Introduction
Acute renal failure (ARF) of the kidney is an
important cause of morbidity and mortality in hospitalized
intensive care unit patients. Renal ischemia is a major cause of
ARF, initiating a complex and interrelated sequence of events,
resulting in injury and the eventual death of renal cells (1,2). The
prognosis is complicated by the fact that reperfusion, although
essential for the survival of ischemic renal tissue, causes
additional damage (reperfusion-injury) (3), contributing to the renal dysfunction
and injury associated with ischemia/reperfusion (I/R) of the kidney
(4). I/R injury is an inevitable
consequence of the procedure of kidney transplantation and has a
negative impact on both short- and long-term graft survival
(5-7).
The pathophysiology of I/R injury is complex, with at least three
major components contributing to the process of reperfusion injury:
Molecular oxygen, neutrophils and components of the activated
complement cascade (8-10).
Long noncoding RNA (lncRNA) is a group of non-coding
RNAs with a length longer than 200 nucleotides (11). LncRNAs are not associated with
protein synthesis and have previously been considered to be ‘noise’
of the transcriptome (11). However,
in recent years, lncRNAs have been indicated to regulate the
expression of protein-coding genes to participate in a number of
pathological and physiological processes (11). It has been well established that
lncRNAs are associated with the progression of a variety of human
diseases, including renal ischemia-reperfusion injury, by
regulating the expression of genes associated with the development
and progression of the disease (12). LncRNA np_5318 has been proved to be
closely correlated with the development of different kidney
diseases such as transforming growth factor (TGF)-β/Smad3 (mothers
against decapentaplegic homolog 3)-mediated renal inflammation
(13), which is common in renal
ischemia-reperfusion (14),
indicating that np_5318 may also be involved in the progression of
renal ischemia-reperfusion injury. However, the role of np_5318 in
renal I/R injury is still unknown.
The current study aimed to explore the expression
and function of np_5318 in renal I/R and investigate the potential
interaction between the np_5318 and TGF-β/Smad signaling pathway in
this disease. In the present study, the roles of np_5318 in renal
I/R were explored by establishing I/R animal and cell models and
the related mechnism was also verified using transfection
experiments. It was demonstrated that lncRNA np_5318 may
participate in the development of renal I/R injury through the
TGF-β/Smad signaling pathway.
Materials and methods
Animal, grouping and model
construction
Specific pathogen free grade female Balb/C mice
(n=20; age, 3 weeks; weight, 20±5 g) were obtained from Jinan
Jinfeng Experimental Animal Breeding Co., Ltd., [license number:
SCXK (lu) 2014-0006]. Animals were reared under specific
pathogen-free conditions (12 h dark/light cycles; 25˚C) with 95%
humidity with access to food and water ad libitum. All
experimental procedures were approved by the Institutional Animal
Care and Use Committee of Yucheng People's Hospital. Mice were
randomly divided into the sham group, renal I/R 4, 9, 24 and 48 h
model groups, 6 mice in each group. Mice were fasted for 12 h
before model construction. Intraperitoneal injection of 4% chloral
hydrate at a dose of 400 mg/kg was performed for anesthesia. An
incision was made along ventral midline to expose the kidneys. Both
renal pedicles were clamped using a non-invasive vessel clip until
the kidneys became purple and black. Clips were removed 45 min
later to restore perfusion. Reperfusion was achieved when kidneys
turned pink. Then, kidneys were collected at 4, 24 and 48 h for
further analysis. Mice in the sham group were treated with the same
procedure but the renal pedicles were not clamped.
Detection and evaluation of renal
function
Serum creatinine (Scr) level and blood urea nitrogen
(BUN) were detected using automatic biochemical test (Hitachi 7180;
Hitachi, Ltd., Tokyo, Japan) at 4, 24 and 48 h after reperfusion.
Renal tissue was collected for hematoxylin and eosin (HE) staining
at room temperature for 24 h after reperfusion to observe the
histopathological changes. A light microscope was used to observe
the staining.
Immunohistochemical staining
Tissues were fixed at 4˚C in fresh 4% (w/v)
formaldehyde solution for 12 h and paraffin-embedding was
performed. A total of 5 paraffin-embedded sections (12 µm) were
selected at each time point. Tissue sections were dewaxed and then
washed with PBS. Tissue sections were blocked in blocking fluid at
room temperature for 20 min to reduce nonspecific background
staining caused by endogenous peroxidase. Tissue sections were then
blocked with 10% serum (Roche Diagnostics) for 10 min at room
temperature, followed by incubation with corresponding primary
antibodies (eBioscience; Thermo Fisher Scientific, Inc.) over night
at 4˚C. After washing with PBS, tissue sections were incubated with
fluorescein isothiocyanate-labeled goat anti-rabbit immunoglobulin
(Ig) G secondary antibody (Chemicon international; Thermo Fisher
Scientific, Inc.) at room temperature for 30 min. After washing
with PBS again, tissue sections were incubated with
streptavidin-peroxidase solution at room temperature for half an
hour. After washing with PBS, DAB color development was performed.
After washing with distilled water and counter staining, the slides
were sealed. A total of 5 fields of view were selected under a
light microscope (x400) and positive cells were counted, and the
average value was calculated to represent microvessel density.
I/R cell model in vitro
Primary human renal cells (PCS-400-012) were
obtained from the American Type Culture Collection (ATCC; Manassas,
VA, USA) and cultured in complete renal epithelial cell growth
media (ATCC) at 37˚C in an incubator with 95% air and 5%
CO2 (15). Primary human
renal cells were plated to six-well plates (105 cells
per well) and cultured for 24 h. Then, these cells were incubated
with hypoxia treatment 0.5% O2 for 15 h, following
replaced with normal culture medium under the atmosphere of
constant oxygen for 6 h. After that, cells were washed and
collected for further reverse transcription-quantitative (RT-q)PCR
or western blotting analysis, respectively.
