Introduction
Facet joint osteoarthritis (FJOA) is a common
degenerative joint disorder that causes the degeneration and
breakdown of cartilage and restricts the movement of joints
(1). It has been reported that
lumbar FJOA occurs at high prevalence and the presence of FJOA is
highly associated with age (2,3). A
community-based cross-sectional study indicated that in populations
aged <50 years, the prevalence of FJOA was <45%, while it was
~75% in populations aged >50 years (4). FJOA generally causes lower back pain
and lower extremity pain and thus largely limits patients' daily
activities and severely impacts their quality of life (5). At present, FJOA is generally treated
with non-steroidal anti-inflammatory drugs to reduce inflammation
and diminish lower back pain (6).
However, the therapeutic effects of FJOA are far from satisfactory.
The identification of novel clinical treatments for FJOA requires a
deeper understanding of FJOA from the cellular and molecular
aspects.
The pathological process of FJOA involves the
degeneration of a variety of tissues, including cartilage, bones,
capsules and synovial tissues (7).
Histomorphological and histomorphometric studies indicated that
patients with FJOA exhibited thinning or even loss of cartilage and
subchondral bones, absence of osteophyte formation, loss of
proteoglycans, increased apoptosis of chondrocytes and clustering
of chondrocytes (8-10).
Histopathological studies of subchondral bone and marrow
compartments of osteoarthritic facet joints revealed that patients
with FJOA possessed extensive de novo bone and blood vessel
formation in subchondral bone tissues and increased inflammatory
cell infiltration, as well as enhanced osteoclast activity in
subchondral marrow spaces (7). A
comparative study of healthy and osteoarthritic lumbar facet joints
demonstrated that the thicknesses and the porosities of subchondral
cortical plates in patients with FJOA were not as good as those in
the healthy population (3).
Furthermore, molecular investigations revealed the infiltration of
inflammatory cells, increased amounts of pro-inflammatory factors
[Including growth-regulated oncogene α, soluble intercellular
adhesion molecule 1, interferon γ, interleukin (IL)-1β, IL-17,
chemokine (C-C motif) ligand 5, tumor necrosis factor α, IL-1α and
IL-17E], and increased amounts of anti-inflammatory cytokines,
including IL-10 and IL-13 in degenerative facet joint capsular
tissues (11).
Although a better understanding of the morphological
and molecular changes underlying FJOA has been obtained, the
dynamic genetic changes in FJOA have not been well elucidated. In a
previous study, by combined use of RNA deep sequencing and Database
for Annotation Visualization and Integrated Discovery
bioinformatics resource, differentially expressed genes (DEGs) in
FJOA were screened and enriched Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways were determined, which revealed that the
Wnt and NF-κ signaling pathways were significantly involved in FJOA
(12). In the present study, the
Ingenuity Pathway Analysis (IPA) software, an advanced
bioinformatics tool with a massive built-in knowledge database, was
used to analyze DEG-associated canonical signaling pathways in
FJOA. Furthermore, the correlations between differentially
expressed genes, diseases and functions (e.g. toxicity functions)
were determined. The interactions between DEGs were also
investigated.
Materials and methods
Human tissue collection and RNA deep
sequencing
Human facet joint tissue collection and RNA deep
sequencing were performed as previously described (12). In brief, healthy facet joint tissues
were gathered from a total of 10 patients with vertebral fracture
who received internal fixation of the lumbar spine. Pathological
facet joint tissues were gathered from a total of 48 patients with
FJOA who received lumbar surgery for neurogenic claudication
between January and December 2017 at the Second Affiliated Hospital
of Nantong University. A total of 21 male patients and 27 female
patients were included. The age range was 46-79 years and the
average age was 64±1.7 years. In total, 20 out of these 48 patients
with FJOA had a facet joint degeneration grading of 2, and 28 out
of these 48 patients had an FJOA grade of 3. The healthy or
pathological facet joint tissues were respectively divided into 3
portions and subjected to RNA deep sequencing using the Illumina
Hiseq X10 platform. RNA deep sequencing outcomes revealed balanced
base compositions of raw reads and even distributions of bases
along reads and on reference genes, indicating that high-quality
data were obtained. Fragments per kilobase of exon per million
fragments mapped were obtained to quantify the expression levels of
mapped genes. All procedures were ethically approved by the Human
Ethics Committee of the Second Affiliated Hospital of Nantong
University (Nantong, China) and documents of informed consent were
signed by the patients.
Bioinformatics analysis
Genes with a log2 fold change >2 or ≤2
and a false discovery rate (adjusted P-value) <0.05 were
considered as differentially expressed and were analyzed by the IPA
software (version 2018; Ingenuity Systems; QIAGEN). RNA deep
sequencing outcomes were uploaded to IPA for core analysis and
jointly analyzed with the global molecular network in the ingenuity
pathway knowledge base (IPKB) (13,14).
Canonical signaling pathways enriched by the differentially
expressed gene were identified and rated according to P-values. The
z-scores and ratios of significantly involved canonical signaling
pathways were also determined. Z-scores were presented by orange or
blue colors and indicated the activation of the canonical signaling
pathways. Ratios were calculated by dividing the numbers of DEGs in
the canonical signaling pathways to the numbers of total genes in
the pathways and indicated the percentages of differentially
expressed genes. The outcomes from the IPA core analysis were
overlaid with the IPKB in order to identify significantly involved
diseases and functions, toxicity (tox) functions and IPA functional
networks in FJOA. Tox functions are used in combination with tox
lists to link experimental data to clinical pathology endpoints,
understand pharmacological response and support mechanism of action
and mechanism of toxicity hypothesis generation in IPA
software.
