Introduction
Cervical cancer is the second most common female
malignancy in China (1), where
Shanxi Province is a particularly high-risk (HR) area; with a
detection rate of 55-100,000 in Jiexiu (2), which is higher compared with the
average incidence of 13.4-100,000 in China (3). Therefore, strategies to prevent the
occurrence of cervical cancer in China, especially in Shanxi, are
urgently required. Cervical cancer generally develops from
pre-existing, non-invasive and squamous precursor lesions, referred
to as cervical intraepithelial neoplasia (CIN) or squamous
intraepithelial lesions (SIL), which also includes low-grade (LSIL)
and high-grade SIL (HSIL). Since SIL develops gradually, early
diagnosis is an important means of preventing cervical cancer.
Although 60% of LSIL cases are transient and spontaneously regress
within 12-24 months, LSIL do persist in 30% of the cases, where
~10% of LSIL develop into HSIL within 2 years (4). Since only a small percentage of
patients with LSIL progress into HSIL, it is therefore preferable
to identify patients with LSIL so that resection can be performed
at an early stage, where they can be safely followed up using
cervix cytology and colposcopy until the lesions regress. Although
loop electrosurgical excision (LEEP) procedures can be performed on
LSIL patients to avoid LSIL progression, this treatment can be
considered excessive in addition to causing an economic burden on
the healthcare system (5).
Therefore, it is of medical and economical relevance to identify
biomarkers that can distinguish patients with LSIL that are at high
risk of progressing into HSIL and cervical cancer (6,7).
Among human papillomavirus (HPV)-infected
individuals, only 1% will progress to cervical cancer (8-11).
Investigations into the relationship between HPV and the host cell
cycle have identified a number of biomarkers, including
cyclin-dependent kinase inhibitor 2A (p16INK4a), marker
of proliferation Ki-67 (Ki-67) and immunohistochemical cocktail
containing antibodies direct against topoisomerase IIα (TOP2A) and
minichromosome maintenance 2 (MCM2) proteins (ProExC), which can be
measured to improve the detection and grading of HPV-associated
SIL, as they were found to be overexpressed in HPV-infected cells
(12). p16INK4a is a
kinase inhibitor and tumor suppressor that regulates cell cycle
progression in the G1-S phase and inhibits cell
proliferation through a reciprocal relationship with another tumor
suppressor protein, retinoblastoma protein (pRb) (13). Ki-67 is a nuclear protein expressed
only during the active phases of the cell cycle, namely in the late
G1, S, G2 and M phases, but not during the
resting phases (G0 and early G1) (14). By contrast, ProExC, a specific marker
of S phase-induced abnormalities, is associated with
transcriptional dysregulation and abnormalities caused by the HPV
E7 oncoprotein through the E2F transcription factor pathway, which
serves an important role in the development and progression of
cervical cancer (15,16). As ProExC is predominantly localized
in the nucleus and has demonstrated high sensitivity and
specificity for HSIL diagnosis (17). Furthermore, it is easier to identify
compared with p16INK4a (18). It is therefore possible that
p16INK4a, Ki-67 and ProExC can be applied as early HSIL
and cervical cancer risk indicators in early cervical lesions.
However, to the best of our knowledge, there have been limited
studies on the predictive value of p16INK4a and Ki-67
for LSIL prognosis with inconsistent results (19-21),
whilst the use of ProExC staining as a biomarker for LSIL prognosis
has not been previously reported.
The central aim of the present study was to
investigate the expression profiles of p16INK4a, Ki-67,
and ProExC in LSIL that progressed into HSIL and those that
regressed or exhibited stable LSIL. Additionally, the prognostic
value of p16INK4a, Ki-67 and ProExC as potential markers
for LSIL progression was evaluated.
Materials and methods
Study design and patient
selection
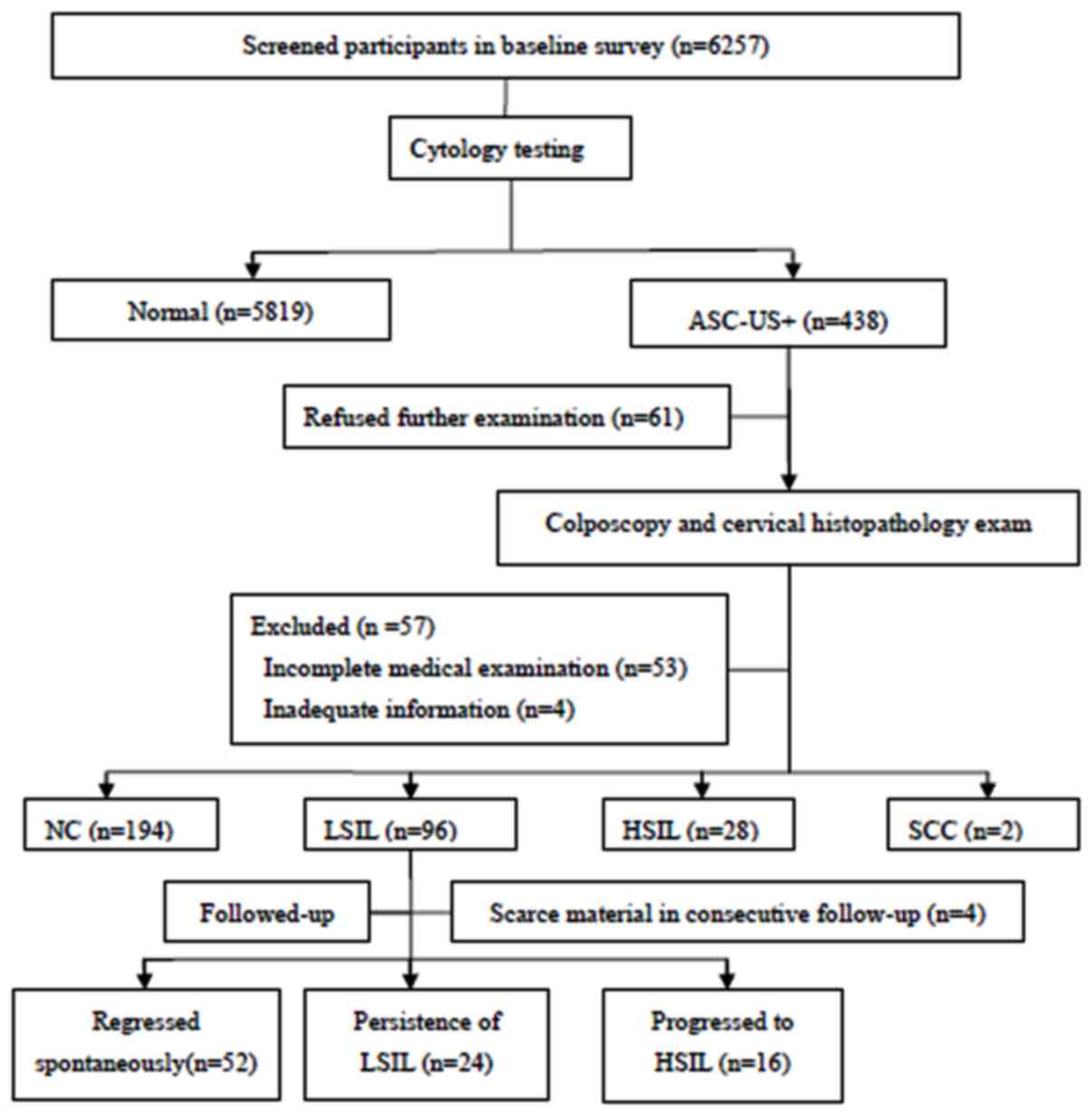
The present study followed a nested case-control
design. Patients were recruited in Jiexiu, Shanxi between October
and December 2014 and were willing to participate in the screening
program. The inclusion criteria were as follows: i) Of Han Chinese
ethnicity; ii) married; iii) had resided in Shanxi for ≥1 year; iv)
had current or past sexual activity; v) not pregnant; vi) no prior
history of cervical cancer or precancerous lesions; vii) no prior
history of treatments associated with the cervix including LEEP,
conization and adnexectomy; viii) agreed to participate in the
present study. A total of 6,257 women aged 19-65 years were
included and completed a demographic characteristic-related
questionnaire and underwent thin-prep cytologic test (TCT) testing.
