Introduction
According to the global cancer statistics in 2018,
ovarian cancer was the 8th most diagnosed cancer type and a leading
cause of cancer-related death for women worldwide (1). Surgical removal and chemotherapy are
two current approaches to treat patients with ovarian cancer;
however, the efficacy of these treatments is limited due to the
development of drug resistance and recurrence of cancer (2,3).
Although numerous previous clinical and experimental studies have
provided novel insight into the molecular mechanisms of ovarian
cancer, patients with advanced-stages of ovarian cancer still have
a poor prognosis (4,5). There is an urgent need to further
understand the molecular mechanisms of ovarian cancer for the
development of targeted therapeutic strategies.
MicroRNAs (miRNAs/miRs) are small, non-coding,
single-stranded nucleotides which are ubiquitously expressed in
eukaryotic cells (6).
Mechanistically, miRNAs directly bind to the 3′untranslated region
(UTR) of their target mRNAs, leading to the degradation of mRNA or
inhibition of translation (7). The
expression of miRNA is tightly controlled in normal cells, for
instance, dysregulation of several key miRNAs resulted in the
disruption of cell signaling networks in human diseases, including
cancer (8–10). With microarray analysis, many
differentially expressed miRNAs are detected between ovarian tumors
and normal tissues (11,12). miRNAs are identified as tumor
suppressors or oncogenes based on their potential prognostic
predictor value (13). For example,
the expression level of miR-21 in serum was considered as a
biomarker for the early detection and prediction of prognosis for
patients with ovarian cancer in 2013 (14). Later experimental studies
demonstrated that miR-21 modulates drug sensitivity by targeting
several key genes (15,16). Most recently, patients have been
accurately diagnosed with ovarian cancer from the expression of 10
miRNAs, which is deemed as a diagnostic model (17). However, how these miRNAs contribute
to the progression of ovarian cancer is unknown.
The v-src avian sarcoma (Schmidt-Ruppin A-2) viral
oncogene homolog (Src) is a well-characterized oncogenic tyrosine
kinase that is frequently upregulated in cancer (18). In ovarian cancer, activation of the
Src signaling pathway is crucial for the epithelial-mesenchymal
transition (EMT) process and in vivo tumor growth in nude
mice (19). Previous studies have
demonstrated that Src is associated with the activity of MAPK/ERK
signaling and PI3K/AKT signaling in cancer cells, which are pivotal
for cell proliferation and survival (20,21). Src
kinase signaling inhibitor 1 (SRCIN1) functions as a tumor
suppressor via inactivation of Src in cancer (22).
In the present study, miR-665 levels were
upregulated in tumor tissues from patients with ovarian cancer
compared with normal tissues. Inhibition of miR-665 inhibited cell
proliferation and colony forming ability of ovarian cancer cells.
SRCIN1 was predicted and validated as a target gene of miR-665.
Silencing of SRCIN1 could reverse the miR-665 inhibitor-induced
cell growth arrest. Moreover, miR-665 levels were negatively
correlated with SRCIN1 mRNA levels in tumor tissues from patients
with ovarian cancer. In conclusion, the present data suggested that
miR-665 functioned as an oncogene in ovarian cancer by directly
repressing the expression of SRCIN1. The present results may
provide novel insight into clinically relevant treatments for
ovarian cancer.
Materials and methods
Collection of tumor and normal
tissues
In total, 40 pairs of tumor tissues and normal
tissues were collected from female patients (aged from 29 to 71
years, with a median age of 53 years) with ovarian cancer who
underwent surgery at The Cancer Hospital Affiliated to Xinjiang
Medical University during June 2015 to July 2018. Written consent
was provided by all the participants before enrollment in the
present study. Patients who received chemotherapy or radiotherapy
prior to surgery were excluded. The Ethic Committee of Xinjiang
Medical University approved the present study. The tumor tissues
and normal tissues were collected during surgery removal, and were
immediately snap-frozen in liquid nitrogen before RNA extraction
and reverse transcription-quantitative PCR (RT-qPCR) were
performed.
Cell culture
The human ovarian cancer cell lines SKOV3 and ES2
were purchased from The Type Culture Collection of The Chinese
Academy of Sciences. All cell lines were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (HyClone; GE Healthcare Life Sciences) in a humidified
incubator with 5% CO2. A normal human ovarian surface
epithelial (HOSE) cell line was established by following a
previously reported method (23).
Fresh ovarian scrapings obtained from patients during the surgery
described above were immortalized with human papilloma virus 16
E6/E7 oncogenes. The cells were maintained in mammary epithelial
cell growth medium (BulletKit™; Clonetics; Lonza Group, Ltd.)
supplemented with 1% FBS (HyClone; GE Healthcare Life
Sciences).
RNA extraction and RT-qPCR
Total RNA was extracted from the tissues of the
patients and SKOV3 and ES2 cells using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. RNA was reverse transcribed to
first-stranded cDNA with PrimeScript™ First Strand cDNA Synthesis
kit (Takara Bio, Inc.). The reverse transcription conditions were
as follows: 37°C for 15 min and 85°C for 5 sec. RT-qPCR was
performed with SYBR Premix Ex Taq (Takara Bio, Inc.) on a CFX96
Touch Real-time PCR Detection System (Bio-Rad Laboratories, Inc.).
The thermocycling conditions were as follows: 95°C for 30 sec,
followed by 35 cycles of 95°C for 5 sec and 60°C for 30 sec. U6 and
GAPDH served as internal controls for miRNA and mRNA, respectively.
The relative expression of genes was calculated using the
2−ΔΔCq method (24). The
primer sequences were as follows: SRCIN1-forward:
5′-GAGGCTCGCAACGTCTTCTAC-3′; SRCIN1-reverse:
5′-GCGATGCGTACACCATCTCTC-3′; GAPDH-forward:
5′-GGAGCGAGATCCCTCCAAAAT-3′; GAPDH-reverse:
5′-GGCTGTTGTCATACTTCTCATGG-3′; stem-loop primer:
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAGGGGCC-3′; miR-665-forward:
5′-GCCGAGACCAGGAGGCUGA-3′; miR-665-reverse:
5′-CTCAACTGGTGTCGTGGA-3′; U6-forward:
5′-GCTTCGGCAGCACATATACTAAAAT-3′; and U6-reverse:
5′-CGCTTCACGAATTTGCGTGTCAT-3′.
Downregulation and upregulation of
miR-665 in ovarian cancer cells
miR-665 inhibitor (5′-AGGGGCCUCAGCCUCCUGGU-3′),
miR-665 mimic (5′-ACCAGGAGGCUGAGGCCCCU-3′) and the corresponding
negative controls (miR-NC; 5′-UCGCUUGGUGCAGGUCGGGAA-3′) were
synthesized and purchased from Shanghai GenePharma Co., Ltd. SKOV3
and ES2 cells were seeded into each well of 24-well plates
(2×105 cells per well) and were transfected with miR-665
inhibitor, miR-665 mimic, miR-NC inhibitor or miR-NC mimic at a
concentration of 50 nM with Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol, and maintained for 48 h before any
subsequent experiments were performed.