Reverse transcription-quantitative
(RT-q)PCR
The same facet joint tissues used for RNA deep
sequencing were subjected to RT-qPCR analysis. Total RNA was
isolated from facet joint tissues by using TRIzol reagent (Thermo
Fisher Scientific, Inc.) and reversely transcribed to complementary
DNA using the Prime-Script reagent kit (Takara Biotechnology Co.,
Ltd.). qPCR was performed by using SYBR Green Premix Ex Taq (Takara
Biotechnology Co., Ltd.) on an Applied Biosystems StepOne real-time
PCR System (Thermo Fisher Scientific, Inc.). The amplification
reaction protocol was as follows: A total of 40 cycles at 95˚C for
5 sec and 60˚C for 10 sec. Relative quantification was performed by
using the comparative 2-∆∆Cq method (15) with GAPDH as the reference gene. The
sequences of all primer pairs are listed in Table I.
 | Table IList of PCR primers. |
Table I
List of PCR primers.
| Gene/primer | Sequence (5' to
3') |
|---|
| ELANE |
|
Forward |
CAGGCATCTGCTTCGGGGAC |
|
Reverse |
AGGGGCGAAGGCATCTGGGT |
| BTG2 |
|
Forward |
CTCCATCTGCGTCTTGTACGA |
|
Reverse |
AGACTGCCATCACGTAGTTCT |
| BRCA1 |
|
Forward |
AGCGCCAGTCATTTGCTCCG |
|
Reverse |
GGACCCAGAGTGGGCAGAGAATG |
| CHAF1A |
|
Forward |
GCAGGCATCTGTCAATTAAG |
|
Reverse |
AGCATCTGACTAGCAACCAC |
| TP63 |
|
Forward |
ACAGGAAGGCGGATGAAGAT |
|
Reverse |
TGTGTGCTGAGGAAGGTACT |
| CXCL8 |
|
Forward |
AATGAAAAGATGAGGGTGCAT |
|
Reverse |
GCTTGTGTGCTCTGCTGTCT |
| GFI1 |
|
Forward |
TCCCTGTCAGTACTGTGGCA |
|
Reverse |
TGGAGCTCTGACTGAAGGCT |
| CCND1 |
|
Forward |
CCCAACAACTTCCTCTCCT |
|
Reverse |
TCCAGAAGGGCTTCAATCTG |
| SPI1 |
|
Forward |
GACACGGATCTATACCAACGCC |
|
Reverse |
CCGTGAAGTTGTTCTCGGCGAA |
| CEBPE |
|
Forward |
AGTACCAAGTGGCACACTGC |
|
Reverse |
GAGAAGGGGACTGCAGGGA |
| GATA1 |
|
Forward |
CAGGGCAGAATCCACAAACT |
|
Reverse |
TCCTCTGCATCAACAAGCC |
| TAL1 |
|
Forward |
AACAACAACCGGGTGAAGAG |
|
Reverse |
CATTCACATTCTGCTGCCTC |
| MCM4 |
|
Forward |
GTATTTTTTGGTAGAGACGGCTTC |
|
Reverse |
GTGACGTGGGTCGGAAAC |
| TREM1 |
|
Forward |
AAGGAGCCTCACATGCTGTT |
|
Reverse |
CACAGTTCTGGGGCTGGTAT |
| GAPDH |
|
Forward |
CCAAGGTCATCCATGACAAC |
|
Reverse |
TGTCATACCAGGAAATGAGC |
Statistical analysis
The PCR results are expressed as the mean ± standard
error of the mean. The differences between the FJOA group and the
healthy control group were assessed by using Student's t-test.
Statistical analysis was performed with GraphPad Prism 5.0
(GraphPad Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of significantly
enriched canonical signaling pathways
In order to investigate significantly involved
signaling pathways involved in FJOA, outcomes from RNA deep
sequencing were subjected to IPA canonical signaling pathway
analysis. DEGs in FJOA were categorized according to these
canonical signaling pathways. The activated canonical signaling
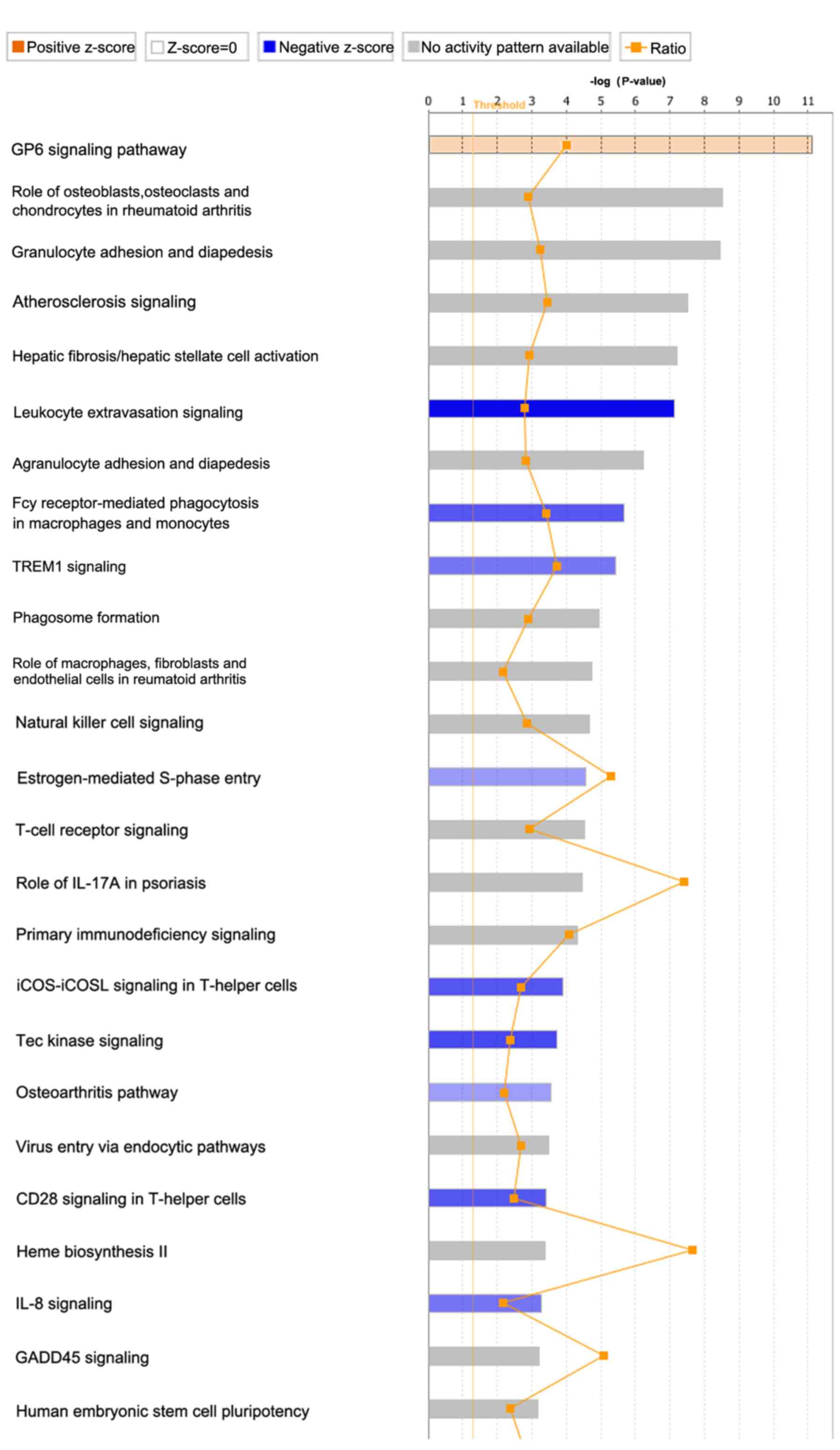
pathways in FJOA were screened and are listed in Table SI. After rating the identified
canonical signaling pathways according to their P-values, the top
25 canonical signaling pathways in FJOA were identified (Fig. 1). The ratios of DEGs to total genes
in these signaling pathways were also listed.
The top enriched canonical signaling pathways were
mainly assigned to the following categories: Cellular stress and
injury [glycoprotein VI platelet (GP6) signaling pathway and