All participants with abnormal cervical cytology results were
referred to colposcopy and histopathological examination. Of the
438 women diagnosed with atypical squamous cells of undetermined
significance and above according to the TCT (ASC-US+), 118 women
were excluded; with 61 due to refused consent, 53 due to incomplete
medical examination and 4 due to inadequate information, including
outliers in the questionnaire regarding age and pregnancy history.
Of a total of 320 women who received the final diagnosis, 194 were
diagnosed with normal cervix, 96 with LSIL, 28 with HSIL and 2 with
SCC. Of the 96 women diagnosed with LSIL, 16 progressed to HSIL and
24 exhibited persistent LSIL which were classified as the case
group, whilst 52 patients whose LSIL regressed spontaneously were
considered the control group. A total of 4 patients were excluded
due to insufficient material in the consecutive follow-up (Fig. 1).
All patients satisfying the inclusion criteria,
including those diagnosed with HSIL, were followed-up every 6
months for 2 years with colposcopy and histopathological
examinations. Any patients diagnosed with HSIL were treated by
professional clinicians using LEEP or cervical conization, where
endocervical curettage was performed simultaneously. and their
follow-up was completed. All subjects provided written informed
consent; the present study was approved by the Ethical Committee of
Shanxi Medical University (Shanxi, China) and performed in
accordance with the Declaration of Helsinki.
Liquid-based cytology and HR-HPV
testing
All subjects were requested to provide two cervical
tissue specimens, obtained by exfoliation using a brush during
gynecological examination. One sample was automatically prepared
for TCT using the Cytyc Thinprep® 2000 (Cytyc
Corporation). Cytological classifications of disease grade were
made by 2 cytopathology physicians under double-blinded conditions
using parameters defined by the current 2001 Bethesda System
(22). The other sample was
processed for HPV genotyping by Hybri-Max™ using an HPV GenoArray
Diagnostic kit (Guangdong Hybribio Biotechnology Co., Ltd.). A
total of 21 types of HPV, including 15 HR-HPV serotypes and 6
low-risk serotypes can be identified by flow-through hybridization
using a SLAN®-96S Real-Time PCR System (cat. no. SN
161403401; Shanghai Hongshi Medical Technology Co., Ltd.) and HHM-2
fast nucleic acid molecule hybridization instrument (cat. no.
20152400604; Guangdong Hybribio Biotechnology Co., Ltd.) (23).
Colposcopy and histopathological
examination
Colposcopy was performed by gynecological
specialists at the Second Hospital of Shanxi Medical University
(Shanxi, China) using the Preventive Oncology International
micro-biopsy protocol of directed and random biopsies (24), which results in ≥4 cervical biopsies
received from patients with or without endocervical curettage
(2). The histology slides were
interpreted and the diagnosis was agreed upon by the two
pathologists.
Immunohistochemical (IHC) detection of
p16INK4a, Ki-67 and ProExC
All specimens were fixed in 10% formalin for 24 h at
