Introduction
Osteosarcoma (OS) is a type of malignant tumor
commonly presenting in children, and adolescents (1) under 20 years of age (2). OS is caused by pathological changes in
mesenchymal tissue in the bone, especially in the metaphysis of
long bones (3). Osteosarcoma cells
easily migrate to other tissues in a short period of time by the
blood, and lungs are the main metastatic organ (4). With advanced chemotherapy techniques,
the 5-year survival rate is approximately 60–70% in OS patients
without distant metastasis (5).
However, the 5-year survival rate is lower than 30% for OS patients
with distant metastasis (6). Thus,
studies concerning the molecular mechanisms underlying OS
carcinogenesis are vital for further clinical treatment.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
consisting of 21–25 nucleotides (nt). miRNAs have been identified
as key regulators of gene expression through binding with target
mRNAs, thus affecting cell proliferation, division, apoptosis and
metabolism (7–9). Dysregulation of miRNA expression is
closely associated with various types of diseases and requires
further research (10). Emerging
research has revealed that miRNAs contribute to human
carcinogenesis as tumor inhibitors or promoters. For example,
miR-10a-5p was found to suppress human cervical cancer
proliferation and division (11).
Similarly, miR-214 plays a crucial role in melanoma tumor
progression by targeting TFAP2C (12). miR-183 was found to contribute to
advanced clinical stage human colorectal cancer (13). Moreover, miR-708 was demonstrated to
suppress tumor cell proliferation in human glioblastoma (14). In order to investigate the
relationship between miRNAs and tumors, we focused on the functions
of miRNAs in OS progression.
miR-496 has been identified to participate in
various pathobiological processes (15,16).
However, the functions of miR-496 in OS have not yet been
elucidated. In the present study, expression of miR-496 was
quantified in both OS tissues and cell lines. Brain derived
neurotrophic factor (BDNF) was identified one of the target
genes of miR-496 in OS development. Based on these previous
findings, the role of miR-496 and BDNF in relation to the
proliferation of OS cells was investigated.
Materials and methods
Tissue samples and cell lines
Pathological tissues and adjacent normal tissues
were collected from 37 OS patients (22 male and 5 female; age,
17.23±7.56 years) during routine therapeutic surgery at the
Department of Orthopedics, Hubei 672 Orthopedics Hospital of
Integrated Chinese and Western Medicine (Wuhan, China) between June
2016 and October 2018. The patients had received no treatment at
the time the tissues were extracted. Normal tissues were examined
to confirm that there were no pathological changes. Liquid nitrogen
was used to store the above tissues for the subsequent experiments.
Written informed consent was obtained from all patients or their
guardians, and the study protocol was approved by the Ethics
Committee of Hubei 672 Orthopedics Hospital of Integrated Chinese
and Western Medicine. The study was carried out in accordance with
the approved guidelines.
OS cell lines
Based on the Enneking-Musculoskeletal Tumor Staging
System (17), five types of OS cell
lines, including hFOB1.19, MG-63, HOS, U2-OS and SAOS-2 were
obtained from the Shanghai Cell Institute of Chinese Academy of
Sciences (Shanghai, China). Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc.) was used to culture
the cell lines. In addition, 10% fetal bovine serum (FBS) and 0.015
mg/ml 5-bromo-2′-deoxyuridine (Gibco; Thermo Fisher Scientific,
Inc.) were added to the DMEM. The cells were incubated at 37°C in
5% CO2.
Prediction of target genes of
miR-496
Targetscan 3.1 (http://www.targetscan.org/mamm_31/) was used to
determine the candidate downstream genes of miR-496. The prediction
results and binding sites were obtained.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted using RNAiso Plus (Takara
Biotechnology, Dalian, China). Prime Script RT Master Mix (Takara
Biotechnology, Co., Ltd.) was used to reverse transcribe the RNA,
and RT-qPCR was performed using SYBR Premix Ex Taq II (Takara
Biotechnology). The following primers were used: GAPDH forward,
5′-GACTCATGACCACAGTCCATGC-3′ and GAPDH reverse,
5′-AGAGGCAGGGATGATGTTCTG-3′; BDNF forward,
5′-GGCTTGACATCATTGGCTGAC-3′ and BDNF reverse,
5′-CATTGGGCCGAACTTTCTGGT-3′. miR-496 expression was quantified with
the All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia, Inc.,
USA), in an Applied Biosystems 7900 Real-time PCR System (Thermo
Fisher Scientific, Inc.).
Transfection
OS cells were transfected with miR-496 mimics
(mimic-miR-496, 5′-CUCUAACCGGUACAUUA-3′), miR-496 inhibitors
(inhibitor-miR-496, 5′-GAGAUUGGCCAUGUAAU-3′) and their negative
controls (mimic-NC, 5′-UUCUCCGAACGUGUCACGU-3′ or inhibitor-NC,
5′-CAGUACUUUUGUGUAGUACAA-3′). LV-NC, LV-miR-496, or
LV-miR-496+LV-BDNF were transfected into MG-63 and HOS cells,
respectively. The lentiviruses used in our study were obtained from
GenePharma (Shanghai, China). Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for cell
transfection based on the manufacturer's protocols. Medium was
replaced with fresh medium with 10% FBS after 6-h transfection.