Silencing of SRCIN1 in ovarian cancer
cells
Control siRNA and SRCIN1 siRNA were synthesized and
purchased from Shanghai GenePharma Co., Ltd. The sequences were as
follows: Control siRNA sequence: 5′-UUCUCCGAACGUGUCACGUTT-3′; and
SRCIN1 siRNA sequence: 5′-CGGGAGAGAGGCAGGCUCUGUCGGAATT-3′. For the
silencing of SRCIN1, 50 nM SRCIN1 siRNA was transfected into SKOV3
and ES2 cells in 24-well plates (2×105 cells per well)
using Lipofectamine® RNAiMax (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. After 48
h, the cells were collected for western blotting.
Western blotting
SRCIN1 (cat. no. ab244527; 1:1,000) and GAPDH (cat.
no. ab8245; 1:5,000) antibodies were bought from Abcam. AKT (cat.
no. 4685; 1:1,000), phosphorylated (p)-AKT (cat. no. 4060;
1:1,000), ERK1/2 (cat. no. 4695; 1:1,000) and p-ERK1/2 (cat. no.
4370; 1:1,000) primary antibodies were purchased from Cell
Signaling Technology, Inc. HRP-conjugated secondary antibodies
against rabbit (cat. no. SA00001-2; 1:10,000) and mouse (cat. no.
SA00001-1; 1:10,000) were products of ProteinTech Group, Inc.
Protein lysates were prepared from cells with RIPA lysis buffer
(Sigma-Aldrich; Merck KGaA). The protein concentration was detected
with a BCA Protein Assay kit (Pierce; Thermo Fisher Scientific,
Inc.). For the western blotting, 20 µg protein was loaded on 8%
gels and separated by SDS-PAGE. After electrophoresis, the proteins
were transferred from SDS-PAGE gels to PVDF membranes. The
membranes were then blocked in 5% non-fat milk at room temperature
for 1 h. The membrane was subsequently incubated with the
appropriate primary antibody at 4°C overnight and the appropriate
secondary antibody at room temperature for 2 h. The blots were
developed with ECL western blotting substrate (Pierce; Thermo
Fisher Scientific, Inc.). The images of blots were analyzed using
ImageJ software version 1.6.0 (National Institutes of Health).
Cell proliferation assay
To determine the proliferation of cells, a Cell
Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) was
used according to the manufacturer's protocol. In total, 10,000
cells were plated in each well of 96-well plates. At 48 h after
transfection with miR-665 inhibitor or miR-NC inhibitor, 10 µl
CCK-8 solution was added into the well and incubated for another 2
h. Afterwards, the medium was transferred to another new 96-well
plate, and the absorbance at 450 nM was detected using a microplate
reader to detect the cell proliferation.
Colony forming assay
The colony forming assay was performed in a standard
procedure. A total of 2,000 cells were plated in each well on
6-well plates. After transfection with miR-665 inhibitor or miR-NC
inhibitor with or without control small interfering RNA (siRNA) or
SRCIN1 siRNA, the cells were incubated for 10 days to form cell
colonies. The culture medium was discarded, and the cells were
washed with PBS. The cells were fixed with 4% paraformaldehyde
(Beijing Solarbio Science & Technology Co., Ltd.) for 1 h at
room temperature. After that, the cell colonies were stained with
Crystal Violet Staining Solution (Beyotime Institute of
Biotechnology) for 20 min at room temperature. The staining
solution was then discarded and the wells were washed with PBS.
Images were captured using an inverted microscope in three random
fields for each well (×10). The colony numbers were counted using
ImageJ version 1.6.0 (National Institutes of Health).
Dual luciferase reporter assay
The full length of SRCIN1 3′UTR was amplified from
SKOV3 cDNA with TransFast® Taq DNA Polymerase (TransGen
Biotech Co., Ltd.) and ligated into a pGL3 plasmid (Promega
Corporation). The PCR conditions were as follows: 94°C for 3 min,
followed by 35 cycles of 94°C for 5 sec; 55°C for 15 sec and 72°C
for 10 sec. The primer sequences were: SRCIN1-forward:
5′-CTCTAGAAAGCCCCTCACCCCGCTG-3′; SRCIN1-reverse:
5′-CTCTAGAGAAGGAGAUCCAGGAGAG-3′. A total of three site mutations
were introduced into the pGL3-SRCIN1-wild-type (WT) plasmid to
establish the pGL3-SRCIN1-mutant (Mut). miR-665 mimic or miR-NC (50
nM) in combination with 2 µg pGL3-SRCIN1-WT or pGL3-SRCIN1-Mut were
transfected into cells with Lipofectamine® 3000 and
incubated for 48 h. The relative luciferase activity was detected
with the Dual Luciferase Reporter assay system (Promega
Corporation) according to the manufacturer's protocol. Firefly
luciferase was normalized to Renilla luciferase.
Bioinformatic analysis
The potential target genes of miR-665 were predicted
using TargetScan software V7.2 (http://www.targetscan.org/vert_72/).
Statistical analysis
All data were analyzed using GraphPad Prism 5.0
(GraphPad Software, Inc.) and are presented as the mean ± SD.
Differences between two groups were compared with Student's t-test,
and differences among three groups were compared with one-way
ANOVA, followed by Newman-Keuls analysis. The association between
SRCIN1 mRNA levels and miR-665 expression was analyzed with Pearson
correlation analysis. P<0.05 was considered to indicate a
statistically significant difference. All experiments were
performed at least three times.