room temperature, embedded in paraffin, cut into continuous 4 -µm
sections and subsequently dewaxed. Following alcohol dehydration,
the sections were incubated in 3% hydrogen peroxide for 20 min at
room temperature to block endogenous peroxidase activity. Antigen
retrieval was then performed in 10 mM citrate buffer (Dako; Agilent
Technologies, Inc.) for 3 min at 100˚C and PBS washing, with this
step was repeated for 3 times. The sections were blocked with 10%
normal goat serum (ZSGB-BIO; OriGene Technologies, Inc.) for 10 min
at room temperature followed by incubation overnight at 4˚C with
the following primary antibodies: Monoclonal mouse
anti-p16INK4a (1:100; cat. no. TA500036; OriGene
Technologies, Inc.), monoclonal mouse anti-Ki-67 (1:100; cat. no.
RMA-0542; Fuzhou Maixin Biotech Co., Ltd.) and ProExC, formed by
mixing mouse monoclonal anti-MCM2 at (1:100; cat. no. sc-373702)
with anti-TOP2A antibody (1:50; cat. no. sc-365916; both Santa Cruz
Biotechnology, Inc.). The following day, the sections were treated
in accordance with manufacturer's protocol of Histostain-SP kit
(cat. no. SP-9002; ZSGB-BIO; OriGene Technologies, Inc.). Briefly,
the sections were incubated with biotinylated goat anti-mouse
immunoglobulin G secondary antibodies for 15 min at room
temperature and horseradish peroxidase-conjugated streptavidin for
15 min at room temperature. The slides were then incubated with
3,3-diaminobenzidine for 1-2 min, washed three times using PBS, and
stained with hematoxylin-eosin for 1 min. Following dehydration,
the sections became transparent and were covered with neutral
balsam (cat. no. G8590; Beijing Solarbio Science & Technology
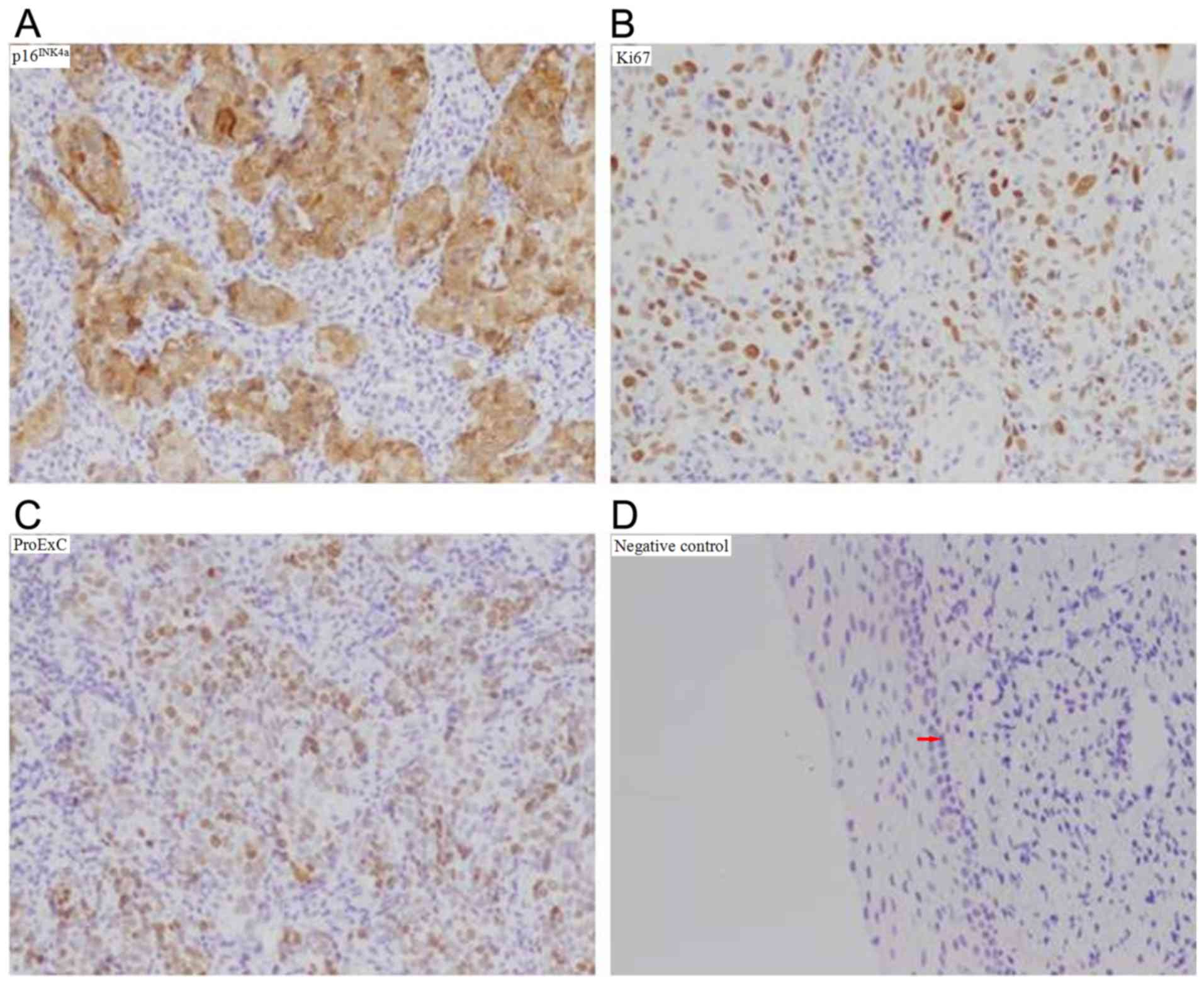
Co., Ltd.). Human prostate carcinoma tissues provided by the
Pathology Department of the Second Hospital of Shanxi Medical
University (Shanxi, China) were applied as positive controls and
PBS solution without primary antibodies was used as negative
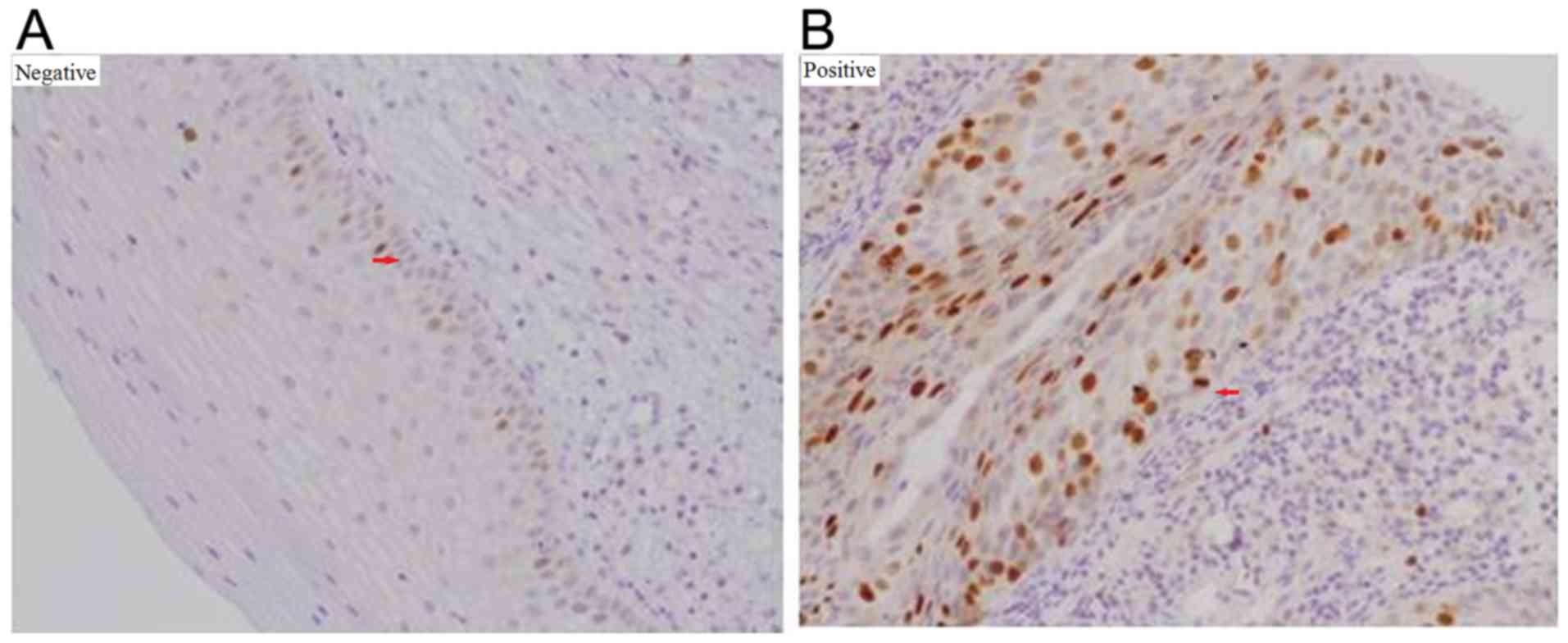
control (Fig. 2).
Light microscopic evaluation of
p16INK4a, Ki-67 and ProExC staining
Nuclear or nuclear plus cytoplasmic staining was
considered to be positive for p16INK4a expression,
whilst cytoplasmic staining alone was recorded as negative.
Staining for Ki-67 and ProExC was exclusively nuclear. For
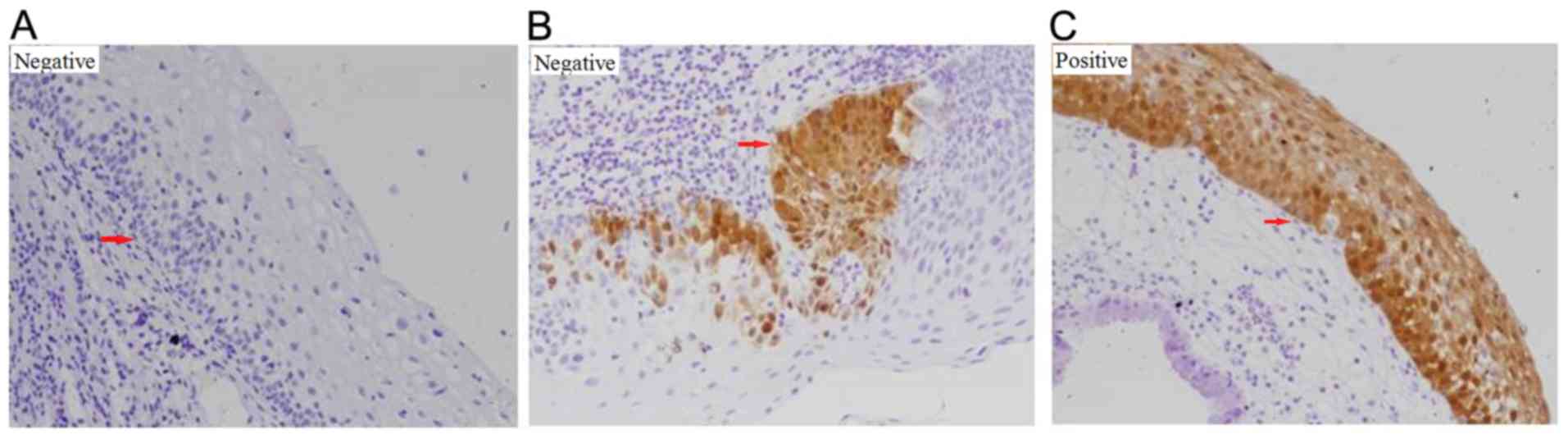
p16INK4a, a lack of staining, staining of isolated cells
or small cell clusters, and a focal staining pattern were
considered negative; whereas continuous, diffuse cellular staining
in the basal and parabasal cell layers was considered positive
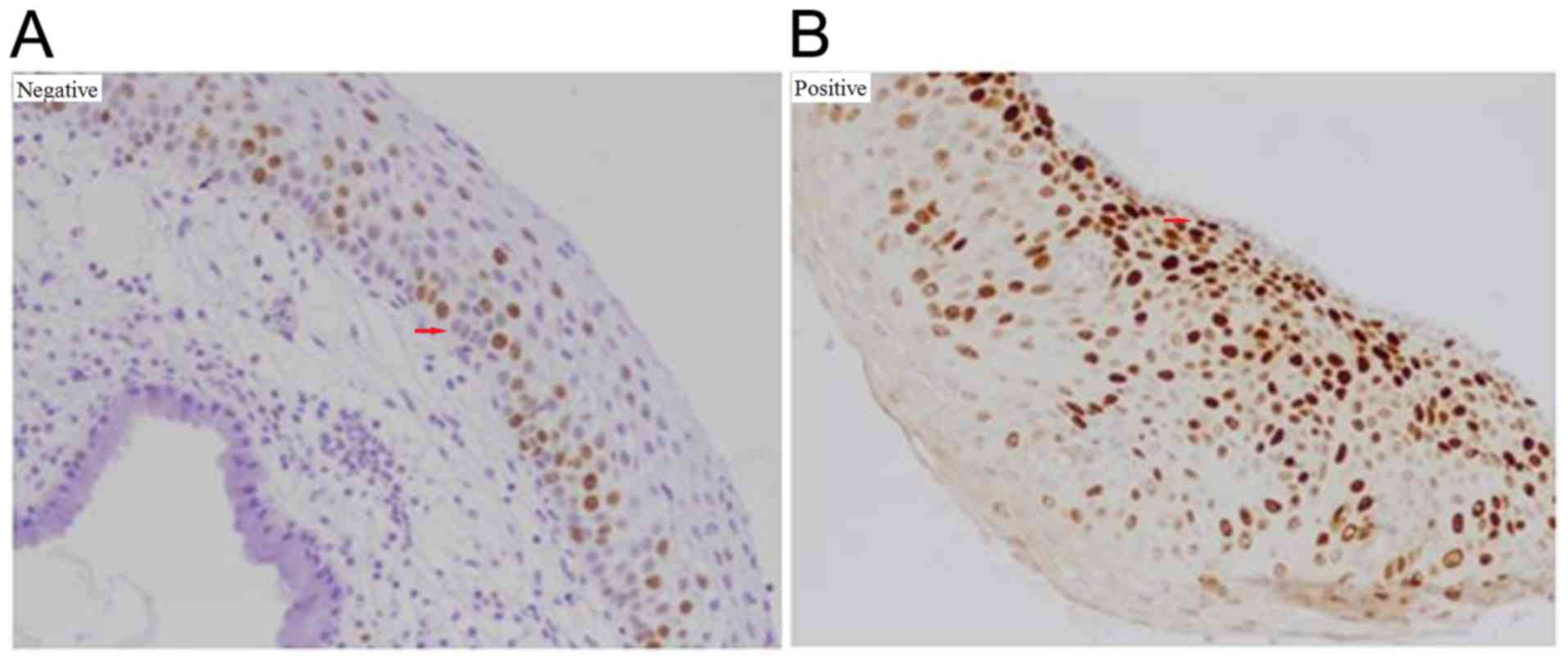
(Fig. 3) (25). For Ki-67, <50% staining or
staining only in the lower half of the epithelium was interpreted
as negative; >50% staining or staining in more than half of the
epithelium was considered positive (Fig.
4) (26). For ProExC, minor
adjustments were made to the protocols previously reported by Shi
et al (27) and Pinto et
al (28), where staining was
assessed in accordance with the distribution of positive cells in