Luciferase activity assay
Dual-Luciferase Reporter Assay System (Promega
Corporation) was used to assay the activity of the luciferase
reporter gene. The wild-type (BDNF 3′UTRWT) or mutant
type (BDNF 3′UTRMUT) binding to miR-496 was subcloned
into the PGL3 Basic vector (Invitrogen; Thermo Fisher Scientific,
Inc.). miR-496 mimics were co-transfected with 10 µg of BDNF
3′UTRWT or BDNF 3′UTRMUT into MG-63 and HOS
cells using Lipofectamine® 2000 reagent, and cell medium
was replaced with fresh medium with 10% FBS after 6 h transfection.
The luciferase activity for each construct was normalized to that
of the Renilla.
Western blot analysis
Proteins were obtained from the MG-63 and HOS cell
lines and their concentration was estimated using the Bradford
protein assay. The proteins (30 µg/lane) were separated by 10%
sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and
transferred to a polyvinylidene difluoride (PVDF) membrane. The
membrane was blocked with 5% defatted milk at room temperature for
1 h and incubated with specific primary antibodies overnight with a
controlled environment at 4°C overnight. After washing in TBST, the
membranes were incubated with goat anti-rabbit horseradish
peroxidase (HRP)-conjugated secondary antibodies (dilution
1:20,000, cat. no. ab205718; Abcam). Anti-BDNF (dilution 1:1,000,
cat. no. ab108319; Abcam) and anti-glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) (dilution 1:10,000, cat. no. ab181602; Abcam)
were used as the primary antibodies. Immunoreactive bands were
developed using an enhanced chemiluminescence kit (cat. no.
170-5061; Bio-Rad Laboratories, Inc.) according to the
manufacturer's instructions. Intensity of bands (BDNF, 15 kDa;
GAPDH, 36 kDa) was quantified using ImageJ 1.49 (National
Institutes of Health).
Cell viability assay
In order to evaluate the cell viability, CCK-8 assay
was employed to assess the cell growth and generate a growth curve.
The transfected 5×103 OS cells were plated into a
96-well plate and incubated at 37°C with 5% CO2. Then,
the cells were digested and counted every 24 h. A cell growth curve
was plotted after five days.
Statistical analysis
R Studio (3.5.1; http://www.r-project.org/) was performed for the
statistical analyses. All data were from three independent
experiments and were expressed as the mean ± standard deviation
(SD). Student paired t-tests were used for the comparison of
miR-496 expression between primary tumors and adjacent noncancerous
tissues. Student's unpaired t-test was used to measure the
differences between quantitative variables. One-way analysis of
variance (ANOVA) was used for the comparisons of the cell viability
in the MG-63LV-NC, MG-63LV-miR-496, and
MG-63LV-miR-496+LV-BDNF groups as well as in
HOSLV-NC, HOSLV-miR-496, and
HOSLV-miR-496+LV-BDNF. The post hoc Tukey's test was
used to analyze the two-group comparisons. P-values<0.05 were
considered statistically significant.
Results
Downregulation of miR-496 expression
in OS tissues and cell lines
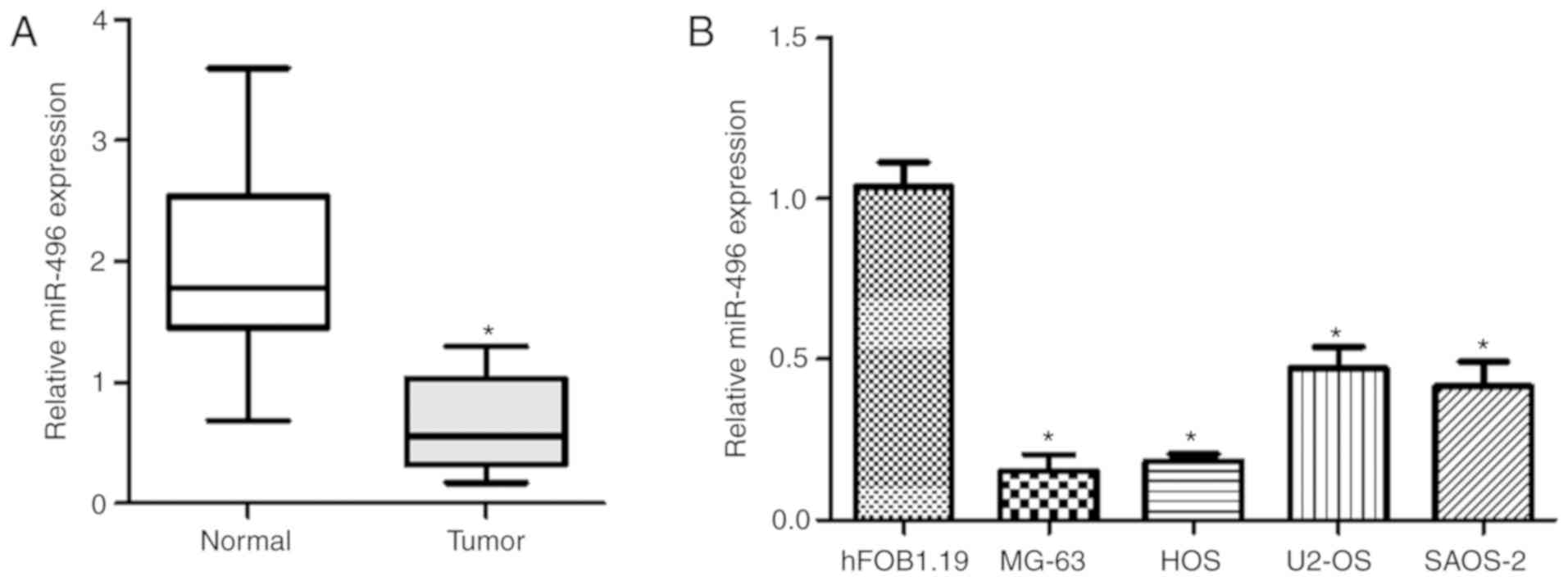
RT-qPCR was employed to determine miR-496 expression
levels in osteosarcoma tissues and five OS cell lines. The results
showed lower miR-496 expression level in the OS tissues than that
in the normal control group (Fig.