Results
miR-665 is upregulated in ovarian
cancer tissues and cell lines
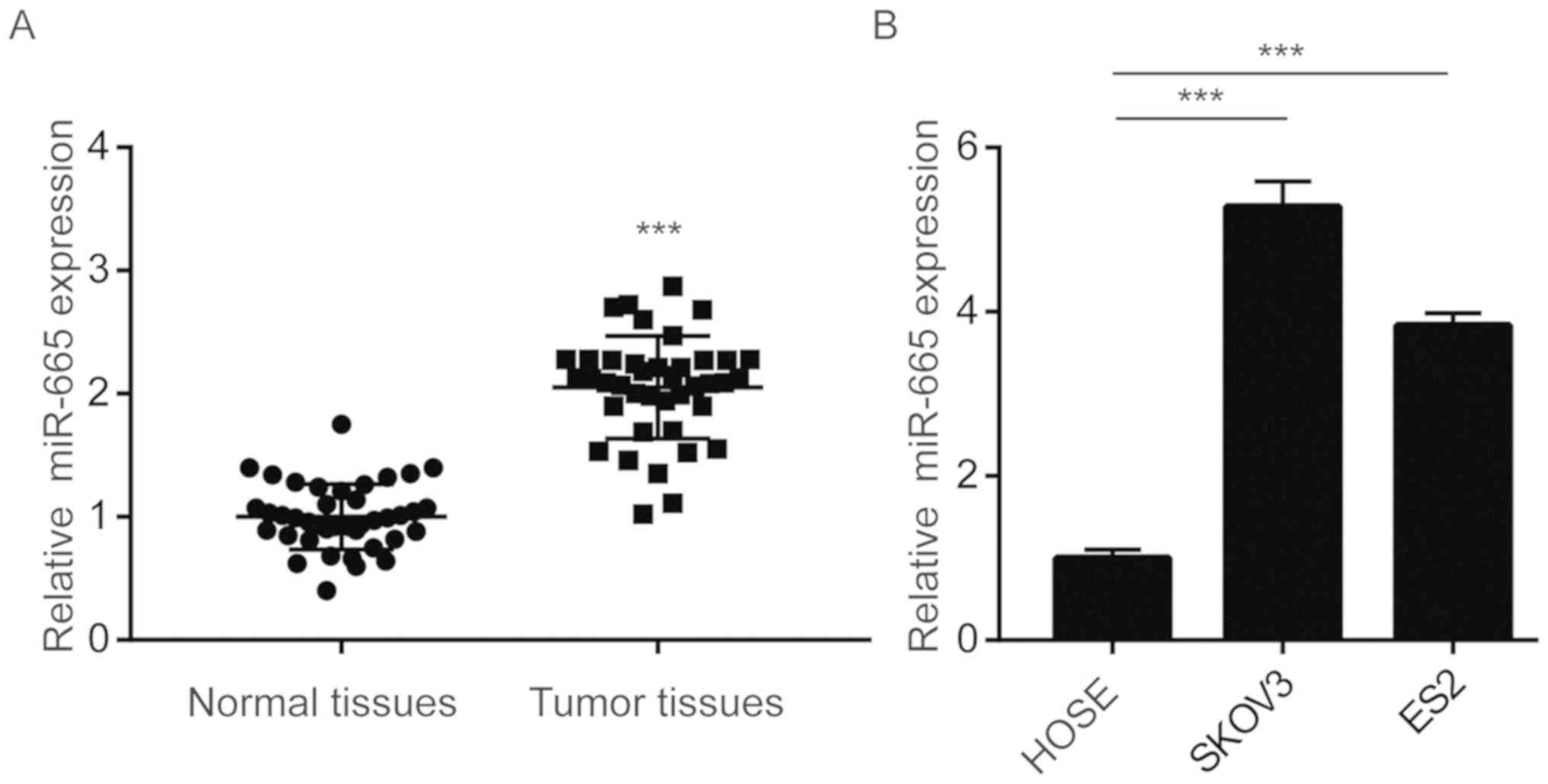
To investigate the potential role of miR-665 in
ovarian cancer, RT-qPCR was performed to detect the expression
level of miR-665 in 40 pairs of tumor and normal tissues from
patients with ovarian cancer. miR-665 expression was significantly
upregulated in tumor tissues compared with normal tissues (Fig. 1A). SKOV3 and ES2 are
well-characterized ovarian cancer cell lines. Both of these
commonly used ovarian cancer cell lines originated from ovarian
clear cell carcinoma (25).
Furthermore, RT-qPCR was used to examine the difference in miR-665
expression level between ovarian cancer cell lines (SKOV3 and ES2)
and the immortal ovarian epithelial HOSE cell line. The present
results demonstrated that miR-665 was significantly upregulated in
SKOV3 and ES2 cells compared with HOSE cells (Fig. 1B).
Downregulation of miR-665 inhibits
cell proliferation and colony forming ability of ovarian cancer
cells
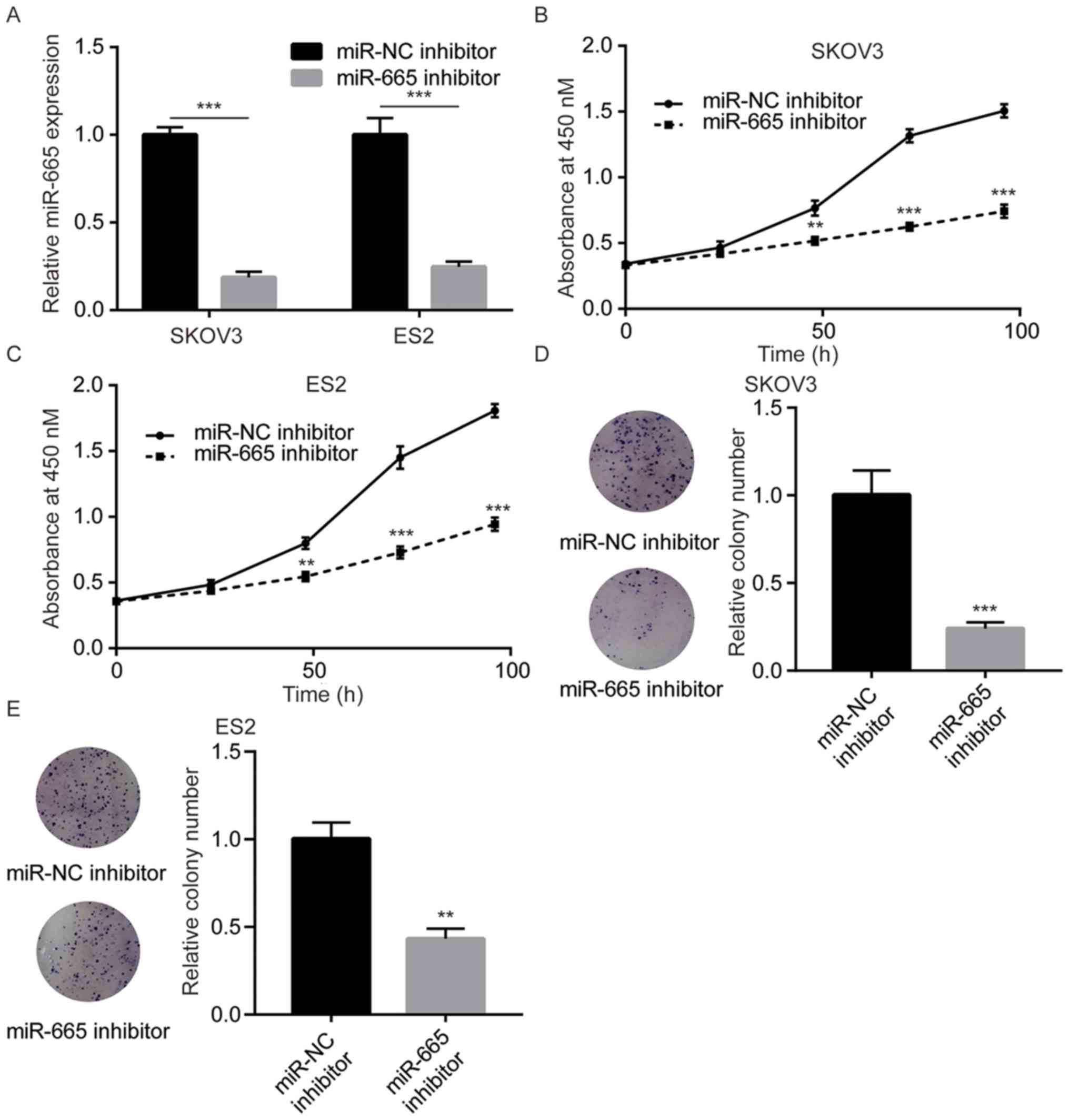
To study the function of miR-665, miR-665 inhibitor
was transfected into SKOV3 and ES2 cells to downregulate the
miR-665 level, which was verified by RT-qPCR (Fig. 2A). Downregulation of miR-665 led to
significant cell growth arrest in SKOV3 cells (Fig. 2B). Consistently, the miR-665
inhibitor also significantly decreased the cell proliferation
ability in ES2 cells (Fig. 2C). In
addition, miR-665 inhibition significantly inhibited the colony
forming ability of SKOV3 and ES2 cells (Fig. 2D and E). The present data suggested
that miR-665 promotes cell proliferation in ovarian cancer
cells.