the vertical plane of the squamous epithelium. No positive cells or
positive cells occupying <33% of the squamous epithelium was
interpreted as negative, whilst positive cells occupying >33% of
the squamous epithelium was interpreted as positive (Fig. 5). One tissue section was analyzed per
patient. Blinded analysis of all sections was conducted by two
pathologists using light microscopy (BX46; Olympus Corporation)
according to the protocol described previously (29).
Statistical analysis
All statistical analyses were performed using the
SPSS software package (version 22.0; IBM Corp.). The data are
expressed as the mean ± standard deviation or percentages. One-way
ANOVA and Pearson's Chi-squared test were applied to compare
continuous and categorical factors, respectively. Sensitivity,
specificity, positive predictive value (PPV), negative predictive
value (NPV), positive likelihood ratio (PLR), negative likelihood
ratio (NLR) and Youden's index (YI) were calculated based on
p16INK4a, Ki-67, and ProExC staining using the formulae
of Galen and Gambino (30):
Sensitivity = true positive-(true positive + false negative);
specificity = true negative-(false positive + true negative); PPV =
true positive-(true positive + false positive); NPV = true
negative-(false negative + true negative); PLR =
sensitivity-(1-specificity); NLR = (1-sensitivity)-specificity; and
YI = sensitivity + specificity-1. Multinomial logistic regression
analysis was used to analyze the association between the potential
predictor variables and LSIL prognosis. Data from the LSIL
regression group served as the reference category. The odds ratios
(OR) with 95% confidence intervals (CI) were calculated based on
Wald Chi-squared statistics. All statistical tests were two-tailed
and P<0.05 was considered to indicate a statistically
significant difference.
Results
Patient characteristics
A total of 92 cases of LSIL were found; of which 16
patients progressed to HSIL, 24 patients were diagnosed with
persistent LSIL and 52 patients exhibited LSIL that regressed
spontaneously. Table I displays the
characteristics of the subjects in the progression, persistence and
regression groups. The median ages in the progression, persistence,
and regression groups were 42.8 (29-55 years), 45.9 (26-60 years)
and 44.9 years (24-60 years), respectively. No significant
differences were observed in age, age at menarche, age at marriage,
age at first intercourse, age at first pregnancy, age at delivery,
TCT, HR-HPV infection, spouse smoking history, gravidity, parity,
menstrual regularity, condom use, intrauterine device use,
vaginitis history, pelvic inflammation disease history, chronic
disease history, or gynecological operation history among the
regression, persistence and progression groups. However, there were
significant differences in menopause status, where significantly
fewer post-menopausal subjects were found in the progression group
compared with those in the persistence and regression groups.
 | Table ICharacteristics among the study
population (n=92). |
Table I
Characteristics among the study
population (n=92).
