version] (Affymetrix; Thermo Fisher
Scientific, Inc.). The platform and series matrix files were
downloaded as CSV files. The dataset information is displayed in
Table I.
 | Table I.Details of GEO endometriosis
data. |
Table I.
Details of GEO endometriosis
data.
| Author (year) | Sample | GEO | Platform | Normal | Endometriosis | (Ref.) |
|---|
| Hull et al
(2008) | Endometrium | GSE11691 | GPL96 | 9 | 9 | (11) |
| Hawkins et
al (2011) | Endometrium | GSE23339 | GPL6102 | 9 | 10 | (12) |
| Crispi et al
(2013) | Endometrium | GSE25628 | GPL571 | 6 | 7 | (13) |
| Herndon et
al (2016) | Endometrium | GSE78851 | GPL6244 | 3 | 5 | (14) |
Data processing
The gene IDs within each gene expression profile was
converted into a gene symbol, and then the data were
log2 transformed and normalized using R 5.3.1
(https://www.r-project.org/). DEGs
between endometriosis and non-endometriosis samples were screened
out under the thresholds of |log2 fold change (FC)|>1
and P<0.05 using the limma package in the Bioconductor 3.9 tool
(http://www.bioconductor.org/packages/release/bioc/html/limma.html).
The volcano map of the DEGs and the heatmap of the top 200 DEGs in
each microarray datasets were obtained using R.
Integration of microarray data
SangerBox 1.0.8 (http://sangerbox.com/) is a computerized and powerful
software for biological information analysis, and is used as a
visualization tool. The robust rank aggregation (RRA) method can be
applied as a useful and general solution for gene list integration
and meta-analysis in an unbiased manner, using a probabilistic
model to make the algorithm parameter free and robust to outliers,
noise and errors, and to assign a significance score to each gene
(15). The RRA method can rank each
item in each list and compare this ranking with the baseline case
where all preference lists are randomly ordered. The P-value can
represent the rank location, with a smaller P-value indicating a
higher gene rank. In the present study, RRA in SangerBox was
performed for comprehensive sorting of DEGs in the four gene
expression profiles. P<0.05 was set as the threshold, and DEGs
that were inconsistent across the four data sets were excluded.
Pathway enrichment analysis
GO analysis (16),
which is composed of biological process (BP), cellular compartment
(CC) and molecular function (MF) terms, is a common method for
large-scale genomic data function annotation. In order to better
understand the mechanism of DEGs involved in the development of
endometriosis, GO and KEGG pathway enrichment analyses were
performed using the DAVID 6.8 (https://david.ncifcrf.gov/) and the KOBAS 3.0
(http://kobas.cbi.pku.edu.cn/) online
analysis tool. P<0.05 was considered to indicate a statistically
significant difference in these analyses.
PPI network construction
The STRING database (http://string-db.org/) was used to identify the
interacting protein pairs among DEGs with the criterion of combined
score of ≥0.4. Upon removal of the isolated and partially connected
nodes, a complex network of DEGs was constructed. The file of
STRING interactions was downloaded and visualized with Cytoscape
3.7.0 (https://cytoscape.org/).
Immunohistochemistry
For immunohistochemical analysis, archival samples
of normal endometrial and endometriosis specimens were used. The
samples had been collected between May 2018 and December 2018 from
patients that underwent surgery at Renmin Hospital of Wuhan
University (Wuhan, China). The age of the females from which these
samples were collected ranged between 20 and 40 years old. The
present study was approved by the Ethics Committee of Renmin
Hospital of Wuhan University, Patients and their families signed an
informed consent form in advance. In short, six normal endometrial
and six endometriosis specimens were confirmed by a pathologist.
The tissue samples were cut into sections of 3 µm in thickness and
3 mm in diameter. Once the samples had been dewaxed, hydrated and
treated with sodium citrate (pH=6), hydrogen peroxide was used to
block any endogenous peroxidase activity. Immunohistochemical
staining was conducted with a rabbit polyclonal primary antibody
against HSPA5 (1:150; cat. no. ab108615; Abcam, Cambridge, MA,
USA), TJP1 (1:150; cat. no. 21773-1-AP; Wuhan Sanying
Biotechnology, Wuhan, China) and ENO2 (1:100; cat. no. ab79757;
Abcam) at 4°C overnight. Subsequently, the samples were incubated
with a horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (1:200; cat. no. AS-1107; Aspen) at 37°C for 50 min, and a
3,3′-diaminobenzidine solution and hematoxylin were then used for
staining and counterstaining at room temperate for 1 min. The
integrated option density was analyzed using the ImageJ software
(version 1.4.6; National Institutes of Health).