1A). Moreover, miR-496 expression levels in OS MG-63, HOS,
U2-OS, and SAOS-2 cell lines were lower compared to the level in
the hFOB1.19 cell line, which acted as a normal control (Fig. 1B). The results showed that miR-496
contributes to OS development.
Effect of the altered expression of
miR-496 on OS cell viability in vitro
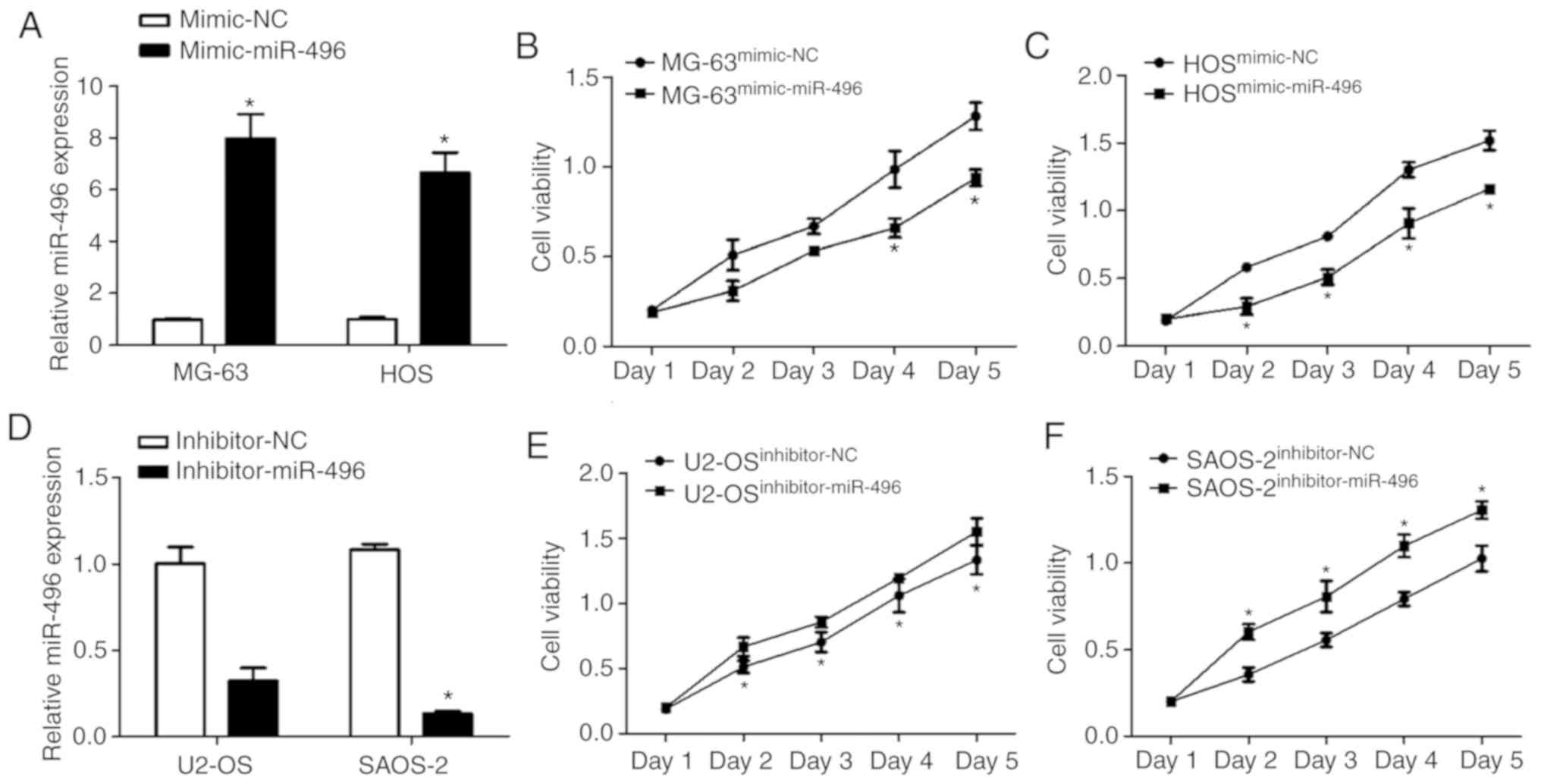
In order to investigate the direct effect of miR-496
on cell viability, mimic-miR-496 and mimic-NC were transfected into
MG-63 and HOS cells, and inhibitor-miR-496 and inhibitor-NC were
transfected into U2-OS and SAOS-2 cells. miR-496 expression was
increased in both MG-63 and HOS cell lines following the
transfection of mimic-miR-496 (Fig.