| Characteristic | Progression
(n=16) | Persistence
(n=24) | Regression
(n=52) |
P-valuea |
|---|
| Age (years) | 42.8±7.3 | 45.9±9.2 | 44.9±9.4 | 0.188 |
| Age at menarche
(years) | 14.6±1.5 | 14.9±2.2 | 14.5±2.1 | 0.794 |
| Age at marriage
(years) | 23.3±2.7 | 23.3±4.4 | 23.1±2.2 | 0.963 |
| Age at first
intercourse (years) | 23.0±2.7 | 22.8±4.3 | 22.9±2.4 | 0.984 |
| Age of first
pregnancy (years) | 23.9±2.6 | 23.0±6.3 | 23.7±2.5 | 0.726 |
| Age at delivery
(years) | 25.2±3.0 | 23.5±6.5 | 24.3±2.5 | 0.445 |
| TCTb (1-2-3-4-5) % |
0-25.0-62.5-12.5-0 |
4.2-33.3-37.5-20.8-4.2 |
4.0-16.0-68.0-10.0-2.0 | 0.461 |
| HR-HPV infection (%
yes) | 81.3 | 62.5 | 48.1 | 0.054 |
| Spouse smoking
history (% yes) | 87.5 | 75 | 59.6 | 0.08 |
| Gravidity (%, <2
times) | 37.5 | 45.8 | 46.2 | 0.822 |
| Parity (%, <3
times) | 75 | 79.2 | 88.5 | 0.348 |
| Menstrual
regularity (% yes) | 68.8 | 87.5 | 90.4 | 0.091 |
| Menopausal status
(% post) | 6.3 | 41.7 | 34.6 | 0.047 |
| Condoms use ever (%
yes) | 6.3 | 8.3 | 11.5 | 0.793 |
| IUD use ever (%
yes) | 50 | 62.5 | 42.3 | 0.261 |
| Vaginitis history
(% yes) | 43.8 | 29.2 | 36.5 | 0.911 |
| Pelvic inflammatory
disease history (% yes) | 12.5 | 0 | 23.1 | 0.113 |
| Chronic disease
history (% yes) | 12.5 | 16.7 | 15.4 | 0.936 |
| Gynecological
operation history (% yes) | 25 | 8.3 | 11.5 | 0.274 |
p16INK4a, Ki-67 and ProExC
staining and analysis of association with HR-HPV infection
According to IHC, positive p16INK4a was
defined as observation of nuclear or nuclear plus cytoplasmic
staining, whereas positive Ki-67 and ProExC expression were defined
as exclusively nuclear staining. The p16INK4a expression
rates in the progression, persistence, and regression groups were
found to be 75.0, 41.7 and 28.8%, respectively. The Ki-67
expression rates in the progression, persistence, and regression
groups were 37.5, 12.5 and 7.7%, respectively. The rates of
positive ProExC staining in the progression, persistence, and
regression groups were 75.0, 41.7 and 32.7%, respectively (Table II). The differences in
p16INK4a, Ki-67 and ProExC staining between the 3 groups
were statistically significant (χ2=10.87, 9.03, 8.98;
P<0.05). Specifically, the rates of p16INK4a, Ki-67,
and ProExC staining were higher in the progression group compared
with those in the persistence and regression groups (P<0.05),
but the differences between the persistence and regression groups
were not statistically significant (P>0.05; Table II).
 | Table IIComparison of p16INK4a,
Ki67, ProExC expression in the progression, persistence and
regression groups. |
Table II
Comparison of p16INK4a,
Ki67, ProExC expression in the progression, persistence and
regression groups.
| A,
p16INK4a |
|---|
| Expression
status | Progression
(n=16) | Persistence
(n=24) | Regression
(n=52) | χ2 | P-value |
|---|
| Negative | 4 (25.0) | 14 (58.3%) | 37 (71.2%) | | 0.004 |
| Positive | 12 (75.0%) | 10 (41.7%) | 15 (28.8%) | 10.87 | |
| B, Ki-67 |
| Expression
status | Progression
(n=16) | Persistence
(n=24) | Regression
(n=52) | χ2 | P-value |
| Negative | 10 (62.5%) | 21 (87.5%) | 48 (92.3%) | | 0.011 |
| Positive | 6 (37.5%) | 3 (12.5%) | 4 (7.7%) | 9.03 | |
| C, ProExC |
| Expression
status | Progression
(n=16) | Persistence
(n=24) | Regression
(n=52) | χ2 | P-value |
| Negative | 4 (25.0%) | 14 (58.3%) | 35 (67.3%) | | 0.011 |
| Positive | 12 (75.0%) | 10 (41.7%) | 17 (32.7%) | 8.98 | |
To analyze the association between HR-HPV infection
and p16INK4a, Ki-67 and ProExC staining, the 92 subjects
were divided into 2 groups in accordance with their HR-HPV status.
The rate of positive p16INK4a expression in the
HR-HPV-positive group was significantly higher compared with that
in the HR-HPV-negative group. Specifically, p16INK4a
expression was found to significantly positively associated with
HR-HPV infection (P=0.001; Table
III). However, no significant associations were found between
Ki-67 or ProExC expression status and HR-HPV infection (P>0.05;
Table III). Of the four LSIL
progression cases with negative ProExC immunostaining, three (3-4,
75%) were tested negative for HR-HPV. By contrast, the majority of
(10-12, 83%) the LSIL progression cases demonstrating positive
ProExC immunostaining were also tested HR-HPV positive (data not
shown).
 | Table IIIAssociation analysis between
p16INK4a, Ki67 and ProExC expression with HR-HPV
infection. |
Table III
Association analysis between
p16INK4a, Ki67 and ProExC expression with HR-HPV
infection.
| A,
p16INK4a |
|---|
| | HR-HPV
infection | | |
|---|
| Expression
status | Negative (%) | Positive (%) | χ2 | P-value |
|---|
| Negative | 31 (79.5) | 24 (45.3) | 10.933 | 0.001 |
| Positive | 8 (20.5) | 29 (54.7) | | |
| B, Ki-67 |
| | HR-HPV
infection | | |
| Expression
status | Negative (%) | Positive (%) | χ2 | P-value |
| Negative | 34 (87.2) | 45 (84.9) | 0.096 | 0.757 |
| Positive | 5 (12.8) | 8 (15.1) | | |
| C, ProExC |
| | HR-HPV
infection | | |
| Expression
status | Negative (%) | Positive (%) | χ2 | P-value |
| Negative | 23 (59.0) | 30 (56.6) | 0.052 | 0.82 |
| Positive | 16 (41.0) | 23 (43.4) | | |
Statistical analysis of the utility of
IHC for p16INK4a, Ki-67, and ProExC for the prediction
of HSIL progression
Patients with progression into HSIL were classified
into the progression group, whereas those with LSIL regression or
persistence were assigned into the non-progression group. The rates
of positive p16INK4a, Ki-67, and ProExC expression in
the progression groups were found to be 75.0, 37.5 and 75.0%,
respectively. The rates of positive expression of
p16INK4a, Ki-67, and ProExC in the non-progression
groups (persistence and regression groups) were found to be 32.9
(25-76), 9.2 (7-76) and 35.5% (27-76), respectively (Table II).
Table IV shows the
sensitivity, specificity, PPV, NPV, PLR, NLR, and YI for
p16INK4a, Ki-67, and ProExC staining for the prediction
of progression to HSIL. The sensitivity and NPV were calculated to
be the highest for p16INK4a (75.0 and 92.7%
respectively), whereas the specificity, PPV, PLR and NLR were the
highest for Ki-67 (90.8%, 46.2%, 4.08 and 0.69, respectively). YI,
which is considered a more comprehensive measure of sensitivity and
specificity (31), was found to be
the highest for p16INK4a (42.1%), followed by ProExC
(39.5%) and Ki-67 (28.3%).
 | Table IVValues of p16INK4a, Ki67
and ProExC positivity in LSIL specimens to predict HSIL. |
Table IV
Values of p16INK4a, Ki67
and ProExC positivity in LSIL specimens to predict HSIL.
| Variable | Sensitivity
(%) | Specificity
(%) | PPV (%) | NPV (%) | PLR | NLR | YI (%) |
|---|
| p16INK4a
positivity | 75.00 | 67.10 | 32.40 | 92.70 | 2.28 | 0.38 | 42.10 |
| Ki-67
positivity | 37.50 | 90.80 | 46.20 | 87.30 | 4.08 | 0.69 | 28.30 |
| ProExC
positivity | 75.00 | 64.50 | 30.80 | 92.40 | 2.11 | 0.39 | 39.50 |
Risk factors affecting LSIL
progression
HR-HPV infection, and staining for
p16INK4a, Ki-67 and ProExC were examined by multivariate
logistic regression analysis (significance level, α=0.05; permitted
error =0.10) to obtain a statistically significant equation.