Results
Differential expression profiles
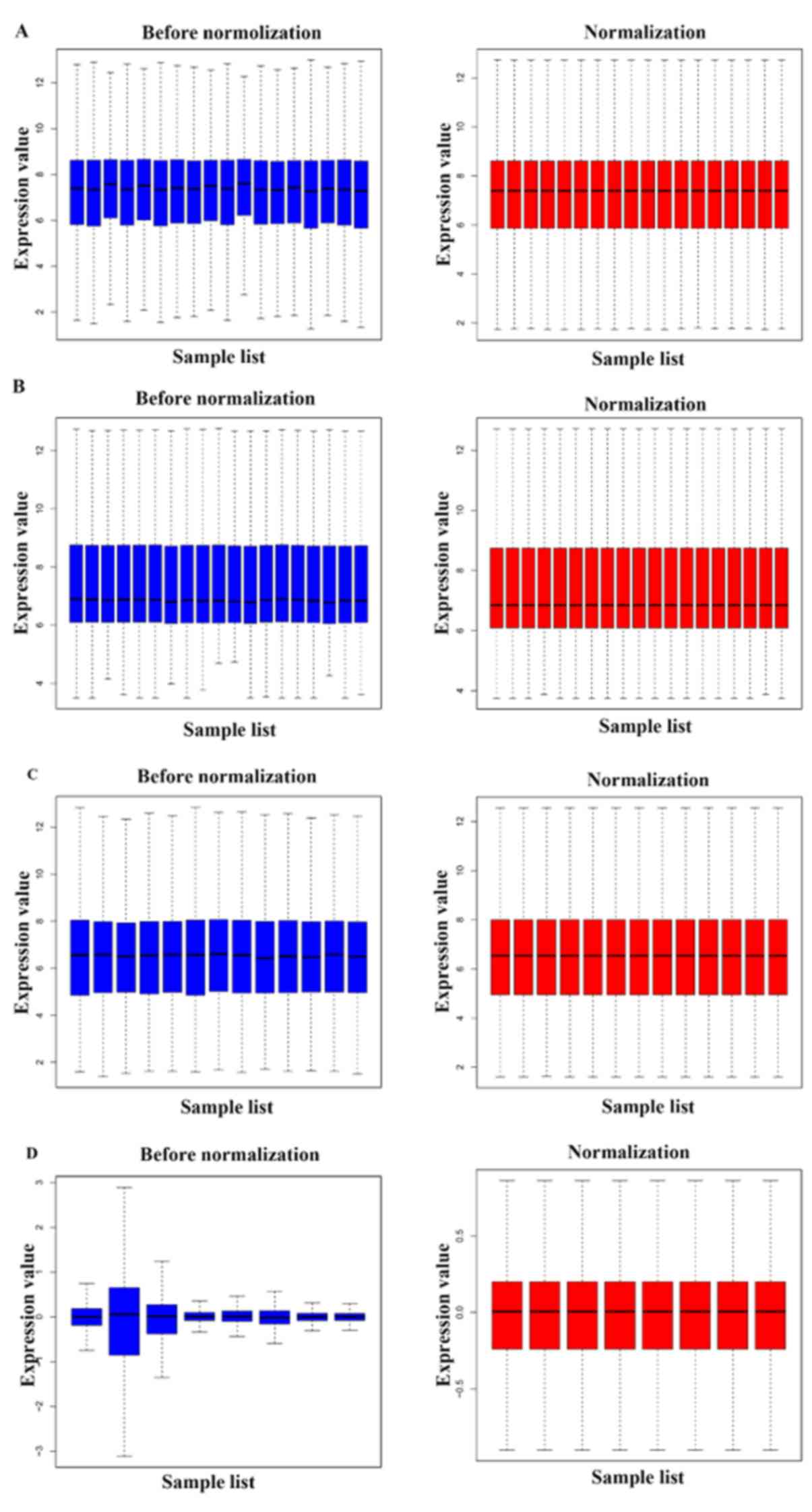
The gene expression profiles of the datasets
GSE11691, GSE23339, GSE25628 and GSE78851 were normalized, as shown
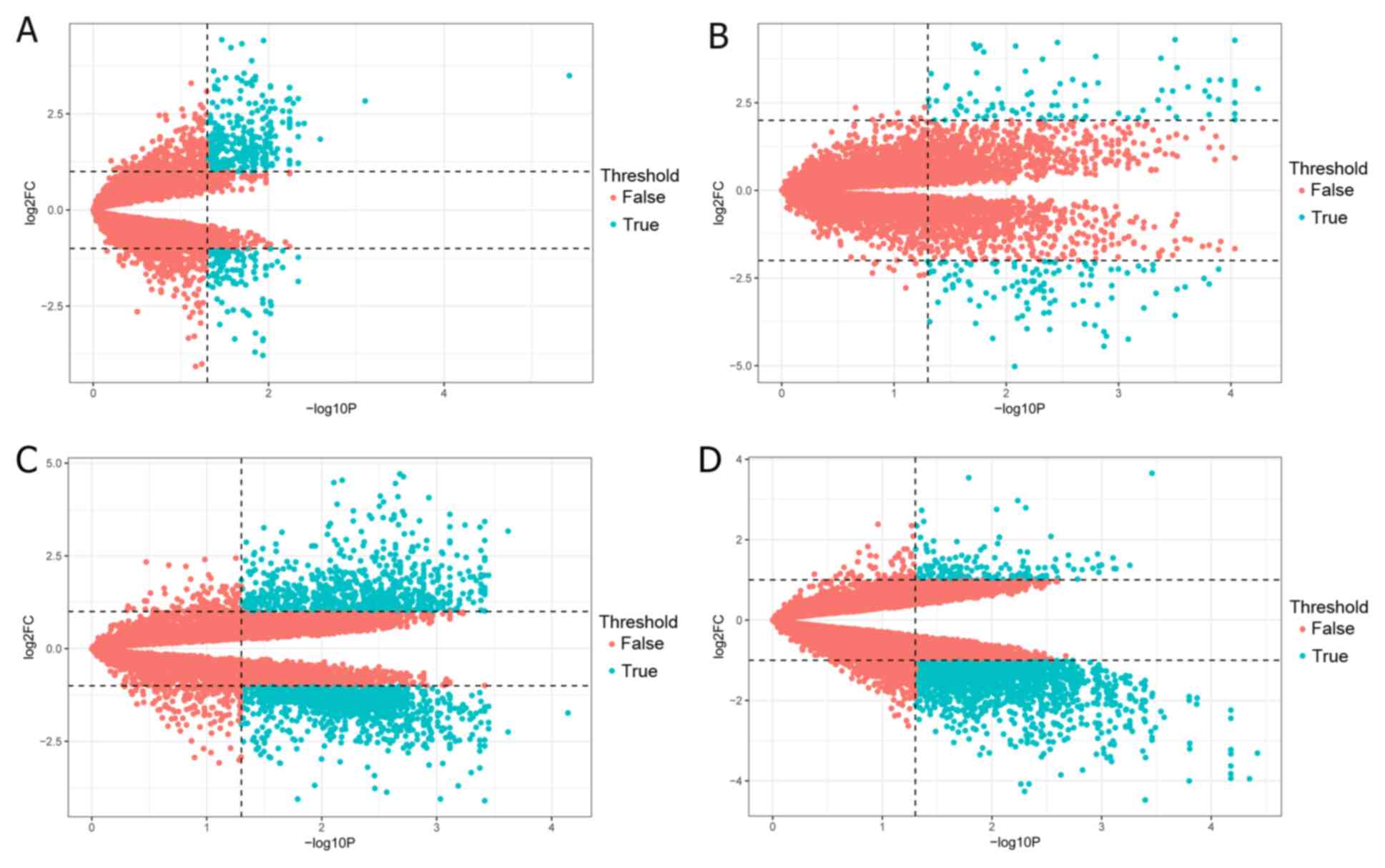
in Fig. 1. According to the criteria
of |log2FC|>1 and P<0.05, a total of 903 DEGs were
identified in GSE11691 using the limma R package, including 575
upregulated and 328 downregulated genes. A total of 1,139 DEGs were
identified from the GSE23339 dataset, including 608 upregulated and
531 downregulated genes. Additionally, 1,731 DEGs were identified
from the GSE25628 dataset, consisting of 708 upregulated and 1,023