osteoarthritis pathway]; disease-specific pathways (role of
osteoblasts, osteoclasts and chondrocytes in rheumatoid arthritis;
atherosclerosis signaling; hepatic fibrosis/hepatic stellate cell
activation; role of macrophages, fibroblasts and endothelial cells
in rheumatoid arthritis; and role of IL-17A in psoriasis); cellular
immune response [granulocyte adhesion and diapedesis; leukocyte
extravasation signaling; agranulocyte adhesion and diapedesis; Fcγ
receptor-mediated phagocytosis in macrophages and monocytes;
triggering receptor expressed on myeloid cells 1 (TREM1) signaling;
phagosome formation; natural killer cell signaling; T cell receptor
signaling; primary immunodeficiency signaling; inducible T-cell
costimulator (iCOS)-iCOS ligand (iCOSL) signaling in T helper
cells; CD28 signaling in T-helper cells; and IL-8 signaling]; cell
cycle regulation (estrogen-mediated S-phase entry and growth arrest
and DNA damage-inducible α signaling); intracellular and second
messenger signaling (Tec kinase signaling); pathogen-influenced
signaling (virus entry via endocytic pathways); biosynthesis (Heme
biosynthesis II); and organismal growth and development (human
embryonic stem cell pluripotency).
Besides the P-values of these canonical signaling
pathways, z-scores and the IPA-calculated indexes of the functional
significances of these canonical signaling pathways were also
determined. Z-scores were presented in orange or blue colors.
Orange color indicated that the results of differentially expressed
gene-associated signaling pathways were consistent with the
original prediction, whereas blue indicated that results were
opposite to the original prediction. The saturation of the color of
each signaling pathway reflected the value of the z-score. It was
observed that the following signaling pathways had z-scores that
indicated their functional importance in FJOA: GP6 signaling
pathway, leukocyte extravasation signaling, Fcγ receptor-mediated
phagocytosis in macrophages and monocytes, TREM1 signaling,
estrogen-mediated S-phase entry, iCOS-iCOSL signaling in T helper
cells, Tec kinase signaling, osteoarthritis pathway, CD28 signaling
in T helper cells and IL-8 signaling.
Leukocyte extravasation signaling
Canonical signaling pathways with high solute values
of z-scores were investigated in detail. Leukocyte extravasation,
also known as diapedesis, is the movement of leukocytes out of the
circulatory system and the migration of leukocytes towards injured
or infected tissue. The process of leukocyte extravasation includes
a first contact between leukocytes and endothelial cells, the
rolling adhesion and tight adhesion of leukocytes along the inner
surface of the vessel wall, as well as the transmigration of
leukocytes through spaces between endothelial cells. A schematic
diagram of the leukocyte extravasation signaling is presented in
Fig. 2, with the DEGs in the
canonical signaling pathway labeled by different colors. Genes
whose expressions were upregulated in FJOA were labeled in red and
genes whose expressions were downregulated in FJOA were labeled in
green. It was observed that various molecules, particularly
numerous cellular adhesion molecules, including integrin α (ITGA)
and ITGB subunits, were downregulated in FJOA.