Menopausal status, which was found to be a significant variable in
the present study (Table I), was
also included as one of the items in this analysis. The logistic
regression models revealed ProExC expression to be an independent
risk factor for LSIL progression after 2 years. The risk of
progression for LSIL patients with positive ProExC staining was
found to be 6.11-fold higher compared with that of patients
negative for ProExC staining (95% CI, 1.438-25.997; Table V).
 | Table VMultivariate analysis of potential
risk factors for LSIL progression or persistence in patients with
different cervical intraepithelial neoplasia prognosis. |
Table V
Multivariate analysis of potential
risk factors for LSIL progression or persistence in patients with
different cervical intraepithelial neoplasia prognosis.
| A, LSIL
persistence |
|---|
| | | | | | | 95% CI |
|---|
| Variable | β | SE | Wald | P-value | OR | Lower | Upper |
|---|
| HR-HPV
infection | 0.607 | 0.549 | 1.223 | 0.269 | 1.834 | 0.626 | 5.376 |
| menopause
status | 0.7 | 0.569 | 1.517 | 0.218 | 2.015 | 0.661 | 6.141 |
| p16INK4a
staining | 0.407 | 0.584 | 0.485 | 0.486 | 1.502 | 0.478 | 4.717 |
| Ki67 staining | 0.57 | 0.867 | 0.432 | 0.511 | 1.767 | 0.323 | 9.664 |
| ProExC
staining | 0.579 | 0.547 | 1.121 | 0.29 | 1.785 | 0.611 | 5.217 |
| B, LSIL
progression |
| | | | | | | 95% CI |
| Variable | β | SE | Wald | P-value | OR | Lower | Upper |
| HR-HPV
infection | 1.502 | 0.844 | 3.167 | 0.075 | 4.489 | 0.859 | 23.465 |
| menopause
status | -0.835 | 1.159 | 0.519 | 0.471 | 0.434 | 0.045 | 4.208 |
| p16INK4a
staining | 1.038 | 0.771 | 1.812 | 0.178 | 2.823 | 0.623 | 12.791 |
| Ki67 staining | 1.526 | 0.91 | 2.808 | 0.094 | 4.598 | 0.772 | 27.386 |
| ProExC
staining | 1.811 | 0.738 | 6.011 | 0.014 | 6.114 | 1.438 | 25.997 |
Discussion
In the present study, IHC was performed to detect
and measure the expression of p16INK4a, Ki-67, and
ProExC in cervical specimens. IHC is a conventional method in
clinical diagnosis where there is no strict requirement for the
size of tissues.
The present nested case-control study demonstrated
that although IHC staining for p16INK4a, Ki-67 and
ProExC can be applied to predict HSIL progression, only ProExC
staining was an independent risk factor for LSIL progression after
2 years. To the best of our knowledge, the present study was the
first to analyze the predictive value of ProExC staining compared
with p16INK4a and Ki-67 for the progression of LSIL
among Han Chinese women.
The p16INK4a tumor suppressor protein
inhibits the cyclin-dependent kinases that regulate progression
through the cell cycle by phosphorylating the retinoblastoma
protein. In most cervical malignancies, the functional inactivation
of pRb by HPV E7 results in the overexpression of
p16INK4a (32). Previous
studies have demonstrated that p16INK4a can be used as a
prognostic marker for disease progression and infection by HR-HPV
(33,34). Consistent with this notion, it was
found in the present study that p16INK4a expression was
associated with HR-HPV infection. The value of p16INK4a
in CIN grading has been previously reported. In 2012, the American
Society for Colposcopy and Cervical Pathology and the Lower
Anogenital Squamous Terminology project recognized the value of
p16INK4a by proposing a new 2-stage classification
system; specifically for LSIL to include CIN1 and
p16INK4a-negative CIN2, and for HSIL to include
p16INK4a-positive CIN2 and CIN3(35). However, studies on the value of
p16INK4a in LSIL progression are limited, with
inconsistent findings. In the past decade, the majority of studies
suggested that patients with p16INK4a-positive LSIL were
at higher risks of developing HSIL (36-38).
Liao et al (19) conducted a
prospective population-based study to evaluate if the
overexpression of p16INK4a in LSIL biopsies can
accurately predict HSIL progression and found that
p16INK4a expression in LSIL on initial diagnosis was
associated with an increased risk of HSIL in 2 years (OR=1.43; 95%
CI, 0.52-3.91). However, Sagasta et al (20) showed that HSIL/CIN2-3 exhibited
higher positive rates for p16INK4a staining compared
with persistent or regressing LSIL/CIN1 lesions (71 vs. 44%), but
found that p16INK4a immunostaining was not associated
with risk of HSIL [hazard ratio 1.6 (95% CI, 0.9-2.6); P=0.095].
Sagasta et al (20)
subsequently concluded that p16INK4a overexpression in
biopsies from women with LSIL was a poor predictor in LSIL
progression, with little or no value as a marker in clinical
practice. Results from the present study were consistent with the
latter study. The p16INK4a expression rate in the
progression group was higher compared with the persistence and
regression groups, and the YI for p16INK4a was the
highest of the markers tested, suggesting that p16INK4a
was the most accurate marker for predicting HSIL progression.
However, p16INK4a expression was not an independent risk
factor for LSIL progression after 2 years, suggesting that
p16INK4a can be used to increase the accuracy of the
HSIL diagnosis but not in predicting LSIL progression.
Ki-67 is a DNA-binding enzyme which is utilized
widely to measure tumor cell proliferation and evaluate the degree
of tumor malignancy and prognosis. A number of studies have
previously reported that Ki-67 expression is positively associated
with the grade of cervical lesions (39-41).
In 2015, the Bethesda guidelines recommended
p16INK4a/Ki-67 double staining as part of the
cytological diagnostic procedure (42). A dual p16/Ki-67 immunocytochemistry
assay is now available for use as an adjunct test for cervical
cancer screening (43). In a
previous study, the p16INK4a/Ki-67 dual staining
strategy was tested in a large, prospective clinical trial
involving 27,456 women; where p16INK4a/Ki-67 dual
staining cytology testing was found to increase the sensitivity of
HSIL diagnosis whilst maintaining high specificity (44). In another study, Kanthiya et
al (45) found that Ki-67
expression was demonstrated in 75.4% of CIN2-3, 22.6% of CIN1 and
11.3% clinical specimens with non-dysplasia. These results suggest
that Ki-67 overexpression can also be used as a marker for the
tendency for progression in early cervical lesions. However, only
one study, which was conducted by Kruse et al (46), included samples from patients with
CIN1 (n=25) and CIN2 (n=65) and investigated Ki-67 IHC in CIN
progression. They found that the prognostic value of the Ki-67
progression-risk model exceeded the value of the histopathological
CIN grade for the prediction of progression to a higher CIN grade.
Although it was found that the Ki-67 expression rate was higher in
the progression group compared with that in the persistence and
regression groups in the present study, Ki-67 expression was not an
independent risk factor for LSIL progression after 2 years.
Therefore, further studies are crucial to confirm the value of
Ki-67 IHC for the prediction of LSIL progression.
ProExC consists of antibodies specific for MCM2 and
TOP2A, both of which are overexpressed during cervical dysplasia
and neoplasia (14). MCM2 is a
member of the DNA licensing factor family and a marker of cell
proliferation, whereas TOP2A is an enzyme that unwinds and
decatenates DNA in preparation for DNA replication, transcription,
chromosome segregation, and cell cycle progression (47,48).