downregulated genes, while there was a total of 2,118 DEGs in the
GSE78851 dataset, including 221 upregulated and 1,897 downregulated
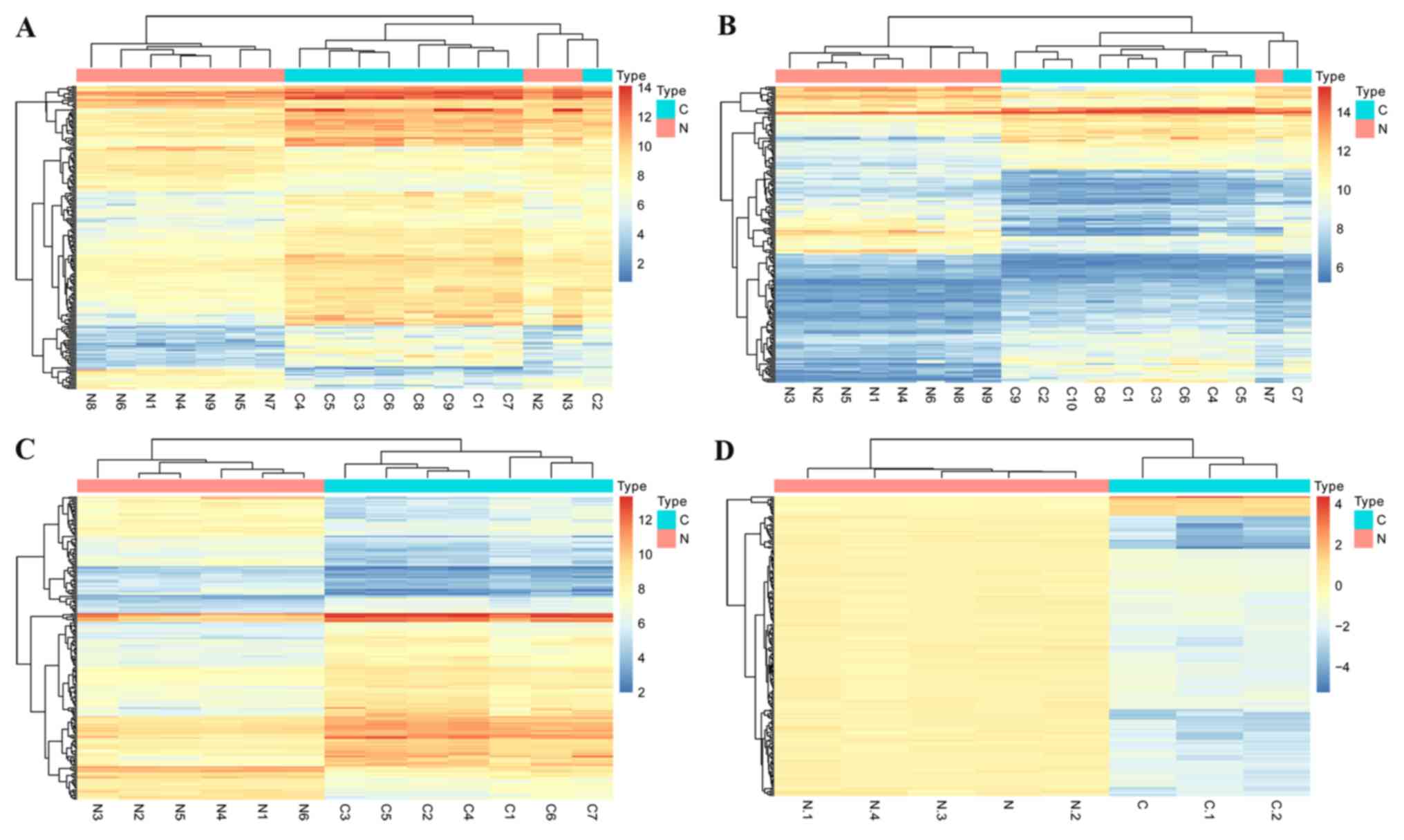
genes. Subsequently, the volcano plots for the identified DEGs and
the cluster heatmaps of the top 200 DEGs in each dataset were
constructed, and are presented in Figs.
2 and 3, respectively.
Identification of DEGs in
endometriosis using integrated bioinformatics analysis
The RRA method assumes that each gene in each
dataset is randomly arranged, which is widely used in integrated
bioinformatics analysis (17,18).