Cell transfection
Vector pc-np_5318 for overexpression of lncRNA
np_5318 in I/R cell was constructed by inserting the coding
oligonucleotides of lncRNA np_5318 into a pcDNA3.1 vector
(Invitrogen: Thermo Fisher Scientific, Inc.). Small interfering
(si)RNAs targeting lncRNA np_5318 (si-np_5318) were constructed for
inhibition of lncRNA np_5318. For cell transfection, I/R cells as
mentioned before were cultured in a six-well plates (105
cells per well) for 24 h and transfected with 10 nM pc-np_5318,
si-np_5318 (5'-CCUGUGCACGUUCGAUUCAUA-3'), and their corresponding
controls [pc-NC and si-NC (5'-GCGUGACGUACGUACGUACGA-3')
respectively] using Lipofectamine 2000® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells continued to be
incubated for another 48 h before subsequent experiments.
MTT assay
Using an MTT colorimetric assay, cell viability was
assessed. In brief, different transfected cells at logarithmic
stage were grown into a 96-well plate. After 24, 48 and 72 h of
transfection, 20 ml MTT was added into each well to incubate cells
for another 4 h. Then 150 ml dimethylsulfoxide was added into each
well to dissolve the formazan precipitates for 10 min. The
absorbance (490 nm) was measured under an absorption
spectrophotometer (Olympus Corporation, Tokyo, Japan). Each
experiment was repeated 3 times.
RT-qPCR
Total RNA was extracted using TRIzol reagent
(Invitrogen, Thermo Fisher Scientific, Inc.). RNA concertation were
determined using NanoDrop™ 2000 Spectrophotometers
(Thermo Fisher Scientific, Inc.) and only the ones with a ratio of
A260/A280 between 1.8 and 2.0 were used for reverse transcription
to synthesize cDNA using ploy (T) as primer and SuperScript III
Reverse Transcriptase kit (Thermo Fisher Scientific, Inc.) through
following conditions: 55˚C for 15 min and 75˚C for 10 min.
SYBR® Green Real-Time PCR Master Mixes (Thermo Fisher
Scientific, Inc.) was used to prepare the PCR reaction system.
Following primers were used: 5'-AACTCGCCACAGAAATCCAC-3' (forward)
and 5'-ACAACCCCAAACAAGCTGTC-3' (reverse) for np_5318;
5'-TGCTGAGTATGTCGTGGAGTCTA-3' (forward) and
5'-AGTGGGAGTTGCTGTTGAAATC-3' (reverse) for GAPDH. PCR reaction
conditions were: 95˚C for 40 sec, followed by 40 cycles of 95˚C for
15 sec and 60˚C for 45 sec. Cq values were processed using
2-ΔΔCq method (16). Relative expression level of each gene
was normalized to endogenous control GAPDH.
ChIP
ChIP was performed using Diagenode's iDeal ChIP-seq
kit for Transcription Factors (Diagenode SA, Seraing, Belgium)
according to the manufacturer's protocol. Mouse embryonic
fibroblasts (MEFs) were purchased from Sigma-Aldrich; Merck KGaA
and were cultivated under conditions described in manufacturer's
protocol. Briefly, Cross-linking was performed at 37˚C for 10 min
and was quenched with glycine. DNA fragments ranged from 300-600 bp
were generated through sonication using a Bioruptor (Diagenode SA).
Then an antibody against Smad3 (cat. no. 06-920, 1:1200; EMD
Millipore) was used for immunoprecipitation with normal IgG as
control. The antibody against IgG was also purchased from EMD
Millipore (1:1200; cat. no. PP64). The following primers were used
in PCR to detect the precipitated DNAs: Smad binding site (SBS) for
np_5318, 5'-CTCTCTCAAACAGCCTGTGG-3' and
5'-GAAATTTGGAGGTGCAATCAA-3'.
Western-blotting
Total protein extraction was performed using a
Qproteome Mammalian Protein Prep kit (Qiagen GmbH, Hilden, German)
according to the manufacturer's protocol. Protein concentration was
measured by bicinchoninic acid method. Protein samples were
denatured and 50 µg of protein was subjected to 10% SDS-PAGE gel
electrophoresis, followed by transmembrane to PVDF membrane. After
blocking with 5% skimmed milk at room temperature for 2 h,
membranes were washed with Tween buffered TBS (TBST; 01% Tween 20).
Membranes were then incubated with primary antibodies overnight at
4˚C. Primary antibodies used were rabbit anti TGF-β1 (1:2,000; cat.
no. ab92486; Abcam), Smad3 (1:2,000; cat. no. ab28379; Abcam),
p-Smad3 (1:2000; cat. no. ab52903; Abcam) and α-tubulin (1:2,000;
cat. no. ab18251; Abcam). After that, membranes were washed three
times with TBST. Membranes were then incubated with secondary
antibody at room temperature for 2 h. The secondary antibody was
HRP-Goat Anti-Rabbit (IgG) secondary antibody (1:2000; cat. no.
ab6721; Abcam). After washing twice with TBST, 15 min for each
time, ECL™ Blotting Reagents GE Healthcare (Sigma-Aldrich; Merck
KGaA) was added to detect the signals. Images were processed using
Bandscan 5.0 software (Nuohebio) to calculate the relative
expression level of each protein.
Statistical analysis
SPSS 10.0 was used for all statistical analysis
(SPSS, Inc., Chicago, IL, USA). Experimental data were expressed as
the mean ± standard deviation. Comparisons within a group were
performed using single factor analysis of variance and comparisons
between two groups were performed using independent t test.