Walts and Bose (12) evaluated the
efficacy of p16INK4a, Ki-67, and ProExC immunostaining,
alone and in combination, for the diagnosis of CIN, and provided
guidance for instances with discordant staining patterns. ProExC
expression was found in 26.0% CIN2-3 and in 6.0% CIN1 cases.
Although these findings confirmed that p16INK4a, Ki-67,
and ProExC are specific and sensitive markers that can be used for
the diagnosis of CIN, no prospective studies had previously
investigated the predictive value of ProExC for LSIL progression.
The present study found that the ProExC staining rate in the
progression group was higher compared with those in the persistence
and regression groups and that the YI for ProExC staining was
higher compared with that for Ki-67, indicating that ProExC IHC was
more suitable for the predicting progression to HSIL. The
overexpression of MCM2 provides the link between oncogenic HPV
infection and the molecular event of cervical dysplasia (49), which is consistent with data from the
present study. Of the four LSIL progression cases with negative
ProExC immunostaining, three (3-4, 75%) were negative for HR-HPV.
By contrast, the majority of (10-12, 83%) the LSIL progression
cases demonstrating positive ProExC immunostaining were also tested
HR-HPV positive. Additionally, only positive ProExC staining was
found to be an independent risk factor for LSIL progression in 2
years, suggesting that ProExC-positive LSIL pose a higher risk of
developing into HSIL.
To conclude, to the best of our knowledge, this is
the first study to report that compared with p16INK4a
and Ki-67, only ProExC staining was an independent risk factor for
LSIL progression over 2 years. This study provides a new insight
into identifying LSIL patients at a higher risk of malignant
progression, potentially facilitating more cost-effective and
efficient interventions.
Acknowledgements
We wish to thank Dr Ying Wang and Dr Xiaoqin Zhang
of The Department of Pathology of the Second Hospital of Shanxi
Medical University, Taiyuan, Shanxi, China for the pathological
analysis of tissue sections.
Funding
This research was supported by the National Natural
Science Foundation of China (grant nos. 81703313, 81702583 and
81473060) and the Special Public Welfare Industry Research of
National Health and Family Planning Commission (grant no.
201402010).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LD, LS, WZ and JW conceived and designed the present
study. XL, WG and ZQ collected the data and performed the
experiments. LD and WZ performed the data analysis and
interpretation. LD and LS wrote the manuscript. All the authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Shanxi Medical
University Science Research Ethics Committee (Shanxi, China), and
written informed consent was obtained from all patients prior to
publication.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wang Z, Wang J, Fan J, Zhao W, Yang X, Wu
L, Li D, Ding L, Wang W, Xu J, et al: Risk factors for cervical
intraepithelial neoplasia and cervical cancer in Chinese women:
Large study in Jiexiu, Shanxi Province, China. J Cancer. 8:924–932.
2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen W, Zheng R, Zhang S, Zhao P, Zeng H,
Zou X and He J: Annual report on status of cancer in China, 2010.
Chin J Cancer Res. 26:48–58. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Schlecht NF, Platt RW, Duarte-Franco E,
Costa MC, Sobrinho JP, Prado JC, Ferenczy A, Rohan TE, Villa LL and
Franco EL: Human papillomavirus infection and time to progression
and regression of cervical intraepithelial neoplasia. J Natl Cancer
Inst. 95:1336–1343. 2003.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Pretet JL, Jacquard AC, Saunier M, Clavel
C, Dachez R, Gondry J, Pradat P, Soubeyrand B, Leocmach Y, Mougin
C, et al: Human papillomavirus genotype distribution in low-grade
squamous intraepithelial lesions in France and comparison with
CIN2-3 and invasive cervical cancer: The EDiTH III study. Gynecol
Oncol. 110:179–184. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Koeneman MM, Kruitwagen RF, Nijman HW,
Slangen BF, Van Gorp T and Kruse AJ: Natural history of high-grade
cervical intraepithelial neoplasia: A review of prognostic
biomarkers. Expert Rev Mol Diagn. 15:527–546. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhao WH, Hao M, Cheng XT, Yang X, Wang ZL,
Cheng KY, Liu FL and Bai YX: c-myc gene copy number variation in
cervical exfoliated cells detected on fluorescence in situ
hybridization for cervical cancer screening. Gynecol Obstet Invest.
81:416–423. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Choi YJ and Park JS: Clinical significance
of human papillomavirus genotyping. J Gynecol Oncol.
27(e21)2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Rodriguez AC, Schiffman M, Herrero R,
Wacholder S, Hildesheim A, Castle PE, Solomon D and Burk R:
Proyecto Epidemiologico Guanacaste G. Rapid clearance of human
papillomavirus and implications for clinical focus on persistent
infections. J Natl Cancer Inst. 100:513–517. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Egawa N, Egawa K, Griffin H and Doorbar J:
Human papillomaviruses; epithelial tropisms, and the development of
neoplasia. Viruses. 7:3863–3890. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Zhao W, Hao M, Wang Y, Feng N, Wang Z,
Wang W, Wang J and Ding L: Association between folate status and
cervical intraepithelial neoplasia. Eur J Clin Nutr. 70:837–842.
2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Walts AE and Bose S: p16, Ki-67, and BD
ProExC immunostaining: A practical approach for diagnosis of
cervical intraepithelial neoplasia. Hum Pathol. 40:957–964.
2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ortega S, Malumbres M and Barbacid M:
Cyclin D-dependent kinases INK4 inhibitors and cancer. Biochim
Biophys Acta. 1602:73–87. 2002.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Takagi M, Sueishi M, Saiwaki T, Kametaka A
and Yoneda Y: A novel nucleolar protein, NIFK, interacts with the
forkhead associated domain of Ki-67 antigen in mitosis. J Biol
Chem. 276:25386–25391. 2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Leone G, DeGregori J, Yan Z, Jakoi L,
Ishida S, Williams RS and Nevins JR: E2F3 activity is regulated
during the cell cycle and is required for the induction of S phase.
Genes Dev. 12:2120–2130. 1998.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Goodwin EC and DiMaio D: Repression of
human papillomavirus oncogenes in HeLa cervical carcinoma cells
causes the orderly reactivation of dormant tumor suppressor
pathways. Proc Natl Acad Sci USA. 97:12513–125138. 2000.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Halloush RA, Akpolat I, Jim Zhai Q,
Schwartz MR and Mody DR: Comparison of ProEx C with p16INK4a and
Ki-67 immunohistochemical staining of cell blocks prepared from
residual liquid-based cervicovaginal material: A pilot study.
Cancer. 114:474–480. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Badr RE, Walts AE, Chung F and Bose S: BD
ProEx C: A sensitive and specific marker of HPV-associated squamous
lesions of the cervix. Am J Surg Pathol. 32:899–906.
2008.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Liao GD, Sellors JW, Sun HK, Zhang X, Bao
YP, Jeronimo J, Chen W, Zhao FH, Song Y, Cao Z, et al: p16 INK4A
immunohistochemical staining and predictive value for progression
of cervical intraepithelial neoplasia grade 1: A prospective study
in China. Int J Cancer. 134:1715–1724. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sagasta A, Castillo P, Saco A, Torné A,
Esteve R, Marimon L, Ordi J and Del PM: p16 staining has limited
value in predicting the outcome of histological low-grade squamous
intraepithelial lesions of the cervix. Mod Pathol. 29:51–59.
2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Silva DC, Goncalves AK, Cobucci RN,
Mendonca RC, Lima PH and Júnior GC: Immunohistochemical expression
of P16, KI67 and P53 in cervical lesions-A systematic review.