2A). The cell viability of MG-63 cells transfected with the
mimic-miR-496 was significantly decreased from day 4 compared to
the mimic-NC-transfected cells (Fig.
2B). In addition, the cell viability of HOS cells transfected
with the mimic-miR-496 was significantly decreased from Day 2
compared to mimic-NC-transfected cells (Fig. 2C).
In contrast, miR-496 expression was decreased in
both U2-OS and SAOS-2 cell lines through the transfection of
inhibitor-miR-496 (Fig. 2D).
However, the cell viability of the U2-OS cells transfected with
inhibitor-miR-496 was significantly increased compared to the
inhibitor-NC-transfected cells (Fig.
2E). The cell viability of SAOS-2 cells transfected with
inhibitor-miR-496 was also significantly decreased compared to the
inhibitor-NC-transfected cells (Fig.
2F). These results indicate that miR-496 acts as a tumor
suppressor in OS development.
BDNF is a direct target of
miR-496
Bioinformatic analysis was used to determine the
downstream genes of miR-496 in OS progression, including ZIC1,
CPEB2, CSMAD3, USD15, MYT1L, SATB2 and EIF4B. We found
that BDNF 3′UTR could precisely bind to miR-496 (Fig. 3A). Furthermore, to verify whether
BNDF is a direct target gene, wild-type (WT) or mutant (MUT) 3′UTR
of BDNF was transfected into the luciferase vector. The vector
co-transfected with miR-496 mimics and BDNF 3′UTRWT had
lower luciferase activity in both MG-63 (Fig. 3B) and HOS cells (Fig. 3D). However, co-transfection of
miR-496 mimics with the luciferase vector containing MUT-BDNF 3′UTR
had the same luciferase activity compared to the mimic negative
control in both MG-63 (Fig. 3C) and
HOS cells (Fig. 3E). This
demonstrated that BDNF is a downstream gene of miR-496.
miR-496 and BDNF expression in OS
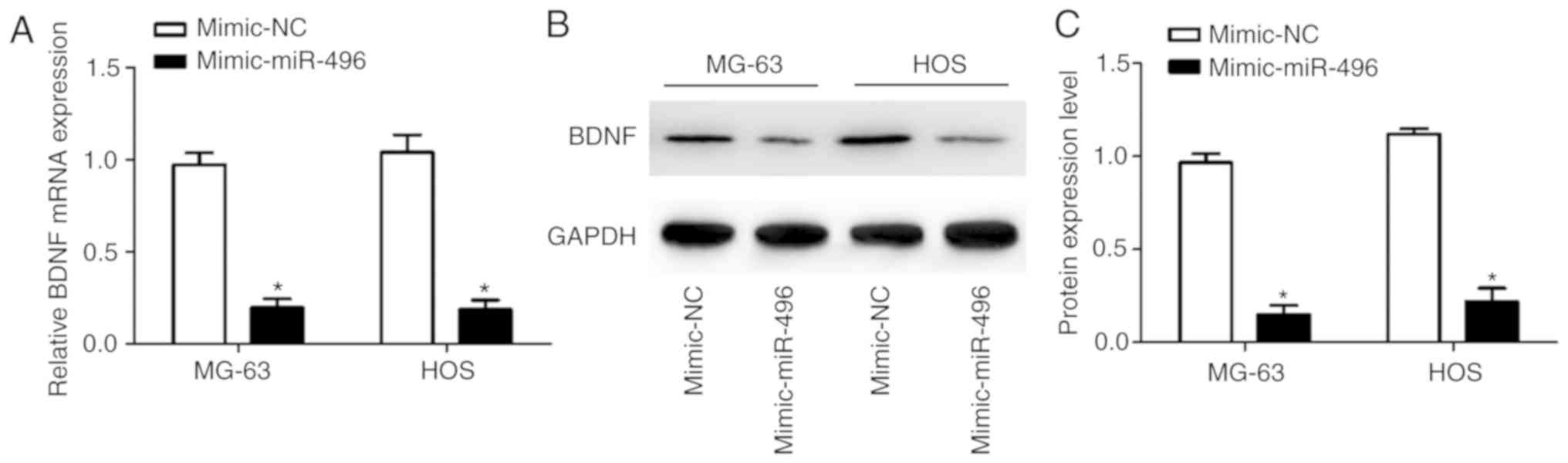
In order to determine the function of miR-496 in OS,
RT-qPCR and western blot analysis were used to measure BDNF mRNA
and protein expression levels. Overexpression of miR-496 suppressed
BDNF in both MG-63 and HOS cells at the mRNA (Fig. 4A) and protein (Fig. 4B and C) levels. This finding thus
confirms that miR-496 negatively modulates BDNF expression.