Multiple comparison corrections were performed using Bonferroni
multiple comparisons test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Renal I/R injury group shows obvious
renal function impairment
Compared with the levels of SCr and BUN in the sham
group (24.56±5.94 and 7.44±3.99 µmol/l, respectively), levels of
SCr and BUN were increased in I/R group at 4 (41.61±4.79 and
14.45±0.83 µmol/l, respectively), 24 (94.42±53.89 and 39.35±15.88
µmol/l, respectively) and 72 h (36.34±4.78 and 17.42±11.48 µmol/l,
respectively) after reperfusion. However, significant differences
were only found at 24 h after reperfusion (P<0.05). Those data
suggest that I/R can increase the levels of SCr and BUN in mice,
which in turn impairs renal function (Table I).
 | Table IComparison of levels of SCr and BUN
among groups. |
Table I
Comparison of levels of SCr and BUN
among groups.
| Groups | SCr (µmol/l) | BUN (µmol/l) |
|---|
| Sham | 24.56±5.94 | 7.44±3.99 |
| I/R 4 h | 41.61±4.79 | 14.45±0.83 |
| I/R 24 h | 94.42±53.89a | 39.35±15.88a |
| I/R 72 h | 36.34±4.78 | 17.42±11.48 |
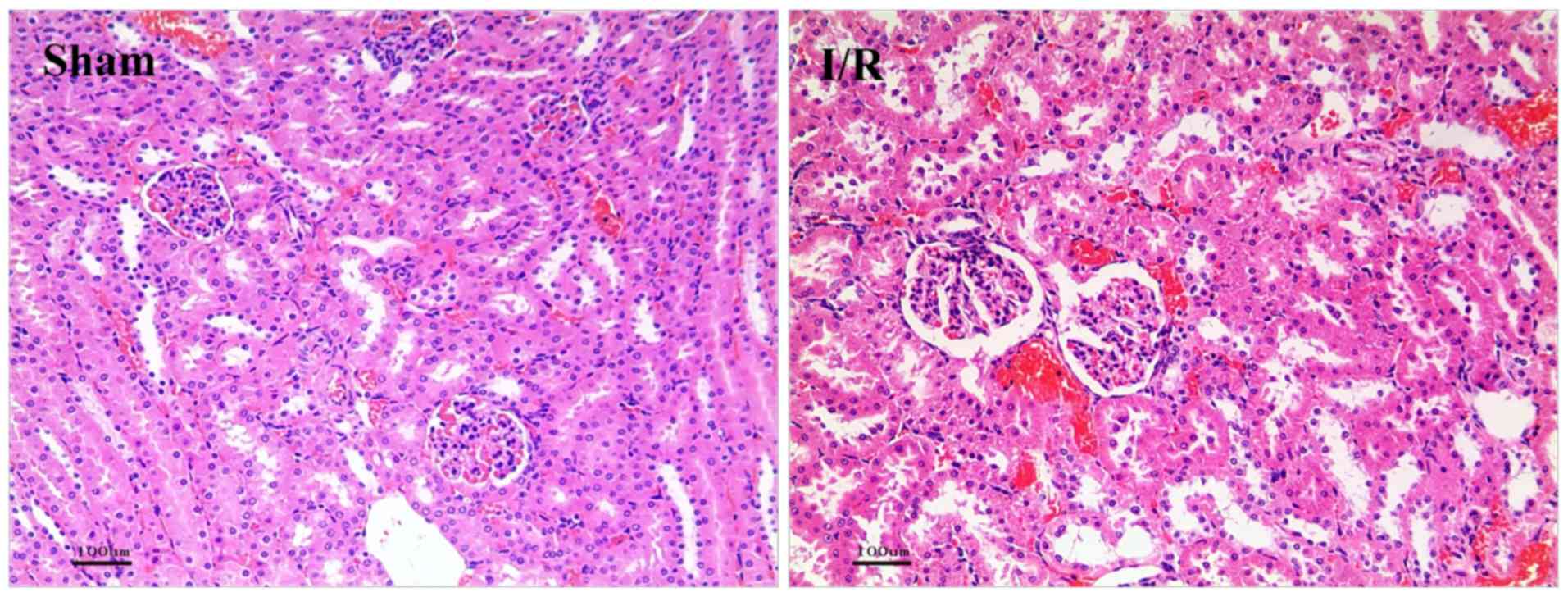
As shown in Fig. 1,
the results of HE staining showed the structure of renal tissue.
The sham group was normal and only local renal tubular epithelial
cell degeneration was observed. Serious tubular epithelial cell
swelling, vacuolar degeneration, deep nuclear staining, cell
necrosis, damaged basement membrane and renal tubular lumen
expansion were observed in I/R groups, indicating the successfully
constructed I/R model.
Changes of microvessel density after
renal I/R injury
Cluster of differentiation (CD)31 mediates the
penetration of leukocytes into blood vessel walls to cause tissue
damage and compensatory hyperplasia (3). The increased staining of CD31 in the
vascular tissue of the sham group and I/R groups was detected by
immunohistochemical staining. Results showed that the signal of
CD31 in I/R groups was significantly increased at 24 and 72 h after
reperfusion compared with the sham group (P<0.05). These data
suggest that renal I/R can stimulate the compensatory hyperplasia
of microvessels (Table II).
 | Table IIComparison of positive rate of CD31
among groups |
Table II
Comparison of positive rate of CD31
among groups
| Groups | CD31 |
|---|
| Sham | 12.04±0.78 |
| I/R 24 h | 21.43±0.22a |
| I/R 72 h | 23.51±0.69a |
Expression level of np_5318 in vivo
and in vitro
To investigate the level of np_5318 in the mice,
RT-qPCR was used in the sham and I/R groups with GAPDH as
endogenous control. As shown in Fig.