Downregulation of miR-665 inactivates
the MAPK/ERK pathway in ovarian cancer cells
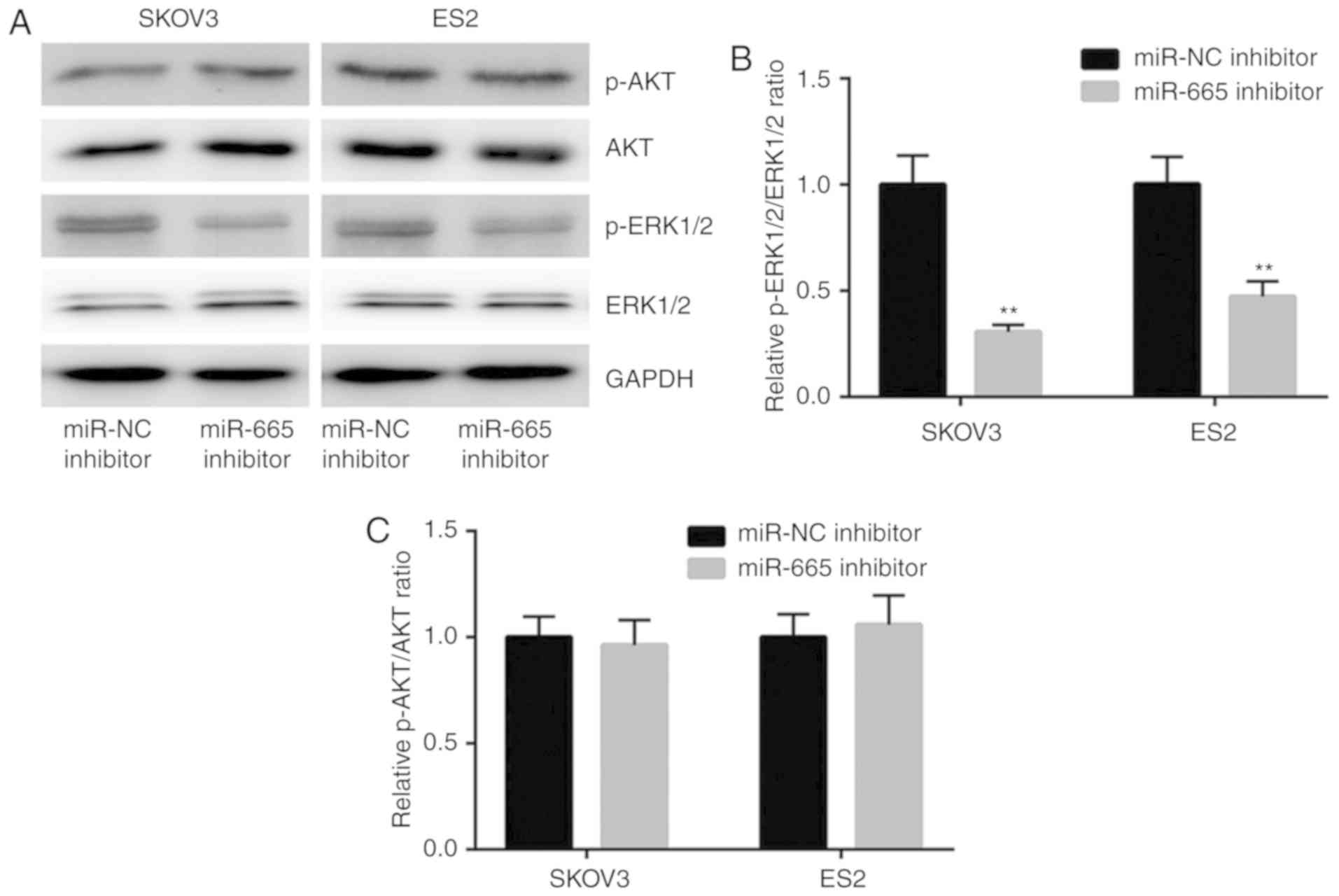
Hyperactivation of the MAPK/ERK and the PI3K/AKT
signaling pathways plays key roles in uncontrolled cell
proliferation of ovarian cancer (20,21). The
results of the western blotting showed that downregulation of
miR-665 decreased the expression of p-ERK1/2 but not p-AKT
(Fig. 3A). Further analysis
demonstrated that the p-ERK1/2 to ERK1/2 ratio was significantly
decreased in cells transfected with miR-665 inhibitor (Fig. 3B), suggesting the inactivation of
MAPK/ERK signaling. However, the p-AKT/AKT ratio was not affected
(Fig. 3C). The present data
suggested that miR-665 might promote ovarian cancer cell
proliferation via activation of MAPK/ERK signaling.