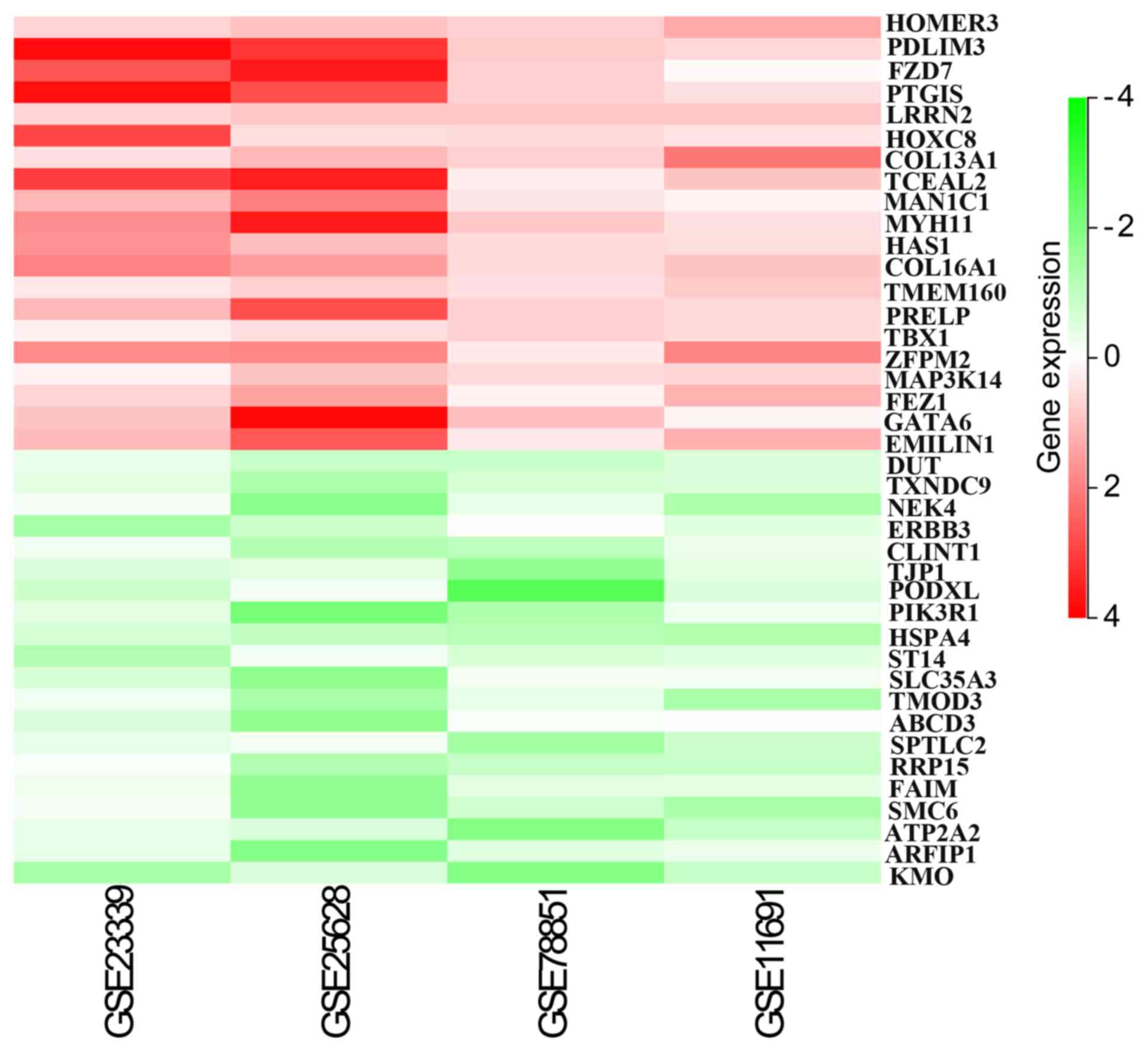
Through rank analysis (corrected P-value of <0.05), 275
integrated DEGs were identified. In order to obtain more reliable
DEGs, genes with inconsistent upregulation and downregulation in
the expression profiles were deleted. In total, 103 integrated
genes were identified, including 47 upregulated and 56
downregulated genes (Table II). The
top 20 upregulated and downregulated genes were represented on
heatmaps using Sanger Box software, as shown in Fig. 4.
 | Table II.Screening DEGs in endometriosis by
integrated microarray. |
Table II.
Screening DEGs in endometriosis by
integrated microarray.