2A, compared with the sham group, the expression level of
np_5318 was increased at 24 h after reperfusion. For the RNA
interference experiment, three siRNA sequences were synthesized and
the transfection efficiency was detected. The sequence was chosen
with the highest transfection efficiency for demonstration
(Fig. 2B). Si-np_5318 significantly
decreased np_5318 level (P<0.05) and pc-np_5318 significantly
increased np_5318 level compared with the Ctrl (control; P<0.01;
Fig. 2B). Furthermore, an I/R cell
model in vitro was constructed and these cells were divided
into six groups, including I/R, I/R+si-NC, I/R+si-np_5318,
I/R+pc-NC, I/R+pc-np_5318, and control groups as shown in Fig. 2B. The expression of np_5318 in these
groups were confirmed by RT-qPCR. Results indicated that np_5318 in
the I/R group was significantly increased compared with the control
group (P<0.05). In addition, the present study confirmed that
si-np_5318 significantly decreased np_5318 level in I/R group and
pc-np_5318 significantly increase np_5318 level in I/R group
(P<0.05). Then, relative cell viability at 24, 48 and 72 h in
these groups were investigated by MTT assay (Fig. 2C). The results of the present study
suggested that cell growth was significantly suppressed in the I/R
groups compared with the control group (P<0.01) and transfection
with pc-np_5318 significantly suppressed the cell growth of I/R
cells at 48 and 72 h (P<0.01). While inhibition of np_5318 by
si-np_5318 significantly increased the cell growth of I/R cells at
48 and 72 h (P<0.01). Those data suggested that the expression
of np_5318 increased in I/R groups and np_5318 may promote renal
I/R injury.
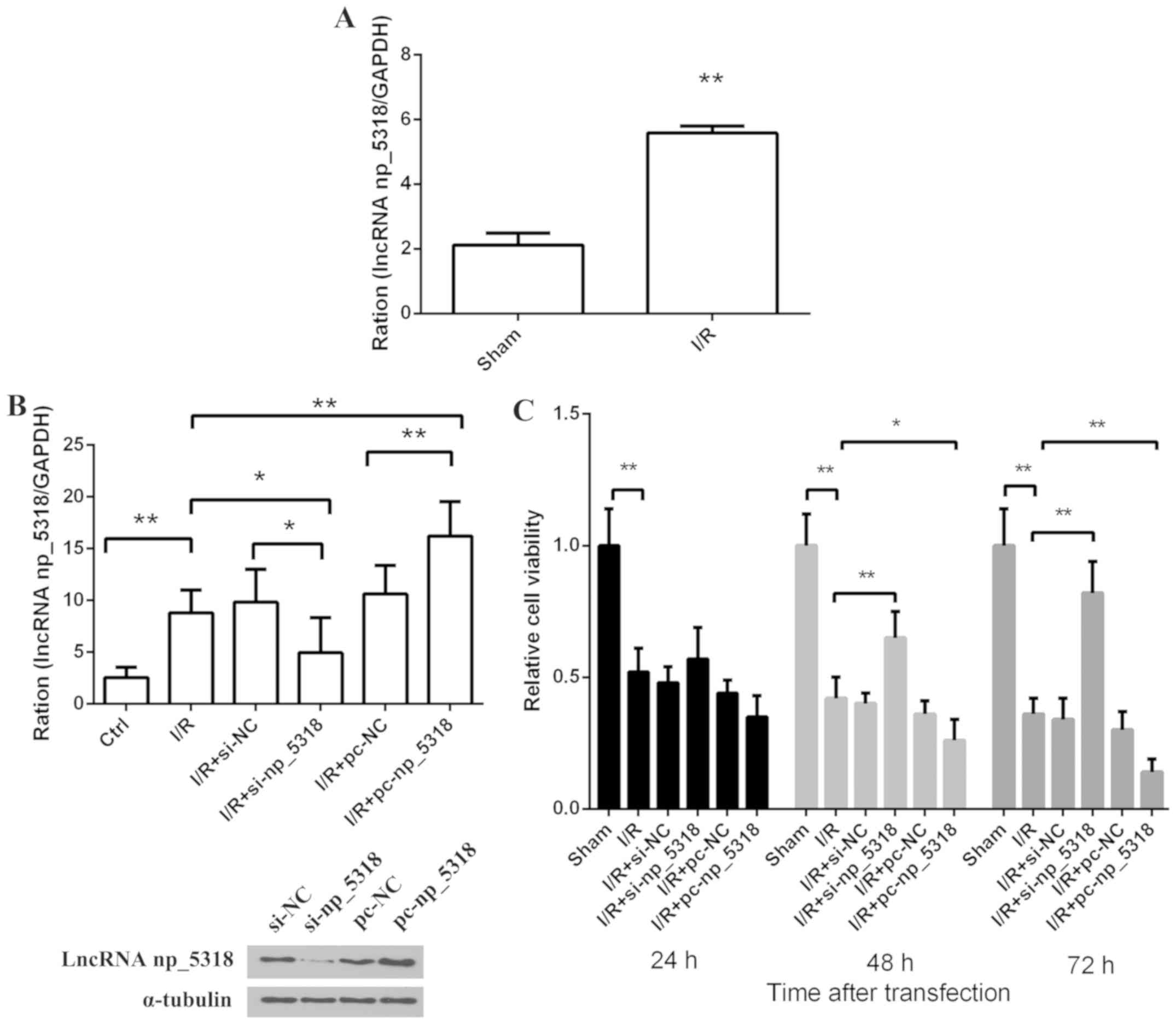 | Figure 2Increased expression level of np_5318
is confirmed in I/R groups and np_5318 may promote renal I/R
injury. (A) The expression of np_5318 in the mice was determined by
RT-qPCR in the sham and I/R groups. (B) The expression of np_5318
in these cell groups (I/R, I/R+si-NC, I/R+si-np_5318, I/R+pc-NC,
I/R+pc-np_5318 and control groups) was confirmed by RT-qPCR. The
transfection efficiency is shown below. Si-np_5318 significantly
decreased np_5318 level and pc-np_5318 significantly increase
np_5318 level. (C) Relative cell viability at 24, 48, 72 h in these
groups (I/R, I/R+si-NC, I/R+si-np_5318, I/R+pc-NC, I/R+pc-np_5318
and control groups) were investigated by MTT assay.