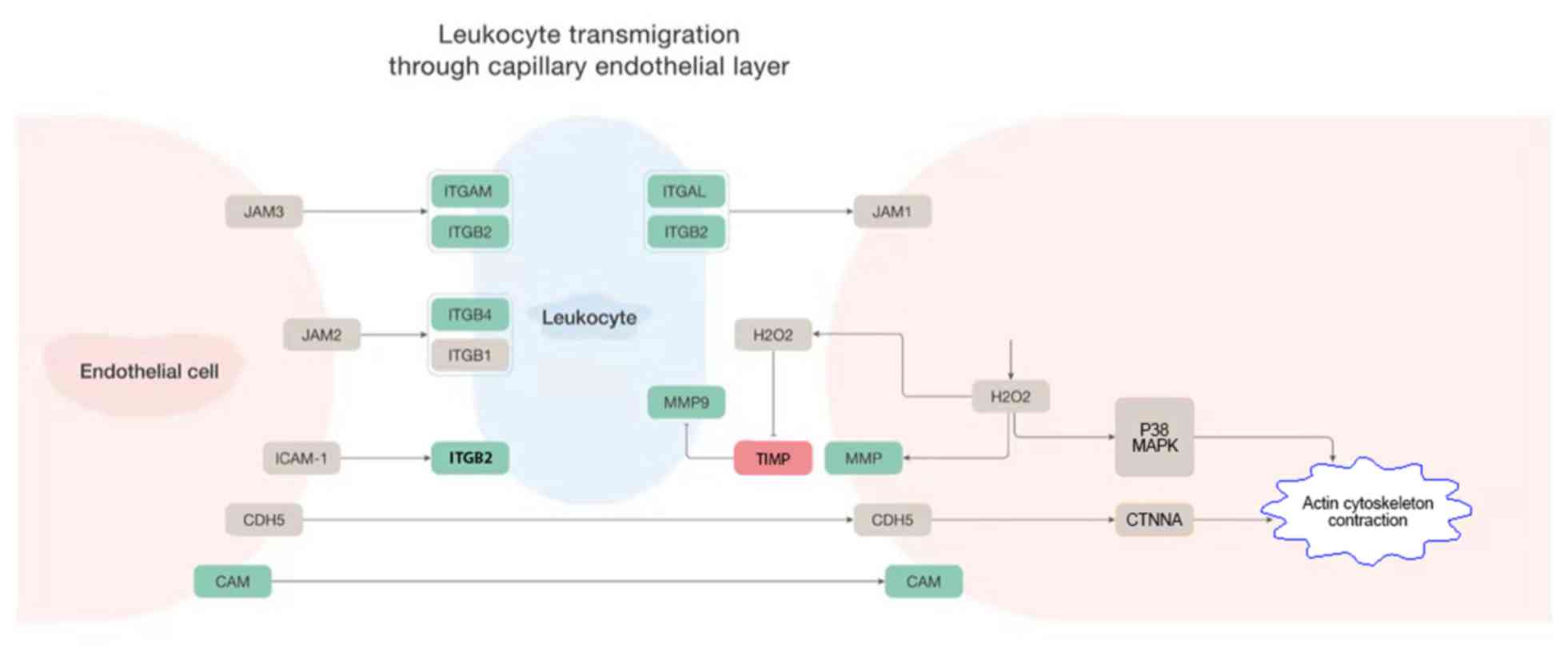 | Figure 2Leukocyte extravasation signaling. A
schematic network of leukocyte extravasation signaling modified
based on ingenuity pathway analysis software. Upregulated genes
were labeled in red and downregulated genes were labeled in green.
CAM, KRIT1 ankyrin repeat containing; CDH5, cadherin 5; CTNNA,
catenin α1; ICAM, intercellular adhesion molecule; ITGAM, integrin
subunit αM; ITGB2, integrin subunit β2; ITGB4, integrin subunit β4;
ITGAL, integrin subunit αL; JAM, junctional adhesion molecule;
MAPK, mitogen-activated protein kinase; MMP9, matrix
metallopeptidase 9; TIMP, TIMP metallopeptidase inhibitor 1. |
Tec kinase signaling
Another canonical signaling pathway with a high
solute value of the z-score, Tec kinase signaling, was also
studied. The tyrosine-protein kinase Tec is an enzyme that belongs
to the Tec family of non-receptor protein-tyrosine kinases. Tec
kinase is activated under various conditions, for instance by the
activation of G protein α subunit, the activation of focal adhesion
kinase, the binding of cytokine to cytokine receptor, the
activation of the death receptor, the activation of receptor
tyrosine kinase, the binding of antigen to the T-cell receptor, the
binding of IgE complex to IgE-loaded high-affinity IgE receptor and
the binding of endotoxin lipopolysaccharide to Toll-like receptor
4. Activated Tec kinase functions as a second messenger, activates
downstream cascades and regulates a large number of physiological
processes, including cell adhesion, cell migration, cell apoptosis,
actin reorganization, gene expression and transformation (Fig. 3).