| Expression | Genes |
|---|
| Upregulated
(n=47) | HOMER3, PDLIM3,
FZD7, PTGIS, LRRN2, HOXC8, COL13A1, TCEAL2, MAN1C1, MYH11, HAS1,
COL16A1, TMEM160, PRELP, TBX1, ZFPM2, MAP3K14, FEZ1, GATA6,
EMILIN1, FCN1, LRRC15, CAMK1G, DPEP2, C7, TRPC1, POU3F3, EHD3,
ROM1, TSSK2, DES, COL11A2, EEF1A2, ITGBL1, LRRC3, LAG3, STAB1,
HS3ST3A1, CDKN1C, ENO2, COL8A2, PRKG1, WWC3, ZFHX4, WISP1, SAP30,
RENBP |
| Downregulated
(n=56) | TSPAN1, CSTF3,
BTBD3, MYO6, HSPA5, TAF15, IER3IP1, MYO5C, NUCKS1, PDZD8, NUPL2,
SNAPC3, TTLL5, PPP1R2, ARFGAP3, NUP88, ADD3, NXT2, POLR1B, EP300,
PKP4, UGDH, PRR11, KMO, ZBTB24, MRPL39, SMAD5, IQGAP1, EXPH5,
SLC5A3, TNC, SUZ12, EIF1AX, NOC3L, MRPS31, TCF12, DUT, SPA17,
TXNDC9, NEK4, ERBB3, CLINT1, TJP1, PODXL, PIK3R1, HSPA4, SLC35A3,
ST14, TMOD3, ABCD3, SPTLC2, RRP15, FAIM, SMC6, ATP2A2,
ARFIP1 |
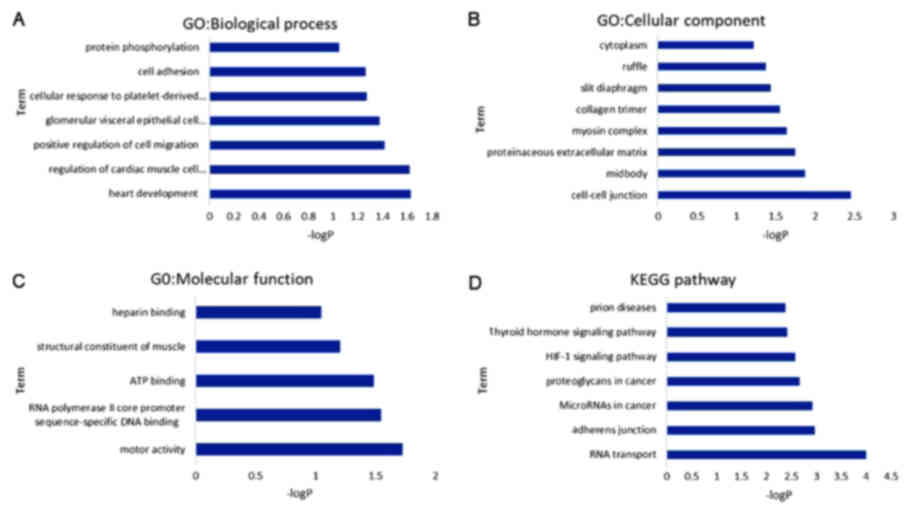
GO functional enrichment analysis
The GO functional analysis was divided into the BP,
MF and CC categories. As displayed in Fig. 5, the DEGs were mainly enriched in
cell adhesion, cell migration, cell-cell junction and heparin
binding in the GO function annotation. Furthermore, according to
the KEGG pathway analysis, the DEGs were mainly involved in
adherens junction and hypoxia-inducible factor (HIF)-1
signaling.
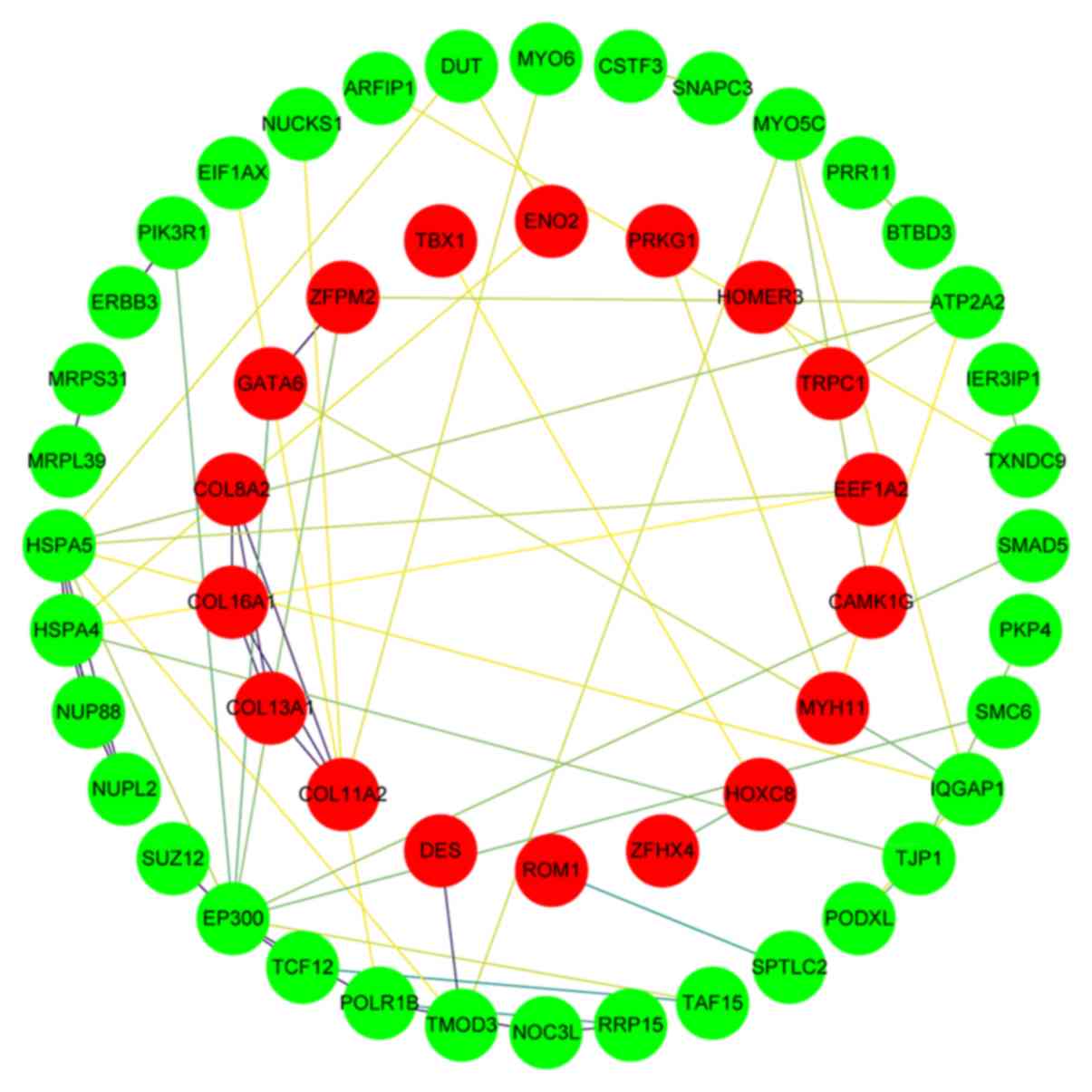
PPI network analysis
A PPI network was subsequently constructed, which
consisted of 54 nodes (proteins) and 62 edges (interactions), as
shown in Fig. 6. The genes showing
the most significant interaction in the network were PIK3R1,
ERBB3, MRPS31, HSPA5, ZFPM2, NUP88, SUZ12, MRPL39, HSPA4, GATA6,
NUPL2, and EP300.