*P<0.05 and **P<0.01. si,
small interfering; I/R, ischaemia/reperfusion; NC, negative
control; RT-qPCR, reverse transcription-quantitative PCR; Ctrl,
control; lnc, long noncoding. |
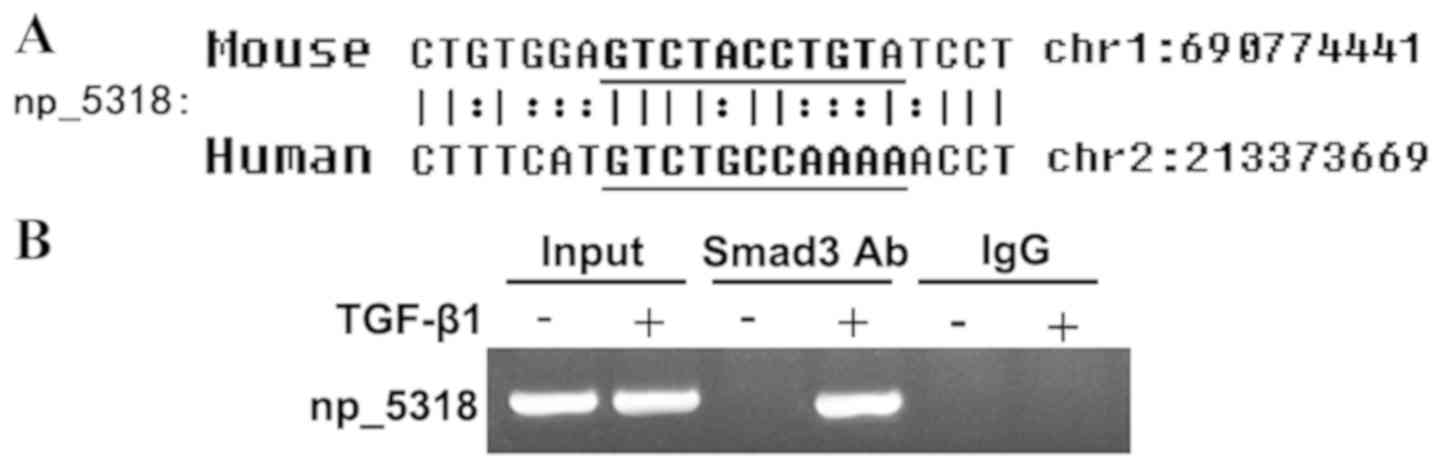
LncRNA np_5318 can bind to Smad3
As shown in Fig. 3A,
np_5318 contained conserved Smad3 binding sites in both human and
mouse genomes. To confirm this finding, a ChIP assay was used to
determine the interaction of Smad3 with the promoter regions of
np_5318 in mouse embryonic fibroblast. The antibody against Smad3
could successfully immunoprecipitate the DNA fragments from MEFs
containing the potential SBSs in the promoter regions of np_17856
and np_5318, supporting that Smad3 could physically interact with
their promoter regions (Fig.
3B).
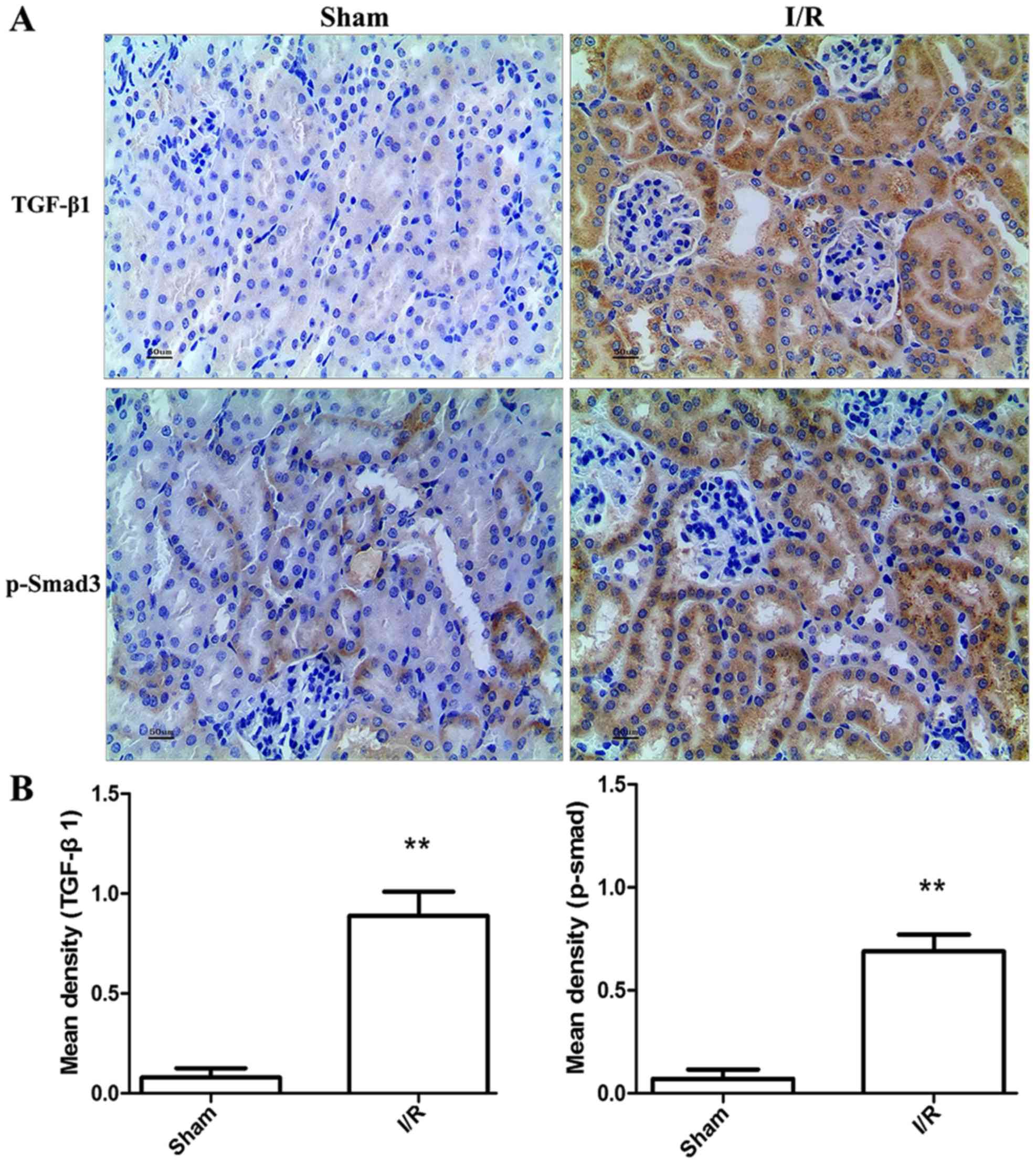
Expression of TGF-β1 and Smad3 in
different groups
To determine TGF-β1 and p-Smad3 expression in the
kidney, immunohistochemical staining was performed. The sham
operated controls did not show any TGF-β1 and p-Smad3 expression.