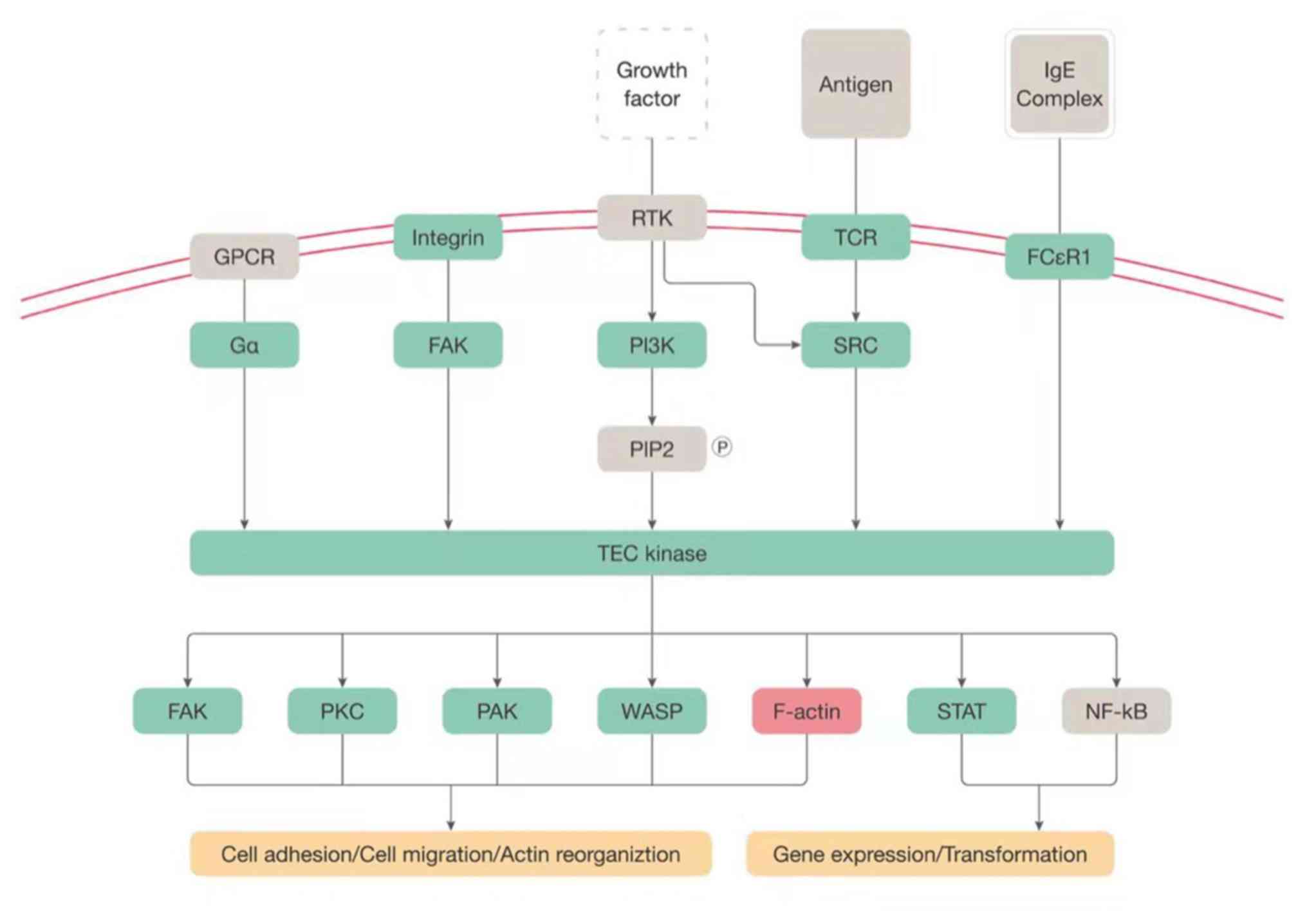 | Figure 3Tec kinase signaling. A schematic
network of Tec kinase signaling modified based on ingenuity pathway
analysis software. Upregulated genes were labeled in red and
downregulated genes were labeled in green. F-actin, filamentous
actin; FAK, protein tyrosine kinase 2; FCεR1, high affinity
immunoglobulin E receptor 1; GPCR, G-protein coupled receptor; IgE,
immunoglobulin E; PAK, serine/threonine protein kinase PAK; PI3K,
phosphatidylinositol-4,5-bisphosphate 3-kinase; PKC, protein kinase
C family; SRC, SRC non-receptor tyrosine kinase family; Tec,
tyrosine protein kinase Tec; WASP, WASP actin nucleation promoting
factor; TCR, T-cell receptor; Ga, α subunit of heterotrimeric G
protein; TEC kinase, Tyrosine-protein kinase; STAT, signal
transducer and activator of transcription protein. |
Osteoarthritis signaling
Besides the two aforementioned canonical signaling
pathways with high solute z-score values, the osteoarthritis
signaling was also examined, since FJOA is a type of osteoarthritis
that specifically occurs in facet joint tissue. Osteoarthritis is a
progressive degradative disease of joints that may be caused by
numerous factors, including inflammation, obesity, injury, aging
and improper or lack of exercise. These risk factors may severely
affect chondrocytes by mediating numerous cytokines and
inflammatory signals, including transforming growth factor β
(TGF-β), IL-1β and IL-8. Certain molecules, e.g. IL-8, activate
transcription factor SOX-9, a protective and stabilizing factor,
whereas TGF-β affects Runt-related transcription factor 2 (a
pro-development, pro-degradation and pro-osteoarthritis factor;
Fig. 4).