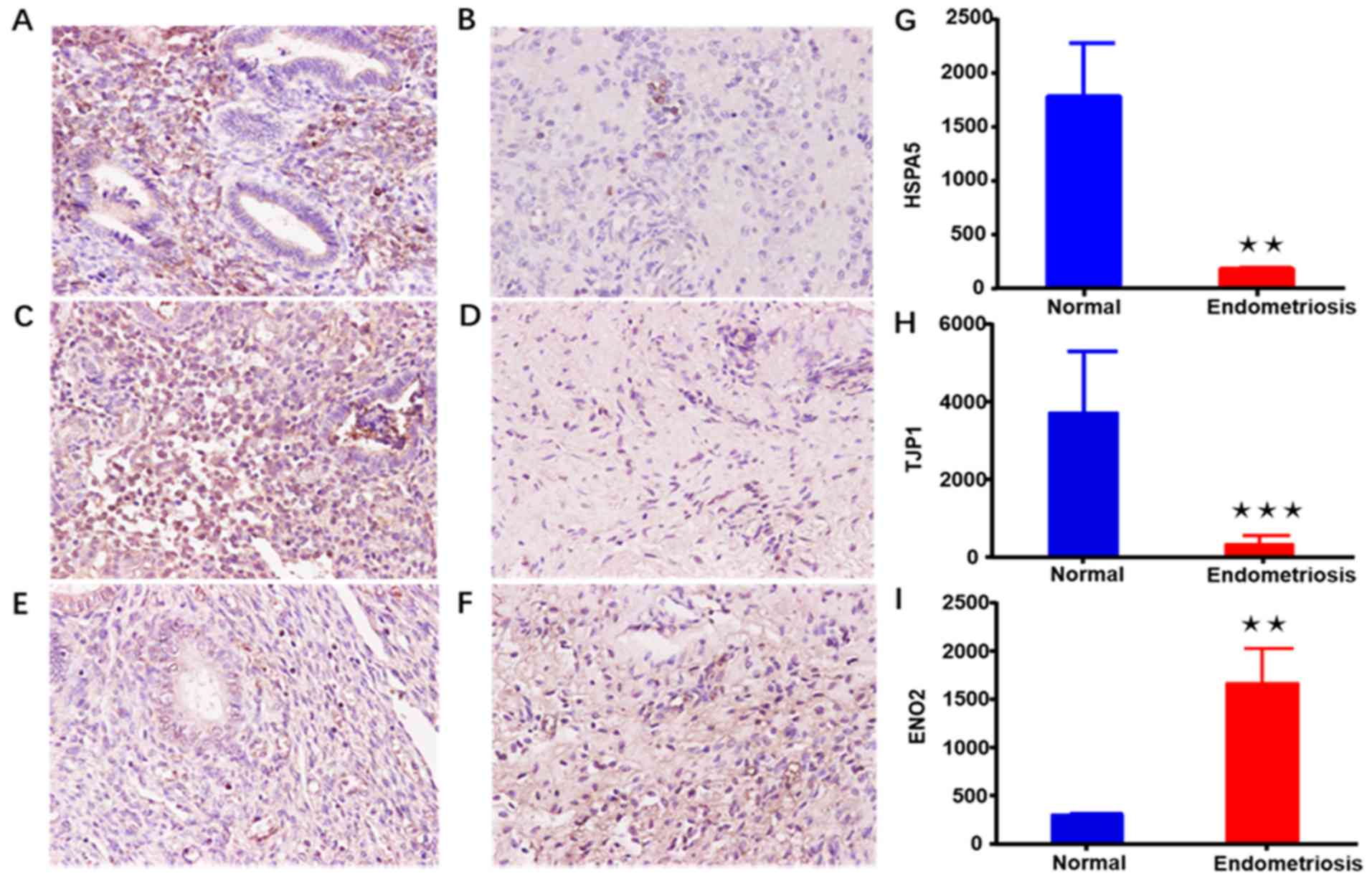
Immunohistochemistry
To further investigate whether the expression of the
identified genes in endometriosis tissues was consistent with the
bioinformatic analysis results, the expression of certain genes in
each pathway was randomly verified. HSPA5, ENO2 and
TJP1 are associated with cell migration, adherens junction
and the HIF-1 signaling pathway, respectively. As shown in Fig. 7, the findings of immunohistochemical
analysis verified that the expression levels of HSPA5 and
TJP1 were evidently reduced in endometriosis as compared
with that in normal tissues. However, ENO2 was significantly
upregulated in endometriosis, which was consistent with the
bioinformatics results.
Discussion
It is estimated that there are 176 million women
with endometriosis worldwide, and this condition seriously affects
10% of women of reproductive age (19). Chronic pelvic pain and infertility
cause great physical pain and mental distress to women with
endometriosis and their partners, greatly reducing the family
happiness index and increasing the domestic burden (20). Accumulating evidence suggests that
the endometrium of patients with endometriosis exhibits abnormal
molecular expression, which gives the tissue the ability to
implant, invade and develop into endometriosis lesions (21,22). In
order to identify more stable and reliable molecular markers, the
present study mapped out the genetic alterations that may be
involved in the development of endometriosis by integrated
bioinformatics analysis.