However, there was significant increase in TGF-β1 and p-Smad3
expression in I/R model (P<0.01) compared with the sham mice
(Fig. 4). Levels of TGF-β1, Smad3
and p-Smad3 were also detected by western blotting in the renal
tissue of the different groups. As shown in Fig. 5A and B, compared with the sham group, levels of
TGF-β1, Smad3 and p-Smad3 were significantly increased in renal
tissue of I/R group (P<0.001).
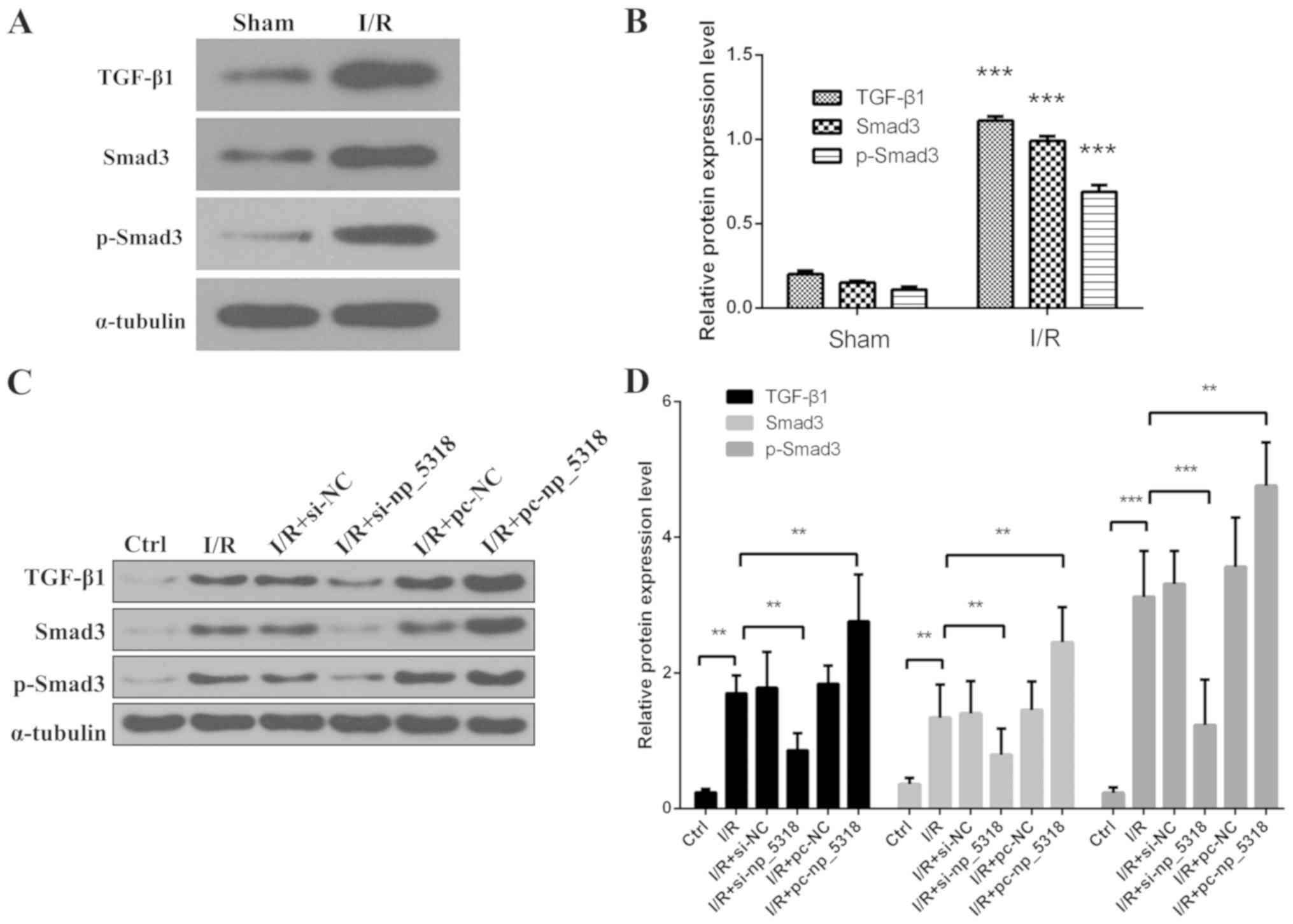 | Figure 5Levels of TGF-β1, Smad3 and p-Smad3
are determined in vivo and in vitro. (A)
Representative results of western blotting for TGF-β1, Smad3 and
p-Smad3 in renal tissue of different group. (B) Normalized
expression levels of TGF-β1, Smad3 and p-Smad3.