Pathol Res Pract. 213:723–729. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Solomon D, Davey D, Kurman R, Moriarty A,
O'connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T Jr, et
al: The 2001 bethesda system: Terminology for reporting results of
cervical cytology. JAMA. 287:2114–2119. 2002.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tao P, Zheng W, Wang Y and Bian ML:
Sensitive HPV genotyping based on the flow-through hybridization
and gene chip. J Biomed Biotechnol. 2012(938780)2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Bornstein J, Bentley J, Bösze P, Girardi
F, Haefner H, Menton M, Perrotta M, Prendiville W, Russell P,
Sideri M, et al: 2011 colposcopic terminology of the international
federation for cervical pathology and colposcopy. Obstet Gynecol.
120:166–172. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Darragh TM, Colgan TJ, Thomas Cox J,
Heller DS, Henry MR, Luff RD, McCalmont T, Nayar R, Palefsky JM,
Stoler MH, et al: The Lower Anogenital Squamous Terminology
Standardization project for HPV-associated lesions: Background and
consensus recommendations from the College of American Pathologists
and the American Society for Colposcopy and Cervical Pathology. Int
J Gynecol Pathol. 32:76–115. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang WC, Wu TT, Chandan VS, Lohse CM and
Zhang L: Ki-67 and ProExC are useful immnohistochemical markers in
esophageal squamous intraepithelial neoplasia. Hum Pathol.
42:1430–1437. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Shi J, Liu H, Wilkerson M, Huang Y,
Meschter S, Dupree W, Schuerch C and Lin F: Evaluation of p16INK4a,
minichromosome maintenance protein 2, DNA topoisomerase IIalpha,
ProEX C, and p16INK4a-ProEX C in cervical squamous intraepithelial
lesions. Hum Pathol. 38:1335–1344. 2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pinto AP, Schlecht NF, Woo TY, Crum CP and
Cibas ES: Biomarker (ProEx C, p16(INK4A), and MiB-1) distinction of
high-grade squamous intraepithelial lesion from its mimics. Mod
Pathol. 21:1067–1074. 2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu C, Ding L, Bai L, Chen X, Kang H, Hou
L and Wang J: Folate receptor alpha is associated with cervical
carcinogenesis and regulates cervical cancer cells growth by
activating ERK1-2-c-Fos-c-Jun. Biochem Biophys Res Commun.
491:1083–1089. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Galen RS and Gambino SR: Beyond Normality:
The Predictive Value and Efficiency of Medical Diagnoses. John
Wiley, Sons, New York, NY, pp1-237, 1975.
|
|
31
|
Youden WJ: Index for rating diagnostic
tests. Cancer. 3:32–35. 1950.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Khleif SN, DeGregori J, Yee CL, Otterson
GA, Kaye FJ, Nevins JR and Howley PM: Inhibition of cyclin
D-CDK4-CDK6 activity is associated with an E2F-mediated induction
of cyclin kinase inhibitor activity. Proc Natl Acad Sci USA.
93:4350–4354. 1996.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Liu HQ, Wang YH, Wang LL and Hao M:
P16INK4A and survivin: Diagnostic and prognostic markers in
cervical intraepithelial neoplasia and cervical squamous cell
carcinoma. Exp Mol Pathol. 99:44–49. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Branca M, Ciotti M, Santini D, Di Bonito
L, Giorgi C, Benedetto A, Paba P, Favalli C, Costa S, Agarossi A,
et al: p16(INK4A) Expression is related to grade of cin and
high-risk human papillomavirus but does not predict virus clearance
after conization or disease outcome. Int J Gynecol Pathol.
23:354–365. 2004.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Darragh TM, Colgan TJ, Cox JT, Heller DS,
Henry MR, Luff RD, McCalmont T, Nayar R, Palefsky JM, Stoler MH, et
al: The lower anogenital squamous terminology standardization
project for HPV associated lesions: Background and consensus
recommendations from the College of American Pathologists and the
American society for colposcopy and cervical pathology. J Low Genit
Tract Dis. 16:205–242. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Cortecchia S, Galanti G, Sgadari C, Costa
S, De Lillo M, Caprara L, Barillari G, Monini P, Nannini R, Ensoli
B and Bucchi L: Follow-up study of patients with cervical
intraepithelial neoplasia grade 1 overexpressing p16Ink4a. Int J
Gynecol Cancer. 23:1663–1669. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
del Pino M, Garcia S, Fuste V, Alonso I,
Fuste P, Torne A and Ordi J: Value of p16(INK4a) as a marker of
progression-regression in cervical intraepithelial neoplasia grade
1. Am J Obstet Gynecol. 201:488.e1–7. 2009.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Hariri J and Oster A: The negative
predictive value of p16INK4a to assess the outcome of cervical
intraepithelial neoplasia 1 in the uterine cervix. Int J Gynecol
Pathol. 26:223–228. 2007.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ozaki S, Zen Y and Inoue M: Biomarker
expression in cervical intraepithelial neoplasia: Potential
progression predictive factors for low-grade lesions. Hum Pathol.
42:1007–1012. 2011.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Hanprasertpong J, Tungsinmunkong K,
Chichareon S, Wootipoom V, Geater A, Buhachat R and Boonyapipat S:
Correlation of p53 and Ki-67 (MIB-1) expressions with
clinicopathological features and prognosis of early stage cervical
squamous cell carcinomas. J Obstet Gynaecol Res. 36:572–580.
2010.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Mitildzans A, Arechvo A, Rezeberga D and
Isajevs S: Expression of p63, p53 and Ki-67 in patients with
cervical intraepithelial neoplasia. Turk Patoloji Derg. 33:9–16.
2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Nayar R and Wilbur DC: The Bethesda System
for Reporting Cervical Cytology Definitions, Criteria, and
Explanatory Note, 3rd (ed). Switzerland: Springer International
Publishing, 103-134, 2015.
|
|
43
|
Šekoranja D and Repše Fokter A: Triaging
atypical squamous cells-cannot exclude high-grade squamous
intraepithelial lesion With p16-Ki67 dual stain. J Low Genit Tract
Dis. 21:108–111. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Tornesello ML, Buonaguro L, Giorgi-Rossi P
and Buonaguro FM: Viral and cellular biomarkers in the diagnosis of
cervical intraepithelial neoplasia and cancer. Biomed Res Int.
2013:519–619. 2013.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Kanthiya K, Khunnarong J, Tangjitgamol S,
Puripat N and Tanvanich S: Expression of the p16 and Ki67 in
cervical squamous intraepithelial lesions and cancer. Asian Pac J
Cancer Prev. 17:3201–3202. 2016.PubMed/NCBI
|
|
46
|
Kruse AJ, Baak JP, Janssen EA, Kjellevold
KH, Fiane B, Lovslett K, Bergh J and Robboy S: Ki67 predicts
progression in early CIN: Validation of a multivariate
progression-risk model. Cell Oncol. 26:13–20. 2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Osaki M, Osaki M, Yamashita H, Shomori K,
Yoshida H and Ito H: Expression of minichromosome maintenance-2 in
human malignant fibrous histiocytomas: Correlations with Ki-67 and
P53 expression, and apoptosis. Int J Mol Med. 10:161–168.
2002.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gibbons D, Fogt F, Kasznica J, Holden J
and Nikulasson S: Comparison of topoisomerase II alpha and MIB-1
expression in uterine cervical squamous lesions. Mod Pathol.
10:409–413. 1997.PubMed/NCBI
|
|
49
|
Ishimi Y, Okayasu I, Kato C, Kwon HJ,
Kimura H, Yamada K and Song SY: Enhanced expression of Mcm proteins
in cancer cells derived from uterine cervix. Eur J Biochem.
270:1089–1101. 2003.PubMed/NCBI View Article : Google Scholar
|