Four gene expression profile datasets from different
groups were integrated in the present study, and R software and
bioinformatics analysis were used to analyze these datasets. A
total of 103 DEGs were identified using the RRA analysis method,
including 47 upregulated and 56 downregulated genes. Furthermore,
through GO and KEGG analyses, these DEGs were found to be closely
associated with cell migration, adherens junction and the HIF-1
signaling pathway. The results revealed that the DEGs associated
with cell migration in endometriosis were PIK3R1, PODXL,
HSPA5 and LRRC15, while the genes IQGAP1, TJP1
and EP300 were involved in adherens junction. Notably, the
DEGs EP300, ENO2 and PIK3R1 were mainly associated
with the HIF-1 signaling pathway.
The most widely accepted theory for the development
of endometriosis is implantation and invasiveness. Accumulating
studies have indicated that the degradation of extracellular matrix
and the alteration of gene expression serve critical roles in the
pathophysiological processes of endometriosis (23,24). In
addition, PI3K/Akt signaling has been reported to be involved in
these processes (25). Rai and
Shivaji indicated that DJ-1 regulated cell proliferation, migration
and invasion in endometriotic epithelial cells via the PI3K/Akt
signaling pathway. In the present study, the findings demonstrated
that the gene PIK3R1 was downregulated and may be involved
in cell migration (26). In
addition, PODXL, HSPA5 and LRRC15 may also have
potential value in this process.
Intercellular junctions (including tight junctions
and adherens junctions) play a critical role in the endometrium.
The development of endometriosis is accompanied with changes in
cell-cell tight junctions (27).
Extensive research has demonstrated that claudin-3, claudin-4,
ZO-3, E-cadherin and α-catenin are downregulated in the ectopic
endometrium as compared with their expression in the corresponding
eutopic endometrium (28–30). In the present study, integrated
bioinformatics analysis revealed that the expression levels of
genes associated with the adherens junction pathway, namely
IQGAP1, TJP and EP300, were significantly reduced in
endometriosis.
In the last decade, researchers have indicated that
the expression of HIF-1α was higher in ectopic endometriosis tissue
as compared with that in eutopic tissue (31,32).
Furthermore, hypoxia can induced the invasion of endometrial
stromal cells and promoted the endometriosis-associated
angiogenesis (33,34). Additionally, the expression of HIF-1α
in the serum was reported to be proportional to the stage of
endometriosis and the severity of pain (32). Indeed, bioinformatics analysis in the
present study deonmonstrated that the expression of genes
associated with HIF-1α, such as ENO2, was upregulated in
endometriosis.
In conclusion, the present study revealed that cell
migration, adherens junction and the HIF-1 signaling pathway may be
involved in the development of endometriosis via integrated
bioinformatics analysis. In addition, these identified DEGs may be
of clinical significance for the diagnosis and treatment of the
endometriosis. However, as the present study is solely based on
data analysis and experimental verification, further studies with
larger samples and clinical trials are required to confirm the
function of the identified genes in endometriosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the China
Graduate School of Graduate Education Fund Project (grant. no.
B2-YX20180302-19) and the Wuhan University People's Hospital
Guidance Fund Project (grant. no. RMYD2018M05).
Availability of data and materials
All data generated or analyzed during this study are
included in the published article.
Authors' contributions
FFD and AYB conceived and designed the research.
XLP, SX and LZ collected the data. YQW, MQY and DYY conducted
literature research. FFD, ZHZ and SYL analyzed the database, and
prepared the diagrams. FFD drafted the manuscript, BL collected the
samples, YXC revised the article and provided funding. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Renmin Hospital of Wuhan University (Hubei, China). Patients who
participated in this research had complete clinical data. The
patients and their families signed an informed consent form in
advance.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jenkins S, Olive DL and Haney AF:
Endometriosis: Pathogenetic implications of the anatomic
distribution. Obstet Gynecol. 67:335–338. 1986.PubMed/NCBI
|
|
2
|
Krawczyk N, Banys-Paluchowski M, Schmidt
D, Ulrich U and Fehm T: Endometriosis-associated malignancy.
Geburtshilfe Frauenheilkd. 76:176–181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Laganà AS, La Rosa VL, Rapisarda AMC,
Valenti G, Sapia F, Chiofalo B, Rossetti D, Ban Frangež H, Vrtačnik
Bokal E and Vitale SG: Anxiety and depression in patients with
endometriosis: Impact and management challenges. Int J Womens
Health. 9:323–330. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fuldeore MJ and Soliman AM: Prevalence and
symptomatic burden of diagnosed endometriosis in the United States:
National estimates from a cross-sectional survey of 59,411 women.
Gynecol Obstet Invest. 82:453–461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu F, Lv X, Yu H, Xu P, Ma R and Zou K:
In search of key genes associated with endometriosis using
bioinformatics approach. Eur J Obstet Gynecol Reprod Biol.
194:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kobayashi H, Imanaka S, Nakamura H and
Tsuji A: Understanding the role of epigenomic, genomic and genetic
alterations in the development of endometriosis (review). Mol Med
Rep. 9:1483–1505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sha G, Wu D, Zhang L, Chen X, Lei M, Sun
H, Lin S and Lang J: Differentially expressed genes in human
endometrial endothelial cells derived from eutopic endometrium of
patients with endometriosis compared with those from patients
without endometriosis. Hum Reprod. 22:3159–3169. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang X, Zhu S, Li L, Zhang L, Xian S, Wang
Y and Cheng Y: Identification of differentially expressed genes and
signaling pathways in ovarian cancer by integrated bioinformatics
analysis. Onco Targets Ther. 11:1457–1474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Zhang Y, Huang Q and Li C:
Integrated bioinformatics analysis reveals key candidate genes and
pathways in breast cancer. Mol Med Rep. 17:8091–8100.