***P<0.001 vs. the sham group.
(C) Representative results of western blotting for TGF-β1, Smad3
and p-Smad3 in these I/R cell groups (I/R, I/R+si-NC,
I/R+si-np_5318, I/R+pc-NC, I/R+pc-np_5318 and control groups). (D)
Normalized expression levels of TGF-β1, Smad3 and p-Smad3.
**P<0.05 and
***P<0.001. p-Smad3,
phosphorylated-mothers against decapentaplegic homolog; TGF,
transforming growth factor; I/R, ischaemia/reperfusion; NC,
negative control. |
Then levels of TGF-β1, Smad3 and p-Smad3 in cells
were detected by western blotting, including I/R, I/R+si-NC,
I/R+si-np_5318, I/R+pc-NC, I/R+pc-np_5318, and the control groups
as shown in Fig. 5C and D. It was demonstrated that the level of
TGF-β1, p-Smad3 and Smad3 was significantly increased in I/R group
compared with the control group, and transfection with pc-np_5318
significantly increased the level of np_5318 compared with in the
I/R group. While inhibition of np_5318 by si-np_5318 significantly
suppressed the level of TGF-β1, p-Smad3 and Smad3 compared with the
I/R group. Those data suggest that I/R can increase the expression
levels of TGF-β1, Smad3 and p-Smad3.
Discussion
Renal IR is a process of the reduced blood supply to
kidneys followed by re-oxygenation and restoration of blood flow
(17). Reperfusion is critical for
the recovery of renal function. However, this procedure sometimes
can cause renal damage. The blocking of renal blood flow is usually
required for some surgical operations, such as nephrolithotomy,
surgical resection of renal tumors and renal transplantation. Renal
reperfusion during those operations can also cause renal damage
(18). BUN and SCr are two
traditional biomarkers of kidney (19). Decreased renal function caused
increases in levels of BUN and SCr (12).
Numerous studies have shown that the onset,
development and progression of ischemia-reperfusion injury are
closely related to the function of different lncRNAs (12-21). Yu et al
(12) showed that, as a HIF-1α
dependent lncRNA, expression level of psoriasis associated
non-protein coding RNA induced by stress (PRINS) was upregulated
under hypoxia conditions caused by I/R and PRINS can interact with
RANTES to promote I/R injury. In another study, lncRNA TapSAKI was
found to be upregulated in the plasma of patients with acute kidney
infarction (AKI) and the expression level of TapSAKI was positively
correlated with the disease severity, indicating that TapSAKI can
serve as a specific biomarker for the prognosis of AKI (20). In contrast, lncRNA AK139328 was found
to be downregulated in mice with liver I/R injury. In addition,
AK139328 knockdown mediated by siRNA silencing was found to be able
to inhibit the activation of caspase-3, reduce the expression of
IP-10, inhibit the activity of nuclear factor-κB signaling and
reduce the expression of various inflammatory cytokines during the
development of liver I/R injury (21). It has been reported that expression
level of lncRNAs np_5318 is increased during renal inflammation
(13). However, to the best of our
knowledge the involvement of lncRNAs np_5318 in renal I/R injury
still hasn't been reported. In the present study, I/R mice models
were successfully constructed and expression of lncRNAs np_5318 was
found to be increased in mice with renal I/R injury compared with
mice in the sham group, and the increased expression of lncRNAs
np_5318 was also confirmed in I/R cells, indicating the possible
involvement of np_5318 in this procedure.
The TGF-β/Smad signaling pathway plays pivotal roles
in various renal diseases. The activation of TGF-β can regulate a
variety of cellular functions including proliferation,
differentiation, apoptosis and inflammation (22). As a member of the TGF-β super family,
TGF-β1 was found to be able to play a profibrotic role by
stimulating the proliferation of fibroblasts, extracellular matrix
synthesis and process of epithelial-to-mesenchymal transition
(23). Members of the Smad family
play central roles in TGF-β signaling pathway (24). In the present study, compared with
mice in the sham group, expression levels of TGF-β1 and Smad3 were
increased in mice of the I/R model group, indicating that I/R
injury can upregulate the expression of TGF-β1 and Smad3. In
addition, the level of p-Smad3, which is the activated form of
Smad3, is also increased in mice with I/R injury. Then the results
in vitro also confirmed that TGF-β1, Smad3 and p-Smad3 was
increased in I/R cells. Those data suggested that I/R can increase
the expression levels of TGF-β1 and Smad3 and induce the activation
of Smad3. Furthermore, it was demonstrated that the inhibition of
lncRNAs np_5318 level in I/R cells could enhance the cell growth of
I/R cells. Additionally, TGF-β1, Smad3 and p-Smad3 levels were also
inhibited by the inhibition of lncRNAs np_5318 level in I/R, while
the increase of lncRNAs np_5318 level provided the opposite
results.
In conclusion, expression level of lncRNA np_5318
was increased in mice with renal I/R and I/R cells. np_5318 could
regulate the expression of Smad3 by binding to its promoter region.
In addition, levels of TGF-β1, Smad3 and p-Smad3 were increased by
renal I/R. Therefore, lncRNA np_5318 may participate in the
development of renal I/R injury through the TGF-β/Smad signaling
pathway. The present study provides new insights into the mechanism
and treatment of I/R injury. However, the present study is still
limited by the small sample size. Further studies with bigger
sample size are still needed to verify the conclusions in this
study.
Acknowledgements
Not applicable.