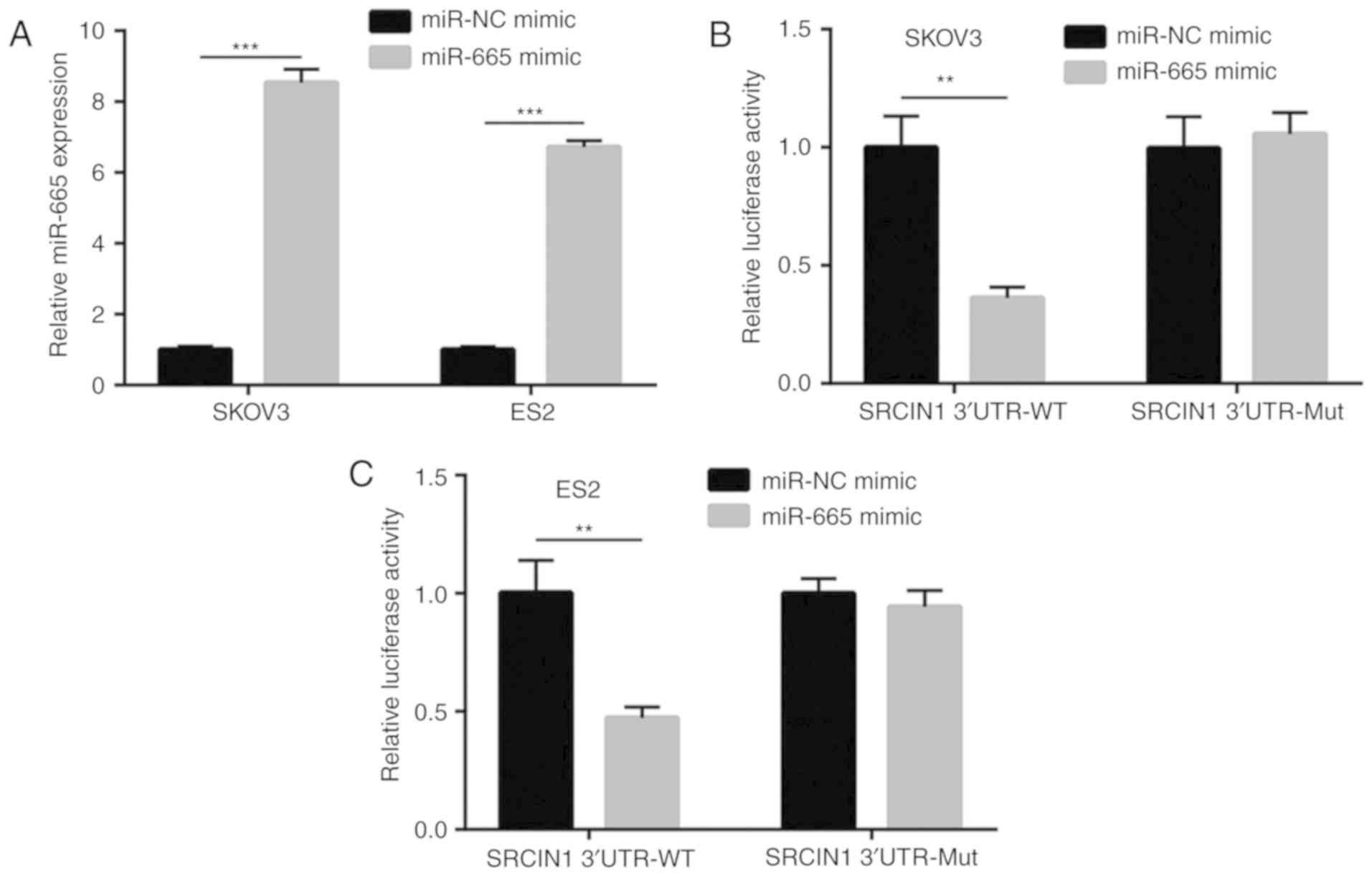
miR-665 represses SRCIN1 expression in
ovarian cancer cells
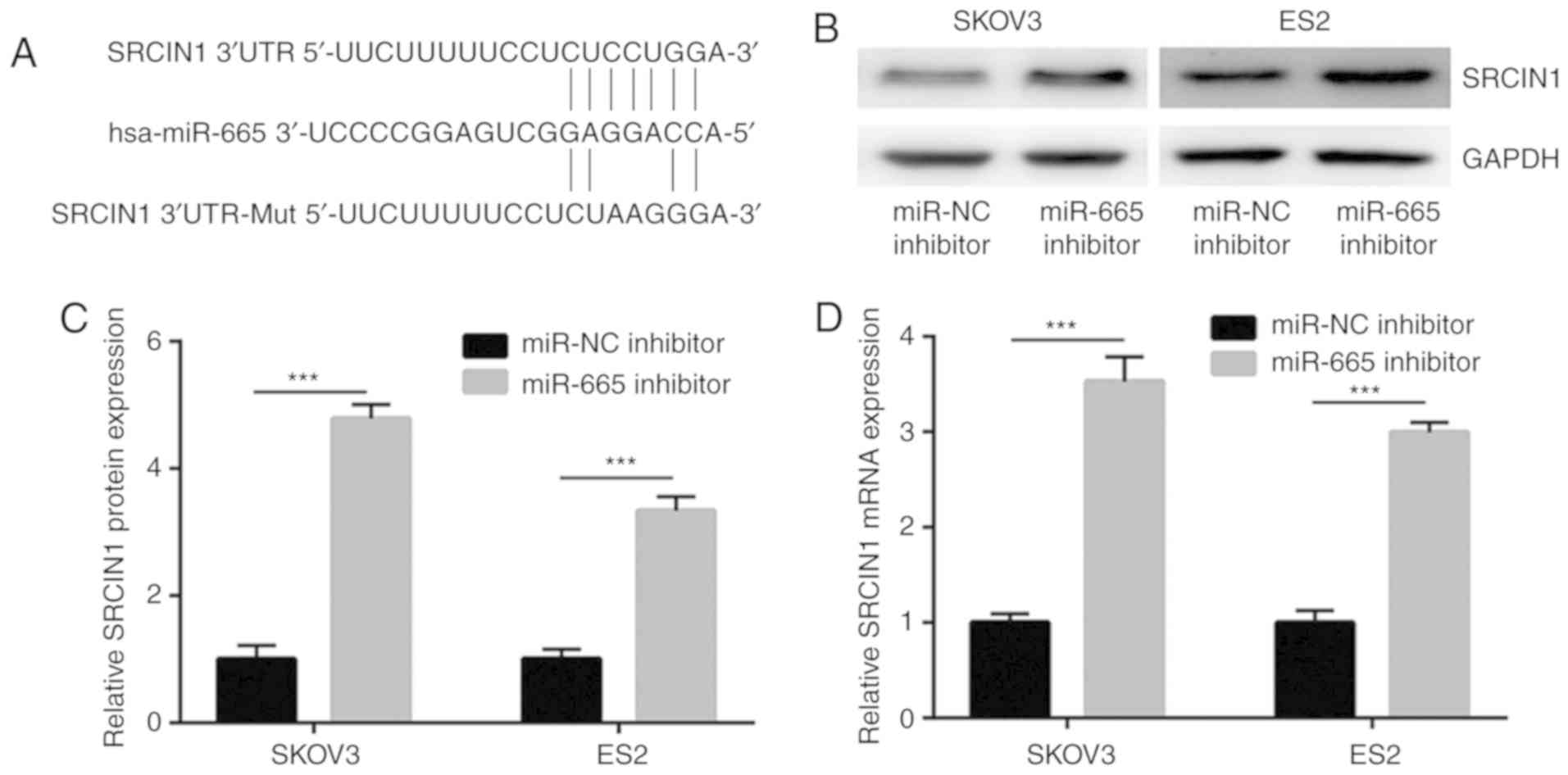
TargetScan was used to predict the potential target
genes of miR-665. Among them, the 3′UTR of SRCIN1, a negative
regulator of MAPK/ERK signaling (22), was identified to harbor binding sites
for miR-665 (Fig. 4A). The western
blotting results demonstrated that the downregulation of miR-665
significantly increased the protein expression level of SRCIN1 in
SKOV3 and ES2 cells (Fig. 4B and C).
Furthermore, the results of RT-qPCR demonstrated that miR-665
inhibition significantly increased the mRNA expression level of
SRCIN1 in SKOV3 and ES2 cells (Fig.