Overexpression of BDNF in OS
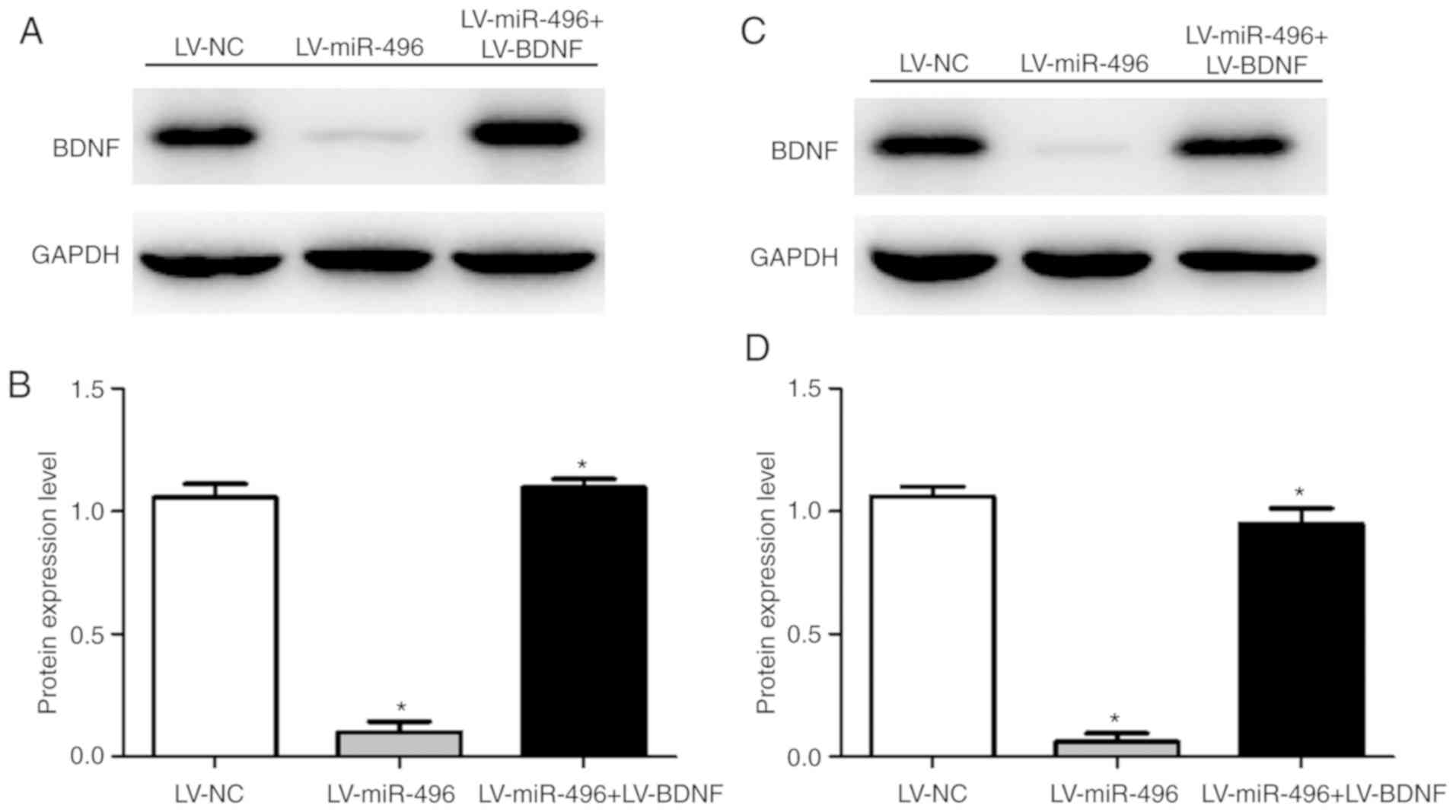
BDNF protein expression and cell viability were
assessed to investigate the function of BDNF in OS through BDNF
upregulation in MG-63 and HOS cells. The BDNF protein expression
level was increased in both MG-63 (Fig.