Funding
No funding received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JS conceived and designed the study. JL and JM
performed the experiments. JL and JM wrote the paper. JL and JS
reviewed and edited the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All experimental procedures were approved by the
Institutional Animal Care and Use Committee of Yucheng People's
Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chhor V and Journois D: Perioperative
acute kidney injury and failure. Nephrol The. 10:121–131. 2014.(In
French). PubMed/NCBI View Article : Google Scholar
|
|
2
|
Seth R, Yang C, Kaushal V, Shah SV and
Kaushal GP: p53-dependent caspase-2 activation in mitochondrial
release of apoptosis-inducing factor and its role in renal tubular
epithelial cell injury. J Biol Chem. 280:31230–31239.
2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Malek M and Nematbakhsh M: Renal
ischemia/reperfusion injury; from pathophysiology to treatment. J
Renal Inj Prev. 4:20–27. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Snoeijs MG, van Heurn LW and Buurman WA:
Biological modulation of renal ischemia-reperfusion injury. Curr
Opin Organ Transplant. 15:190–199. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Salvadori M, Rosso G and Bertoni E: Update
on ischemia-reperfusion injury in kidney transplantation:
Pathogenesis and treatment. World J Transplant. 5:52–67.
2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ponticelli C: Ischaemia-reperfusion
injury: A major protagonist in kidney transplantation. Nephrol Dial
Transplant. 29:1134–1140. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Snoeijs MG, Vink H, Voesten N, Christiaans
MH, Daemen JW, Peppelenbosch AG, Tordoir JH, Peutz-Kootstra CJ,
Buurman WA, Schurink GW and van Heurn LW: Acute ischemic injury to
the renal microvasculature in human kidney transplantation. Am J
Physiol Renal Physiol. 299:F1134–F1140. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Rodriguez F, Bonacasa B, Fenoy FJ and
Salom MG: Reactive oxygen and nitrogen species in the renal
ischemia/reperfusion injury. Curr Pharm Des. 19:2776–2794.
2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Lorenzen JM, Kaucsar T, Schauerte C,
Schmitt R, Rong S, Hübner A, Scherf K, Fiedler J, Martino F,
Kumarswamy R, et al: MicroRNA-24 antagonism prevents renal ischemia
reperfusion injury. J Am Soc Nephrol. 25:2717–2729. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Eldaif SM, Deneve JA, Wang NP, Jiang R,
Mosunjac M, Mutrie CJ, Guyton RA, Zhao ZQ and Vinten-Johansen J:
Attenuation of renal ischemia-reperfusion injury by
postconditioning involves adenosine receptor and protein kinase C
activation. Transpl Int. 23:217–226. 2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang X, Gao L, Guo X, Shi X, Wu H, Song F
and Wang B: A network based method for analysis of lncRNA-disease
associations and prediction of lncRNAs implicated in diseases. PloS
One. 9(e87797)2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yu TM, Palanisamy K, Sun KT, Day YJ, Shu
KH, Wang IK, Shyu WC, Chen P, Chen YL and Li CY: RANTES mediates
kidney ischemia reperfusion injury through a possible role of
HIF-1α and LncRNA PRINS. Sci Rep. 6(18424)2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhou Q, Chung AC, Huang XR, Dong Y, Yu X
and Lan HY: Identification of novel long noncoding RNAs associated
with TGF-β/Smad3-mediated renal inflammation and fibrosis by RNA
sequencing. Am J Pathol. 184:409–417. 2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Thurman JM: Triggers of inflammation after
renal ischemia/reperfusion. Clin Immunol. 123:7–13. 2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Huang X, Gao Y, Qin J and Lu S: The
mechanism of long noncoding RNA MEG3 for hepatic
ischemia‐reperfusion: Mediated by miR‐34a/Nrf2 signaling pathway. J
Cell Biochem. 119:1163–1172. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wang H, Yan Z, Qiu L, Hu Z, Qian W and Xu
L: Dynamic changes of platelet endothelial cell adhesion molecule-1
(PECAM-1/CD31) on pulmonary injury induced by ischemia-reperfusion
in rats. Ir J Med Sci. 180:483–488. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Daemen M, de Vries B and Buurman W:
Apoptosis and inflammation in renal reperfusion injury.
Transplantation. 73:1693–1700. 2002.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Vaidya VS, Ozer JS, Dieterle F, Collings
FB, Ramirez V, Troth S, Muniappa N, Thudium D, Gerhold D, Holder
DJ, et al: Kidney injury molecule-1 outperforms traditional
biomarkers of kidney injury in preclinical biomarker qualification
studies. Nat Biotechnol. 28:478–485. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
20
|
Lorenzen JM, Schauerte C, Kielstein JT,
Hübner A, Martino F, Fiedler J, Gupta SK, Faulhaber-Walter R,
Kumarswamy R, Hafer C, et al: Circulating long noncoding RNA
TapSAKI is a predictor of mortality in critically ill patients with
acute kidney injury. Clin Chem. 61:191–201. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen Z, Jia S, Li D, Cai J, Tu J, Geng B,
Guan Y, Cui Q and Yang J: Silencing of long noncoding RNA AK139328
attenuates ischemia/reperfusion injury in mouse livers. PloS One.
8(e80817)2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chung AC, Huang XR, Meng X and Lan HY:
miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc
Nephrol. 21:1317–1325. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Massagué J and Wotton D: Transcriptional
control by the TGF-beta/Smad signaling system. EMBO J.
19:1745–1754. 2000.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Savage C, Das P, Finelli AL, Townsend SR,
Sun CY, Baird SE and Padgett RW: Caenorhabditis elegans genes
sma-2, sma-3, and sma-4 define a conserved family of transforming
growth factor beta pathway components. Proc Natl Acad Sci USA.
93:790–794. 1996.PubMed/NCBI View Article : Google Scholar
|