4D).
SRCIN1 is a target gene of miR-665 in
ovarian cancer cells
To further demonstrate SRCIN1 as a target gene of
miR-665, a dual luciferase reporter assay was performed. As
presented in Fig. 5A, the
transfection of miR-665 mimic increased miR-665 expression in
ovarian cancer cells. Overexpression of miR-665 reduced the
relative luciferase activity of SKOV3 cells that were transfected
with SRCIN1 3′UTR-WT (Fig. 5B). A
consistent result was observed in ES2 cells (Fig. 5C). The present data demonstrated that
SRCIN1 was a target gene of miR-665 in ovarian cancer cells.
miR-665 regulates cell proliferation
via regulation of SRCIN1 in ovarian cancer cells
SRCIN1 siRNA was used to study the role of SRCIN1 in
miR-665 mediated cell proliferation of ovarian cancer cells. SRCIN1
siRNA significantly decreased SRCIN1 protein expression in SKOV3
cells (Fig. 6A and B). Additionally,
transfection of miR-665 inhibitor significantly increased the
SRCIN1 protein level in SKOV3 cells, which was significantly
decreased upon transfection of SRCIN1 siRNA (Fig. 6C and D). The cell proliferation assay
showed that miR-665 downregulation significantly inhibited cell
proliferation of SKOV3 cells, which was reversed after SRCIN1
silencing (Fig. 6E). In the colony
forming assay, the miR-665 inhibitor significantly repressed the
colony forming ability, which was reversed after SRCIN1 silencing
in SKOV3 cells (Fig. 6F), suggesting
that SRCIN1 is involved in miR-665-mediated cell proliferation of
ovarian cancer cells.
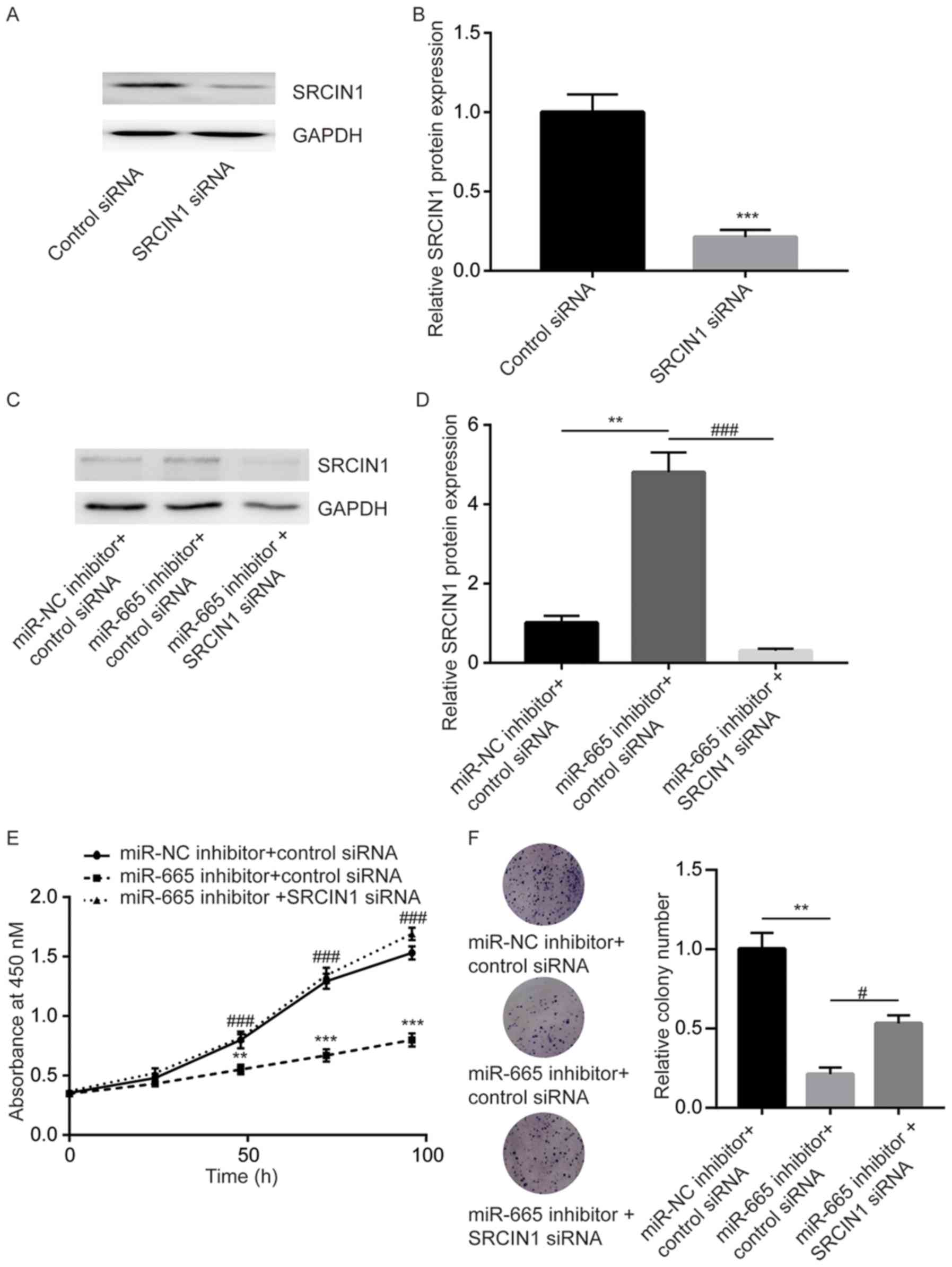 | Figure 6.miR-665 regulates cell proliferation
mainly through SRCIN1 in ovarian cancer cells. (A) In SKOV3 cells,
transfection of SRCIN1 siRNA decreased SRCIN1 protein expression.
(B) Quantitative analysis of SRCIN1 expression following
transfection with SRCIN1 siRNA. ***P<0.001 vs. control siRNA.
(C) In SKOV3 cells, transfection of miR-665 inhibitor increased the
protein level of SRCIN1, which was downregulated after transfection
with SRCIN1 siRNA. (D) Quantitative analysis of SRCIN1 expression
following multiple transfections. (E) In SKOV3 cells, transfection
of miR-665 inhibitor inhibited cell proliferation, which was
reversed after transfection SRCIN1 siRNA. (F) In SKOV3 cells,
transfection of miR-665 inhibitor inhibited colony formation, which
was reversed after transfection with SRCIN1 siRNA. **P<0.01 vs.
miR-NC inhibitor + control siRNA; ***P<0.001 vs. miR-NC
inhibitor + control siRNA, control siRNA; #P<0.05 vs.
miR-665 inhibitor + SRCIN1 siRNA; ###P<0.001 vs.