5A and B) and HOS cells (Fig. 5C and
D) following co-transfection of LV-miR-496+LV-BDNF, compared to
the transfection with LV-miR-496 only. Moreover, cell viability was
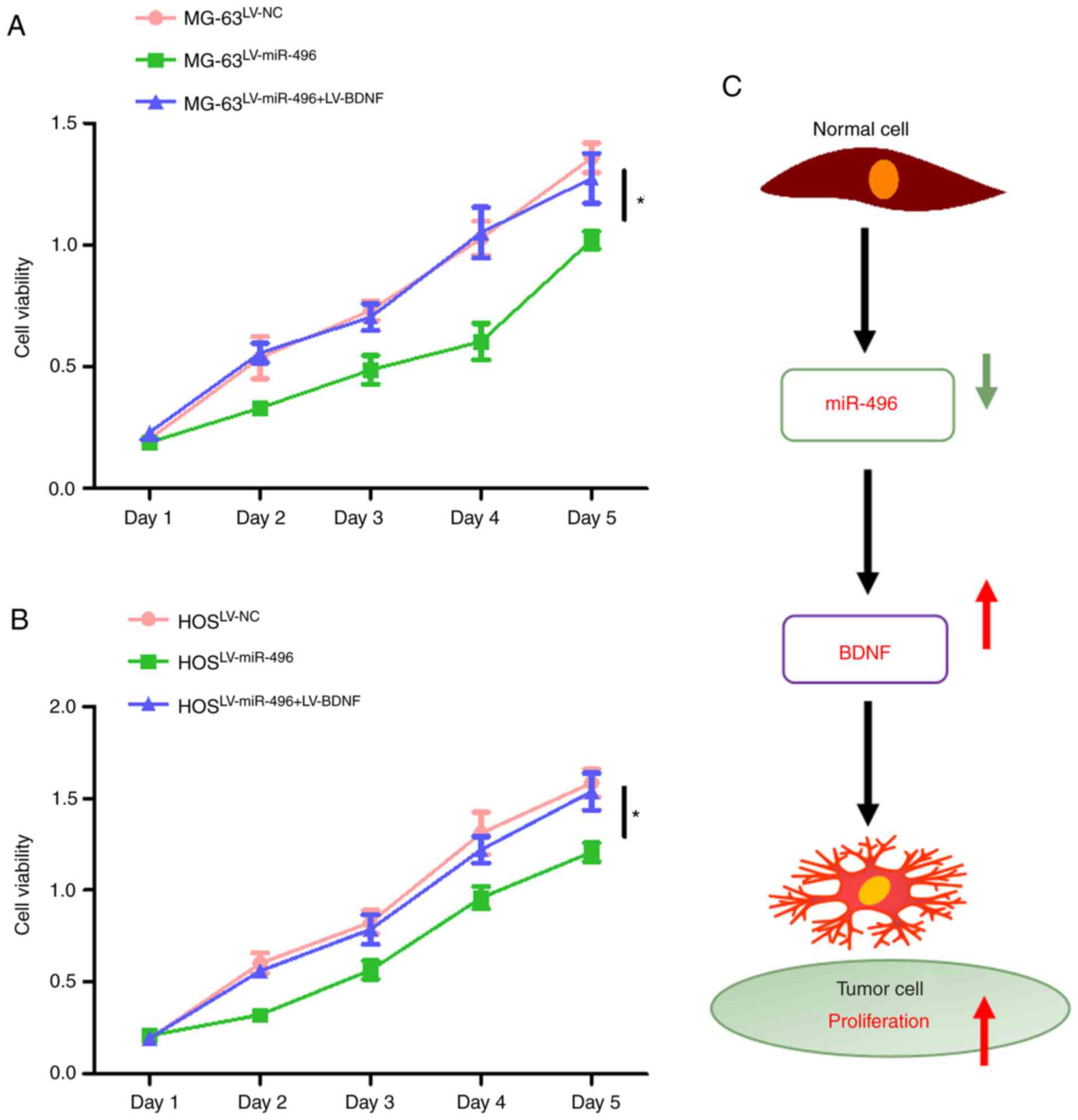
increased in both MG-63 (Fig. 6A)
and HOS cells (Fig. 6B) following
LV-miR-496+LV-BDNF co-transfection. This indicated that
overexpression of BDNF reversed the suppressive effect of miR-496
on OS cell viability (Fig. 6C). BDNF
thus acts as a promoter of OS progression.
Discussion
Several studies have identified microRNAs as
oncogenes or tumor suppressors in various types of cancer,
including human osteosarcoma (OS) (18–20). For
example, miR-181a was found to promote cell proliferation and
invasion in OS (21). miR-504 was
found to induce cell proliferation and suppress apoptosis as well
as G1 phase arrest in OS via negatively regulating p53 (22). Moreover, miR-758 suppressed the
malignant phenotype of OS cells by directly targeting HMGA1
(23). miR-214 was found to
accelerate the progression of OS by modulating the Wnt/β-catenin
signaling pathway (24). miR-143-3p
was demonstrated to inhibit the proliferation and migration in OS
by targeting FOSL2 (25). The
different mechanisms of miRNAs in OS provide a possible orientation
for the clinical diagnosis and treatment of OS (26). Therefore, comprehensive research is
needed to explore the targets of miRNAs in the clinical therapy of
OS; specifically drawing our attention to miR-496.