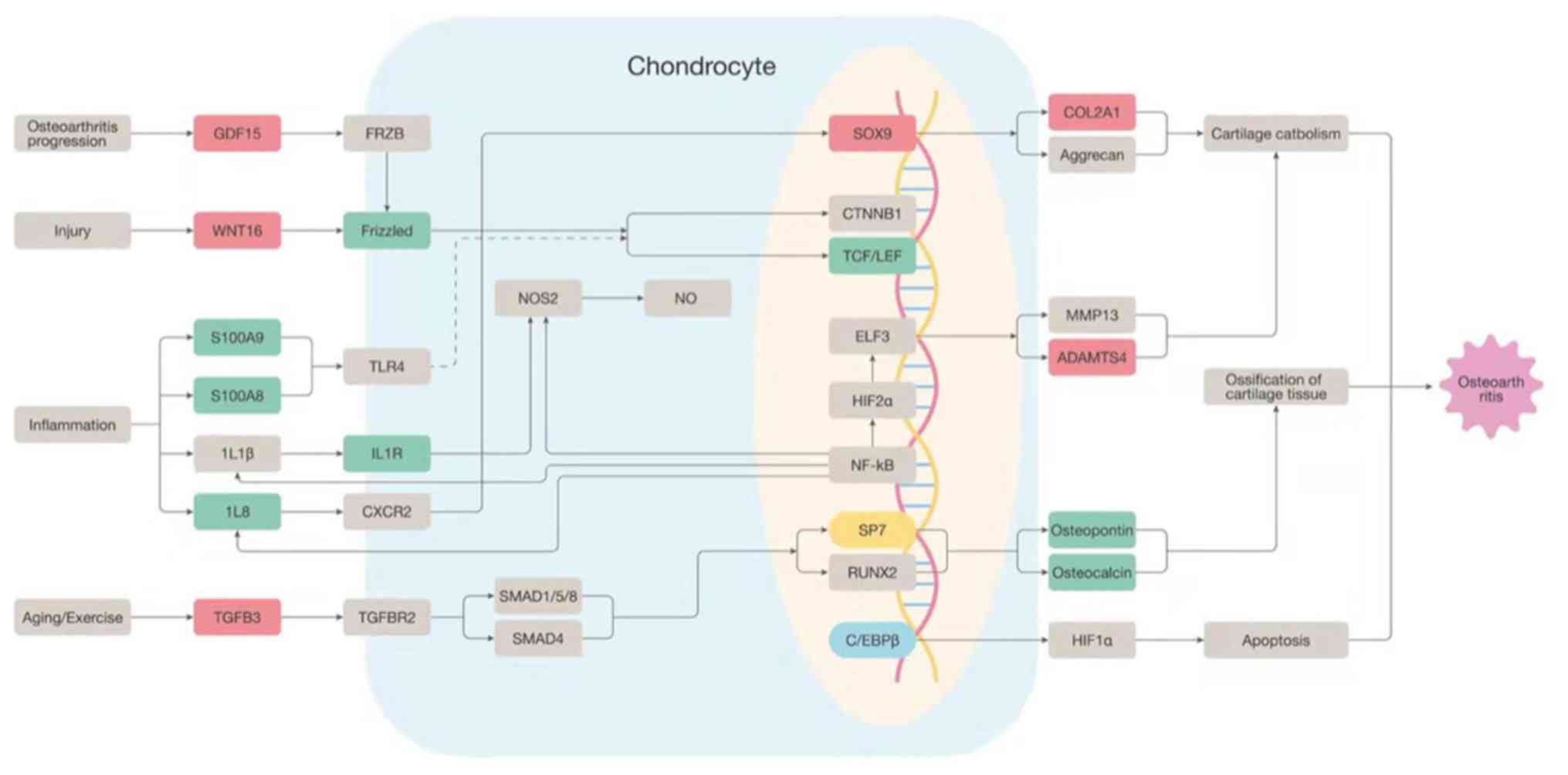 | Figure 4Osteoarthritis signaling. A schematic
network of osteoarthritis signaling modified based on ingenuity
pathway analysis software. Upregulated genes were labeled in red
and downregulated genes were labeled in green. GDF15, Growth
Differentiation Factor 15; WNT16, Wnt Family Member 16; Frizzled,
frizzled gene family; S100A9, S100 Calcium Binding Protein A9;
S100A8, S100 Calcium Binding Protein A8; IL8, Interleukin 8; IL1R,
Interleukin 1 Receptor Type 1; TGFB3, Transforming Growth Factor
Beta 3; SOX9, SRY-Box Transcription Factor 9; TCF/LEF,
Transcription Factor/Lymphoid Enhancer Binding Factor; SP7, Sp7
Transcription Factor; COL2A1, Collagen Type II Alpha 1 Chain;
ADAMTS4, ADAM Metallopeptidase With Thrombospondin Type 1 Motif
4. |
Identification of significantly
involved diseases and functions and tox functions
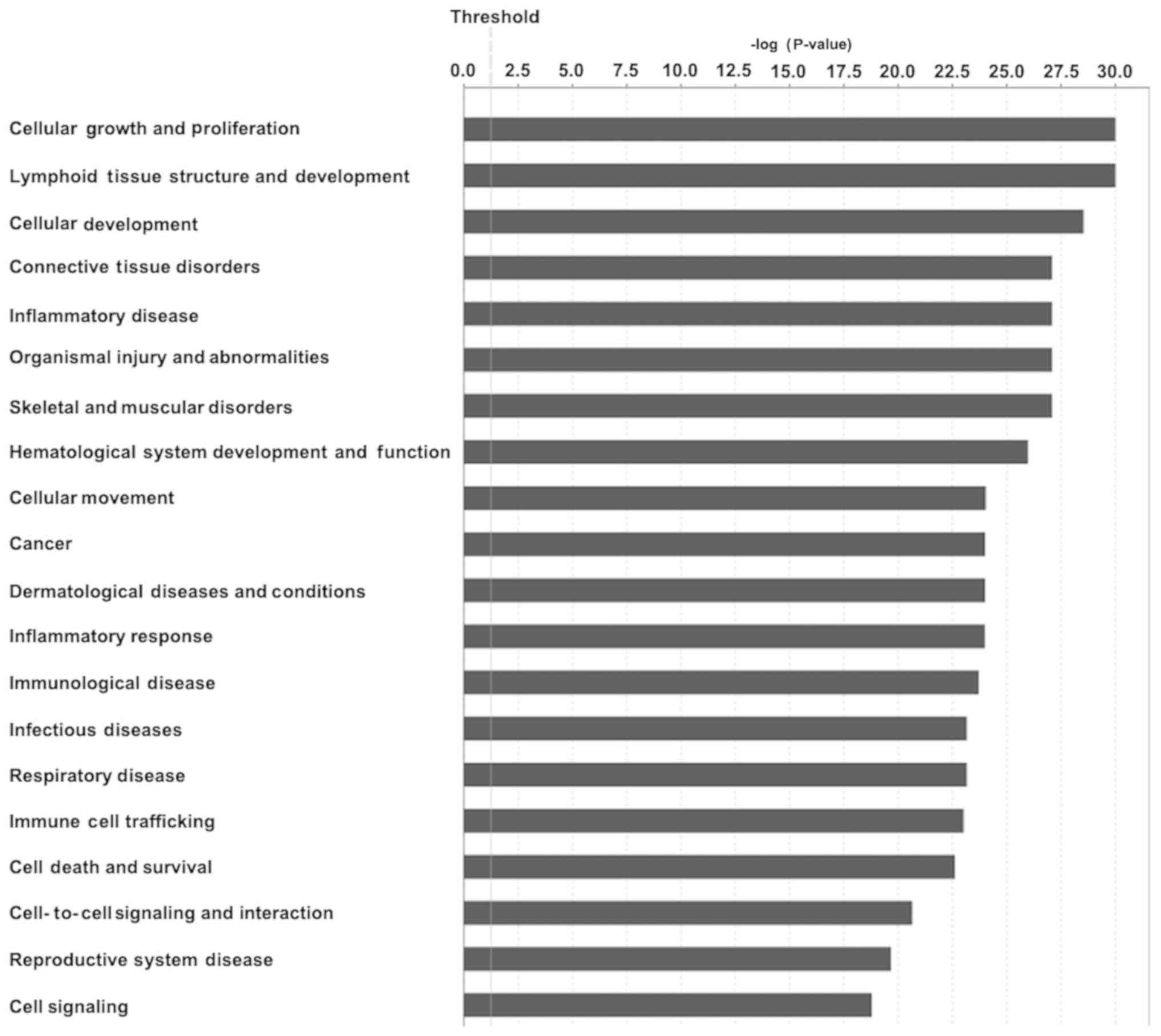
DEGs were shown to be further associated with
diseases and functions by using the IPA core analysis. A total of
60 associated diseases and functions were identified in FJOA. These
associated diseases and functions were then rated according to
their significance (P-value) and the top 20 enriched categories of
diseases and functions in FJOA were selected (Fig. 5).
It was demonstrated that cell and tissue
growth-associated biological functions, including cellular growth
and proliferation, lymphoid tissue structure and development,