2018.PubMed/NCBI
|
|
10
|
Ni M, Liu X, Wu J, Zhang D, Tian J, Wang
T, Liu S, Meng Z, Wang K, Duan X, et al: Identification of
candidate biomarkers correlated with the pathogenesis and prognosis
of non-small cell lung cancer via integrated bioinformatics
analysis. Front Genet. 9:4692018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hull ML, Escareno CR, Godsland JM, Doig
JR, Johnson CM, Phillips SC, Smith SK, Tavaré S, Print CG and
Charnock-Jones DS: Endometrial-peritoneal interactions during
endometriotic lesion establishment. Am J Pathol. 173:700–715. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hawkins SM, Creighton CJ, Han DY, Zariff
A, Anderson ML, Gunaratne PH and Matzuk MM: Functional microRNA
involved in endometriosis. Mol Endocrinol. 25:821–832. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Crispi S, Piccolo MT, D'Avino A, Donizetti
A, Viceconte R, Spyrou M, Calogero RA, Baldi A and Signorile PG:
Transcriptional profiling of endometriosis tissues identifies genes
related to organogenesis defects. J Cell Physiol. 228:1927–1934.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herndon CN, Aghajanova L, Balayan S,
Erikson D, Barragan F, Goldfien G, Vo KC, Hawkins S and Giudice LC:
Global transcriptome abnormalities of the eutopic endometrium from
women with adenomyosis. Reprod Sci. 23:1289–1303. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kolde R, Laur S, Adler P and Vilo J:
Robust rank aggregation for gene list integration and
meta-analysis. Bioinformatics. 28:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiong DD, Dang YW, Lin P, Wen DY, He RQ,
Luo DZ, Feng ZB and Chen G: A circRNA-miRNA-mRNA network
identification for exploring underlying pathogenesis and therapy
strategy of hepatocellular carcinoma. J Transl Med. 16:2202018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao X, Chen Y, Chen M, Wang S, Wen X and
Zhang S: Identification of key candidate genes and biological
pathways in bladder cancer. PeerJ. 6:e60362018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rogers PA, D'Hooghe TM, Fazleabas A,
Gargett CE, Giudice LC, Montgomery GW, Rombauts L, Salamonsen LA
and Zondervan KT: Priorities for endometriosis research:
Recommendations from an international consensus workshop. Reprod
Sci. 16:335–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Simoens S, Dunselman G, Dirksen C,
Hummelshoj L, Bokor A, Brandes I, Brodszky V, Canis M, Colombo GL,
DeLeire T, et al: The burden of endometriosis: Costs and quality of
life of women with endometriosis and treated in referral centres.
Hum Reprod. 27:1292–1299. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mehasseb MK, Panchal R, Taylor AH, Brown
L, Bell SC and Habiba M: Estrogen and progesterone receptor isoform
distribution through the menstrual cycle in uteri with and without
adenomyosis. Fertil Steril. 95:2228–2235, 2235 e1. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aghajanova L, Velarde MC and Giudice LC:
Altered gene expression profiling in endometrium: Evidence for
progesterone resistance. Semin Reprod Med. 28:51–58. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kobayashi H: Invasive capacity of
heterotopic endometrium. Gynecol Obstet Invest. 50 (Suppl
1):S26–S32. 2000. View Article : Google Scholar
|
|
24
|
Saare M, Krigul KL, Laisk-Podar T,
Ponandai-Srinivasan S, Rahmioglu N, Lalit Kumar PG, Zondervan K,
Salumets A and Peters M: DNA methylation alterations-potential
cause of endometriosis pathogenesis or a reflection of tissue
heterogeneity? Biol Reprod. 99:273–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng T and Yang J: Differential
expression of EWI-2 in endometriosis, its functional role and
underlying molecular mechanisms. J Obstet Gynaecol Res.
43:1180–1188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rai P and Shivaji S: The role of DJ-1 in
the pathogenesis of endometriosis. PLoS One. 6:e180742011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grund S and Grümmer R: Direct cell-cell
interactions in the endometrium and in endometrial pathophysiology.
Int J Mol Sci. 19(pii): E22272018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan XY, Li X, Weng ZP and Wang B: Altered
expression of claudin-3 and claudin-4 in ectopic endometrium of
women with endometriosis. Fertil Steril. 91:1692–1699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sohler F, Sommer A, Wachter DL, Agaimy A,
Fischer OM, Renner SP, Burghaus S, Fasching PA, Beckmann MW,
Fuhrmann U, et al: Tissue remodeling and nonendometrium-like
menstrual cycling are hallmarks of peritoneal endometriosis
lesions. Reprod Sci. 20:85–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shaco-Levy R, Sharabi S, Benharroch D,
Piura B and Sion-Vardy N: Matrix metalloproteinases 2 and 9,
E-cadherin, and beta-catenin expression in endometriosis, low-grade
endometrial carcinoma and non-neoplastic eutopic endometrium. Eur J
Obstet Gynecol Reprod Biol. 139:226–232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhan L, Wang W, Zhang Y, Song E, Fan Y and
Wei B: Hypoxia-inducible factor-1alpha: A promising therapeutic
target in endometriosis. Biochimie. 123:130–137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang F, Liu XL, Wang W, Dong HL, Xia YF,
Ruan LP and Liu LP: Expression of MMIF, HIF-1α and VEGF in serum
and endometrial tissues of patients with endometriosis. Curr Med
Sci. 38:499–504. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiong W, Zhang L, Xiong Y, Liu H and Liu
Y: Hypoxia promotes invasion of endometrial stromal cells via
hypoxia-inducible factor 1α upregulation-mediated β-catenin
activation in endometriosis. Reprod Sci. 23:531–541. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Becker CM, Rohwer N, Funakoshi T, Cramer
T, Bernhardt W, Birsner A, Folkman J and D'Amato RJ:
2-methoxyestradiol inhibits hypoxia-inducible factor-1{alpha} and
suppresses growth of lesions in a mouse model of endometriosis. Am
J Pathol. 172:534–544. 2008. View Article : Google Scholar : PubMed/NCBI
|