miR-665 inhibitor + SRCIN1 siRNA. miR-665, microRNA-665; miR-NC,
microRNA negative control; siRNA, small interfering RNA; SRCIN1,
Src kinase signaling inhibitor 1. |
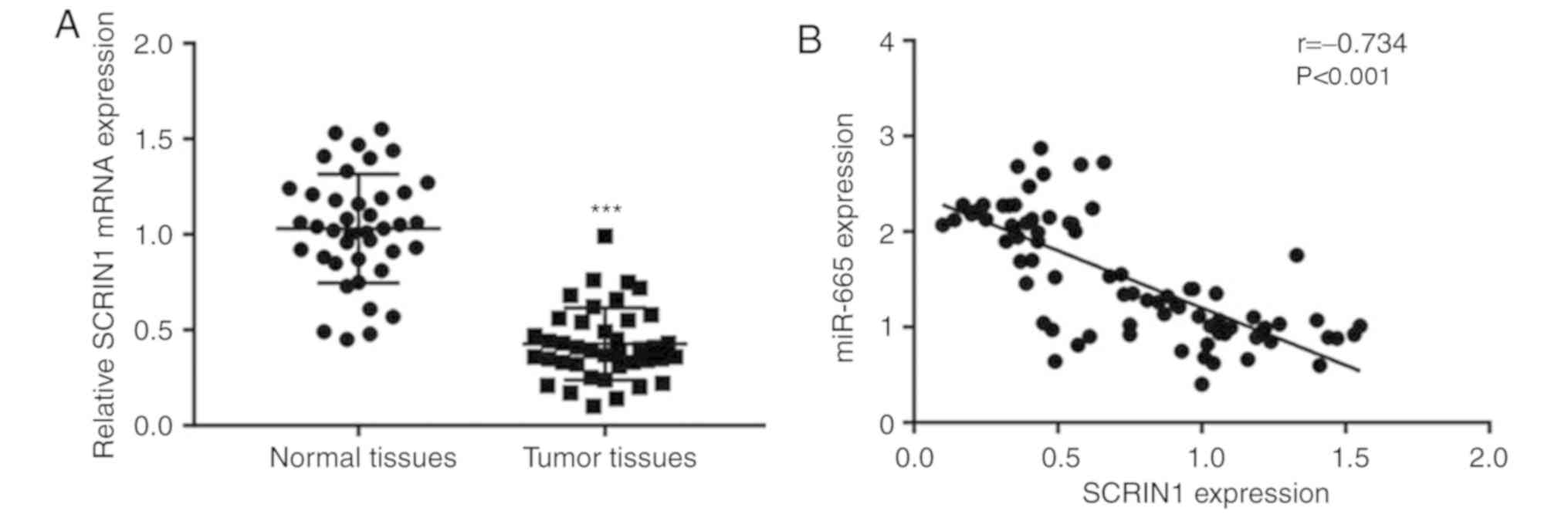
Negative correlation between miR-665
expression and SRCIN1 mRNA levels in tumor tissues from patients
with ovarian cancer
To study the clinical association between miR-665
and SRCIN1, RT-qPCR was applied for the detection of SRCIN1 mRNA
levels in 40 pairs of normal and tumor tissues collected from
patients with ovarian cancer. SRCIN1 mRNA levels were significantly
decreased in tumor tissues compared with normal tissues (Fig. 7A). Notably, a significant negative
correlation was observed between SRCIN1 mRNA levels and miR-665
expression (Fig. 7B).
Discussion
Due to their involvement in the regulation of
sustained cell growth signaling, miRNAs are considered as potential
biomarkers and therapeutic targets for cancer (26). By analyzing the comprehensive miRNA
profiles of samples from volunteers and patients with ovarian
cancer, 10 miRNAs were identified as accurate predictors for the
early detection of ovarian cancer (17). Among them, miR-320a was identified as
a tumor suppressor of ovarian cancer by targeting twist family bHLH
transcription factor 1 and MAPK1 (27,28). In
the present study, miR-665 promoted ovarian cancer cell
proliferation by targeting SRCIN1, which activated MAPK/ERK
signaling.
The role of miR-665 in cell proliferation is cell
context dependent. During the development of intervertebral disc
degeneration, the miR-665 level gradually increased to repress the
expression of growth differentiation factor 5, thus promoting the
cell proliferation of nucleus pulposus cells (29). In osteosarcoma cells, ectopic
expression of miR-665 suppressed cell proliferation, migration and
invasion by targeting Ras-related protein Rab-23 (30). The expression of miR-665 in tumor
tissues and normal tissues from patients with ovarian cancer was
analyzed, and was upregulated in tumor tissues compared with the
normal tissues, which was also observed in two ovarian cancer cell
lines compared with the immortal ovarian cancer cell line. These
results of miR-665 are consistent with its role in discriminating
patients with non-epithelial ovarian cancer from non-cancer
controls (17). Furthermore,
inhibition of miR-665 significantly inhibited cell proliferation
and colony formation of ovarian cancer cells, which further
validated the oncogenic role of miR-665 in ovarian cancer.
MAPK/ERK signaling is a well-studied driver of
cancer initiation and development (31). In ovarian cancer, sustained
activation of MAPK/ERK signaling is associated with strong cell
proliferation, metastasis and stemness ability (32). Dysregulation of positive and negative
regulators is responsible for the uncontrolled activation of the
MAPK/ERK pathway (33). The present
western blotting results showed that the MAPK/ERK pathway was
inactivated after miR-665 inhibition, suggesting that miR-665 might
promote cell proliferation via activation of MAPK/ERK
signaling.
Among several potential target genes of miR-665
predicted using TargetScan, SRCIN1, a negative regulator of
MAPK/ERK signaling, was identified. Previous studies demonstrated
that SRCIN1 was involved in the progression of cancer; including
gastric cancer and breast cancer; SRCIN1 was targeted and repressed
by miR-374a and miR-346, respectively (34,35). The
present study demonstrated that miR-665 acted as a new miRNA
regulator of SRCIN1 in ovarian cancer, which was verified by the
following results: Inhibition of miR-665 increased SRCIN1 at the
mRNA and protein levels in ovarian cancer; and miR-665 mimic
significantly reduced the luciferase activity of cells transfected
with SRCIN1 3′UTR-WT. The present study further demonstrated that
silencing of SRCIN1 could reverse miR-665-inhibitor-induced cell
growth arrest, suggesting that SRCIN1 may be important for the
function of miR-665 in ovarian cancer.
The current study showed overexpression and the role
of miR-665 in ovarian cancer in vitro. The molecular
mechanism by which miR-665 is aberrantly expressed in ovarian
cancer requires further investigation using both in vitro
and in vivo models.