cellular development, cellular development and hematological system
development and function were significant in FJOA. Cellular
movement and cell death and survival, two biological functions of
cellular behavior, were also enriched. Besides these critical
biological processes, it was indicated that the DEGs in FJOA were
significantly involved in numerous diseases, including connective
tissue disorders, organismal injury and abnormalities, skeletal and
muscular disorders, cancer, dermatological diseases and conditions,
respiratory disease and reproductive system disease. Furthermore,
it was demonstrated that diseases and functions associated with
inflammation and immune responses, including inflammatory disease,
inflammatory response, immunological disease, infectious disease
and immune cell trafficking, were also significantly involved in
FJOA.
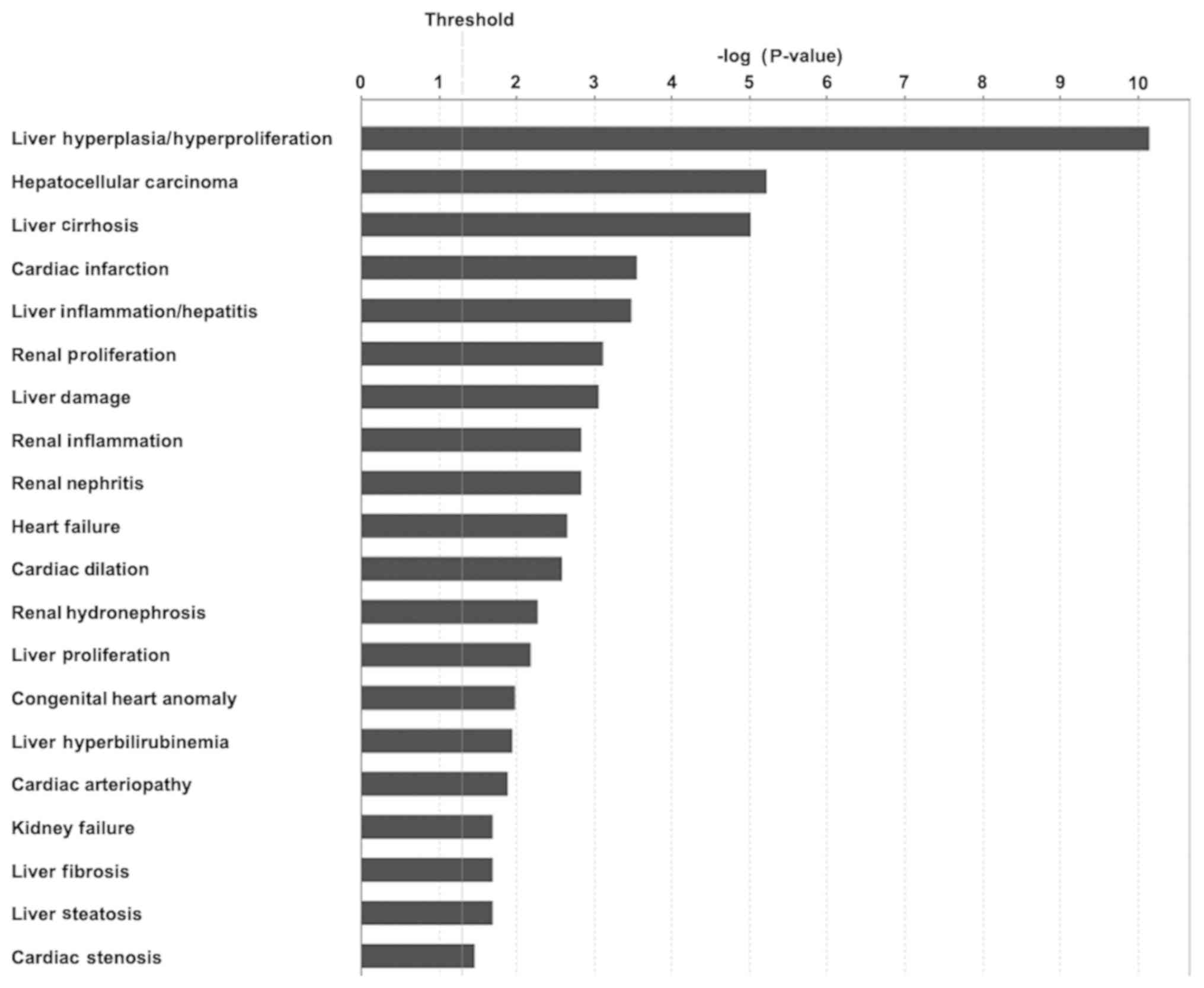
Next, the associations between DEGs and IPA toxicity
(tox) functions were determined. Following examination of toxicity
phenotypes, a total of 47 tox functions were identified. Among the
47 tox functions, 22 exhibited a P<0.05. The top enriched tox
function, liver hyperplasia/hyperproliferation, had the lowest
P-value of <7x10-11. The top 20 tox functions in FJOA
are listed (Fig. 6). The tox
function analysis, from the genetic aspect, indicated the possible
associations of FJOA with other diseases.