Previous studies have indicated that miR-496 is
associated with several human pathological changes, including
oropharyngeal cancer (27), cerebral
ischemia/reperfusion injury (28),
type 2 diabetes mellitus and obesity (29), and osteogenesis of human bone marrow
(30). These findings suggest that
miR-496 serves as a suppressor in human pathological changes, which
is consistent with our findings. In the present study, miR-496
expression was downregulated in OS cell lines. Overexpression of
miR-496 reduced cell viability in both MG-63 and HOS cell lines,
and loss of miR-496 promoted cell viability in both U2-OS and
SAOS-2 cell lines. To note, miR-496 expression in the MG-63 and HOS
cell lines was the lowest, which is the main reason why the MG-63
and HOS cell lines were chosen to perform the functional
experiments.
Brain-derived neurotrophic factor (BDNF) gene
is related to nervous system development (31). Previous research has shown that BDNF
contributes to cancer development. For example, the BDNF/leptin
axis was correlated with cancer remission and inhibition (32). BDNF was found to act as an inhibitor
in breast cancer development and metastasis in mice (33). miR-10a-5p was found to suppress human
cervical cancer progression by targeting BDNF (11). In the present study, BDNF was
proposed as the target gene of miR-496 in OS progression. We found
that miR-496 negatively modulated BDNF expression in OS cells.
Overexpression of BDNF promoted cell proliferation of the OS cells.
This indicated that BDNF is a promoter of OS development.
Recently, a similar study revealed that
overexpression of miR-496 suppressed human OS cell proliferation,
invasion, and migration by targeting eIF4E (34). These previous results reinforced our
finding that miR-496 acts as a suppressor of human OS cell
proliferation.
To conclude, miR-496 expression was downregulated in
OS tissues and cell lines, whereas the overexpression of miR-496
reduced cell viability and suppressed cell proliferation in OS cell
lines. BDNF is directly targeted by miR-496. Upregulation of BDNF
promoted cell proliferation in the OS cell lines. Thus, the
miR-496/BDNF axis provides a novel therapeutic target for OS
(Fig. 6C). miR-496 may be one of the
epigenetic modifications to regulate the expression of BDNF. Other
mechanisms such as DNA methylation, histone modifications,
kinase-associated phosphorylation may also affect BDNF expression.
A larger sample size with more pathological information and
functional studies with a focus on cell apoptosis, cell migration
and cell invasion as well as in vivo experiments are
necessary to further confirm our results.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JT conceived and designed the study. JY, WX, YZ and
GJ performed the experiments. JY and JT wrote the paper. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients or their guardians, and the study protocol was approved by
the Ethics Committee of Hubei 672 Orthopedics Hospital of
Integrated Chinese and Western Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interest.
References
|
1
|
Yang J and Zhang W: New molecular insights
into osteosarcoma targeted therapy. Curr Opin Oncol. 25:398–406.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu W, Zhu J, Wang Y, Wang J, Fang W, Xia
K, Shao J, Wu M, Liu B, Liang C, et al: A review and outlook in the
treatment of osteosarcoma and other deep tumors with photodynamic
therapy: From basic to deep. Oncotarget. 8:39833–39848.
2017.PubMed/NCBI
|
|
3
|
Berner K, Johannesen TB, Berner A,
Haugland HK, Bjerkehagen B, Bøhler PJ and Bruland OS: Time-trends
on incidence and survival in a nationwide and unselected cohort of
patients with skeletal osteosarcoma. Acta Oncol. 54:25–33. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Taran SJ, Taran R and Malipatil NB:
Pediatric osteosarcoma: An updated review. Indian J Med Paediatr
Oncol. 38:33–43. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Faisham WI, Mat Saad AZ, Alsaigh LN, Nor
Azman MZ, Kamarul Imran M, Biswal BM, Bhavaraju VM, Salzihan MS,
Hasnan J, Ezane AM, et al: Prognostic factors and survival rate of
osteosarcoma: A single-institution study. Asia Pac J Clin Oncol.
13:e104–e110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang S, Zheng S, Hu K, Sun H, Zhang J,
Rong G, Gao J, Ding N and Gui B: A predictive model to estimate the
pretest probability of metastasis in patients with osteosarcoma.
Medicine (Baltimore). 96:e59092017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sonkoly E: The expanding microRNA world in
psoriasis. Exp Dermatol. 26:375–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mellis D and Caporali A: MicroRNA-based
therapeutics in cardiovascular disease: Screening and delivery to
the target. Biochem Soc Trans. 46:11–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Acunzo M and Croce CM: MicroRNA in cancer
and cachexia-A mini-review. J Infect Dis. 212 (Suppl 1):S74–S77.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhai L, Li Y, Lan X and Ai L:
MicroRNA-10a-5p suppresses cancer proliferation and division in
human cervical cancer by targeting BDNF. Exp Ther Med.