In conclusion, the present study showed that miR-665
functioned as an oncogene by targeting SRCIN in ovarian cancer
cells, providing rationale for using the miR-665 level as a
predictor of ovarian cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TX and LY collected the clinical samples. JY
designed and supervised the study. PZ, TX, LY and JY acquired and
analyzed the data. JY prepared and edited the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent
before enrollment in the present study and The Ethic Committee of
Xinjiang Medical University approved the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bristow RE: Surgical standards in the
management of ovarian cancer. Curr Opin Oncol. 12:474–480. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Harries M and Gore M: Part II:
Chemotherapy for epithelial ovarian cancer-treatment of recurrent
disease. Lancet Oncol. 3:537–545. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Trimble EL, Wright J and Christian MC:
Treatment of platinum-resistant ovarian cancer. Expert Opin
Pharmacother. 2:1299–1306. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen L, Cheng X, Tu W, Qi Z, Li H, Liu F,
Yang Y, Zhang Z and Wang Z: Apatinib inhibits glycolysis by
suppressing the VEGFR2/AKT1/SOX5/GLUT4 signaling pathway in ovarian
cancer cells. Cell Oncol (Dordr). 42:679–690. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma J, Li Y, Yao L and Li X: Analysis of
MicroRNA expression profiling involved in MC-LR-induced
cytotoxicity by high-throughput sequencing. Toxins (Basel).
9:E232017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma J and Li X: High-throughput sequencing
provides an insight into the hepatotoxicity mechanism of MC-LR in
HepG2 cells. Toxin Reviews. 37:1–10. 2017. View Article : Google Scholar
|
|
11
|
Zhang L, Volinia S, Bonome T, Calin GA,
Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K,
et al: Genomic and epigenetic alterations deregulate microRNA
expression in human epithelial ovarian cancer. Proc Natl Acad Sci
USA. 105:7004–7009. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Davidson B, Tropé CG and Reich R: The
clinical and diagnostic role of microRNAs in ovarian carcinoma.
Gynecol Oncol. 133:640–646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu YZ, Xi QH, Ge WL and Zhang XQ:
Identification of serum microRNA-21 as a biomarker for early
detection and prognosis in human epithelial ovarian cancer. Asian
Pac J Cancer Prev. 14:1057–1060. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cappellesso R, Tinazzi A, Giurici T,
Simonato F, Guzzardo V, Ventura L, Crescenzi M, Chiarelli S and
Fassina A: Programmed cell death 4 and microRNA 21 inverse
expression is maintained in cells and exosomes from ovarian serous
carcinoma effusions. Cancer Cytopathol. 122:685–693. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xie Z, Cao L and Zhang J: miR-21 modulates
paclitaxel sensitivity and hypoxia-inducible factor-1α expression
in human ovarian cancer cells. Oncol Lett. 6:795–800. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yokoi A, Matsuzaki J, Yamamoto Y, Yoneoka
Y, Takahashi K, Shimizu H, Uehara T, Ishikawa M, Ikeda SI, Sonoda
T, et al: Integrated extracellular microRNA profiling for ovarian
cancer screening. Nat Commun. 9:43192018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu W, Yue F, Zheng M, Merlot A, Bae DH,
Huang M, Lane D, Jansson P, Lui GY, Richardson V, et al: The
proto-oncogene c-Src and its downstream signaling pathways are
inhibited by the metastasis suppressor, NDRG1. Oncotarget.
6:8851–8874. 2015.PubMed/NCBI
|
|
19
|
Fang D, Chen H, Zhu JY, Wang W, Teng Y,
Ding HF, Jing Q, Su SB and Huang S: Epithelial-mesenchymal
transition of ovarian cancer cells is sustained by Rac1 through
simultaneous activation of MEK1/2 and Src signaling pathways.
Oncogene. 36:1546–1558. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Le XF and Bast RC Jr: Src family kinases
and paclitaxel sensitivity. Cancer Biol Ther. 12:260–269. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang LQ, Lv RW, Qu XD, Chen XJ, Lu HS and
Wang Y: Aloesin suppresses cell growth and metastasis in ovarian
cancer SKOV3 cells through the inhibition of the MAPK signaling
pathway. Anal Cell Pathol (Amst). 2017:81582542017.PubMed/NCBI
|
|
22
|
Kennedy S, Clynes M, Doolan P, Mehta JP,
Rani S, Crown J and O'Driscoll L: SNIP/p140Cap mRNA expression is
an unfavourable prognostic factor in breast cancer and is not
expressed in normal breast tissue. Br J Cancer. 98:1641–1645. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gregoire L, Rabah R, Schmelz EM, Munkarah
A, Roberts PC and Lancaster WD: Spontaneous malignant
transformation of human ovarian surface epithelial cells in vitro.
Clin Cancer Res. 7:4280–4287. 2001.PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fogh J, Wright WC and Loveless JD: Absence
of HeLa cell contamination in 169 cell lines derived from human
tumors. J Natl Cancer Inst. 58:209–214. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pal MK, Jaiswar SP, Dwivedi VN, Tripathi
AK, Dwivedi A and Sankhwar P: MicroRNA: A new and promising
potential biomarker for diagnosis and prognosis of ovarian cancer.
Cancer Biol Med. 12:328–341. 2015.PubMed/NCBI
|
|
27
|
Li C, Duan P, Wang J, Lu X and Cheng J:
miR-320 inhibited ovarian cancer oncogenicity via targeting TWIST1
expression. Am J Transl Res. 9:3705–3713. 2017.PubMed/NCBI
|
|
28
|
Xu Y, Hu J, Zhang C and Liu Y: MicroRNA320
targets mitogenactivated protein kinase 1 to inhibit cell
proliferation and invasion in epithelial ovarian cancer. Mol Med
Rep. 16:8530–8536. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan H, Zhao L, Song R, Liu Y and Wang L:
microRNA-665 promotes the proliferation and matrix degradation of
nucleus pulposus through targeting GDF5 in intervertebral disc
degeneration. J Cell Biochem. 119:7218–7225. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dong C, Du Q, Wang Z, Wang Y, Wu S and
Wang A: MicroRNA-665 suppressed the invasion and metastasis of
osteosarcoma by directly inhibiting RAB23. Am J Transl Res.
8:4975–4981. 2016.PubMed/NCBI
|
|
31
|
Samatar AA and Poulikakos PI: Targeting
RAS-ERK signalling in cancer: Promises and challenges. Nat Rev Drug
Discov. 13:928–942. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu Z, Ye S, Hu G, Lv M, Tu Z, Zhou K and
Li Q: The RAF-MEK-ERK pathway: Targeting ERK to overcome obstacles
to effective cancer therapy. Future Med Chem. 7:269–289. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen R, Liao JY, Huang J, Chen WL, Ma XJ
and Luo XD: Downregulation of SRC kinase signaling inhibitor 1
(SRCIN1) expression by MicroRNA-32 promotes proliferation and
epithelial-mesenchymal transition in human liver cancer cells.
Oncol Res. 26:573–579. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu X, Wang W, Su N, Zhu X, Yao J, Gao W,
Hu Z and Sun Y: miR-374a promotes cell proliferation, migration and
invasion by targeting SRCIN1 in gastric cancer. FEBS Lett.
589:407–413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang F, Luo LJ, Zhang L, Wang DD, Yang SJ,
Ding L, Li J, Chen D, Ma R, Wu JZ and Tang JH: miR-346 promotes the
biological function of breast cancer cells by targeting SRCIN1 and
reduces chemosensitivity to docetaxel. Gene. 600:21–28. 2017.
View Article : Google Scholar : PubMed/NCBI
|