14:6147–6151. 2017.PubMed/NCBI
|
|
12
|
Penna E, Orso F, Cimino D, Tenaglia E,
Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De
Pittà C, et al: microRNA-214 contributes to melanoma tumour
progression through suppression of TFAP2C. EMBO J. 30:1990–2007.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou T, Zhang GJ, Zhou H, Xiao HX and Li
Y: Overexpression of microRNA-183 in human colorectal cancer and
its clinical significance. Eur J Gastroenterol Hepatol. 26:229–233.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo P, Lan J, Ge J, Nie Q, Mao Q and Qiu
Y: miR-708 acts as a tumor suppressor in human glioblastoma cells.
Oncol Rep. 30:870–876. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rubie C, Kölsch K, Halajda B, Eichler H,
Wagenpfeil S, Roemer K and Glanemann M: microRNA-496-A new,
potentially aging-relevant regulator of mTOR. Cell Cycle.
15:1108–1116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu J, Liu T, Zhang Z, Xu Y and Zhu F:
Oxidized low-density lipoprotein promotes vascular endothelial cell
dysfunction by stimulating miR-496 expression and inhibiting the
Hippo pathway effector YAP. Cell Biol Int. 43:528–538. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cates JMM: Simple staging system for
osteosarcoma performs equivalently to the AJCC and MSTS systems. J
Orthop Res. 36:2802–2808. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Acunzo M, Romano G, Wernicke D and Croce
CM: MicroRNA and cancer-a brief overview. Adv Biol Regul. 57:1–9.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Palmini G, Marini F and Brandi ML: What is
new in the miRNA world regarding osteosarcoma and chondrosarcoma?
Molecules. 22(pii): E4172017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jianwei Z, Fan L, Xiancheng L, Enzhong B,
Shuai L and Can L: MicroRNA 181a improves proliferation and
invasion, suppresses apoptosis of osteosarcoma cell. Tumour Biol.
34:3331–3337. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Lv C, Zhu X, Lin W, Wang L, Huang
Z, Yang S and Sun J: MicroRNA-504 modulates osteosarcoma cell
chemoresistance to cisplatin by targeting p53. Oncol Lett.
17:1664–1674. 2019.PubMed/NCBI
|
|
23
|
Ren J, Yang M, Xu F and Chen J:
microRNA-758 inhibits the malignant phenotype of osteosarcoma cells
by directly targeting HMGA1 and deactivating the Wnt/β-catenin
pathway. Am J Cancer Res. 9:36–52. 2019.PubMed/NCBI
|
|
24
|
Zhu XB, Zhang ZC, Han GS, Han JZ and Qiu
DP: Overexpression of miR-214 promotes the progression of human
osteosarcoma by regulating the Wnt/β-catenin signaling pathway. Mol
Med Rep. 15:1884–1892. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun X, Dai G, Yu L, Hu Q, Chen J and Guo
W: miR-143-3p inhibits the proliferation, migration and invasion in
osteosarcoma by targeting FOSL2. Sci Rep. 8:6062018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miao J, Wu S, Peng Z, Tania M and Zhang C:
MicroRNAs in osteosarcoma: Diagnostic and therapeutic aspects.
Tumour Biol. 34:2093–2098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mason D, Zhang X, Marques TM, Rose B,
Khoury S, Hill M, Deutsch F, Lyons JG, Gama-Carvalho M and Tran N:
Human papillomavirus 16 E6 modulates the expression of miR-496 in
oropharyngeal cancer. Virology. 521:149–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yao X, Yao R, Yi J and Huang F:
Upregulation of miR-496 decreases cerebral ischemia/reperfusion
injury by negatively regulating BCL2L14. Neurosci Lett.
696:197–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rubie C, Zimmer J, Lammert F, Gross JC,
Weber SN, Kruse B, Halajda B, Wagner M, Wagenpfeil S and Glanemann
M: MicroRNA-496 and mechanistic target of rapamycin expression are
associated with type 2 diabetes mellitus and obesity in elderly
people. Ann Nutr Metab. 74:279–286. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang J and Chen L: IL-1β inhibits
osteogenesis of human bone marrow-derived mesenchymal stem cells by
activating FoxD3/microRNA-496 to repress wnt signaling. Genesis.
55:2017. View Article : Google Scholar
|
|
31
|
Mariga A, Mitre M and Chao MV:
Consequences of brain-derived neurotrophic factor withdrawal in CNS
neurons and implications in disease. Neurobiol Dis. 97:73–79. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao L, Liu X, Lin EJ, Wang C, Choi EY,
Riban V, Lin B and During MJ: Environmental and genetic activation
of a brain-adipocyte BDNF/leptin axis causes cancer remission and
inhibition. Cell. 142:52–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, McMurphy T, Xiao R, Slater A, Huang
W and Cao L: Hypothalamic gene transfer of BDNF inhibits breast
cancer progression and metastasis in middle age obese mice. Mol
Ther. 22:1275–1284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qi NN, Tian S, Li X, Wang FL and Liu B:
Up-regulation of microRNA-496 suppresses proliferation, invasion,
migration and in vivo tumorigenicity of human osteosarcoma cells by
targeting eIF4E. Biochimie. 163:1–11. 2019. View Article : Google Scholar : PubMed/NCBI
|