Identification of essential functional
networks
DEGs in different diseases and functions were
further connected with each other to build IPA functional networks.
A total of 25 IPA networks were identified. These IPA networks were
ranked based on their scores (Table
II). The top-scoring IPA network included a total of 34 DEGs
and had a score of 33, indicating that the possibility that genes
in the IPA network were not connected was not lower the 10 and not
higher than 3310. Even the IPA network with the lowest score had a
score of 15, suggesting that these genes were highly connected.
 | Table IIIngenuity pathway analysis networks
in facet joint osteoarthritis. |
Table II
Ingenuity pathway analysis networks
in facet joint osteoarthritis.
| Score | Top diseases and
functions |
|---|
| 33 | Cell-to-cell
signaling and interaction, hematological system development and
function, immune cell trafficking |
| 33 | Cardiovascular
disease, developmental disorder, hematological disease |
| 31 | Cell cycle,
cellular movement, cancer |
| 31 | DNA replication,
recombination, and repair, cell cycle, cellular assembly and
organization |
| 31 | Connective tissue
disorders, dermatological diseases and conditions, developmental
disorder |
| 27 | Antimicrobial
response, inflammatory response, cell-to-cell signaling and
interaction |
| 27 | Cardiovascular
disease, cell death and survival, connective tissue disorders |
| 27 | Cancer, connective
tissue disorders, organismal injury and abnormalities |
| 27 | Cell-to-cell
signaling and interaction, hematological system development and
function, cellular development |
| 25 | Hematological
system development and function, cell-to-cell signaling and
interaction, cell morphology |
| 25 | Cell cycle,
reproductive system development and function, cell death and
survival |
| 25 | Cell signaling,
molecular transport, vitamin and mineral metabolism |
| 23 | Cell-to-cell
signaling and interaction, hematological system development and
function, immune cell trafficking |
| 23 | Cell cycle,
organismal injury and abnormalities, reproductive system
disease |
| 23 | Hematological
system development and function, humoral immune response, lymphoid
tissue structure and development |
| 22 | Cellular movement,
immune cell trafficking, hematological system development and
function |
| 20 | Cellular movement,
cell-to-cell signaling and interaction, hematological system
development and function |
| 20 | Cell cycle,
cellular assembly and organization, DNA replication, recombination
and repair |
| 20 | Cancer, organismal
injury and abnormalities, reproductive system disease |
| 19 | Cellular
development, cellular movement, cell morphology |
| 19 | DNA replication,
recombination, and repair, nucleic acid metabolism, small molecule
biochemistry |
| 17 | Cell-to-cell
signaling and interaction, hematological system development and
function, immune cell trafficking |
| 17 | Cellular movement,
hematological system development and function, immune cell
trafficking |
| 15 | Cellular assembly
and organization, DNA replication, recombination and repair,
cancer |
| 15 | Infectious
diseases, cell signaling, molecular transport |
A total of 5 IPA networks exhibited a score of
>30 (network 1 and 2: Score of 33; network 3, 4 and 5: Score of
31). Numerous significantly enriched diseases and functions were
also enriched in these IPA networks. It was indicated that cell and
tissue growth-associated diseases and functions, including
hematological system development and function, developmental
disorder, cellular movement, DNA replication, recombination,
repair, cell cycle, and cellular assembly and organization were
involved in these top 5 IPA networks. These outcomes suggested that
the dynamic changes of cellular development were essential
characteristics in FJOA.
These highly significant IPA networks were further
outlined in detail (Fig. 7). It was
observed that in IPA network 1, most genes were connected with
C-X-C motif chemokine ligand 8 (CXCL8). Furthermore, elastase,
neutrophil expressed (ELANE), growth factor independent 1
transcriptional repressor (GFI1), Spi-1 proto-oncogene (SPI1),
CCAAT/enhancer binding protein epsilon (CEBPE) and GATA binding
protein 1 (GATA1) were also connected with many other DEGs
(Fig. 7A). In IPA network 2, hub
genes were TAL BHLH transcription factor 1, erythroid
differentiation factor (TAL1), minichromosome maintenance complex
component 4 (MCM4), BTG anti-proliferation factor 2 (BTG2) and
BRCA1, DNA repair associated (BRCA1) (Fig. 7B). In IPA network 3, the most
connected gene was cyclin D1 (CCND1) (Fig. 7C). Chromatin assembly factor 1
subunit A (CHAF1A) and histones were hub genes in IPA network 4
(Fig. 7D), whereas TREM1 and tumor
protein P63 (TP63) were hub genes in IPA network 5 (Fig. 7E).
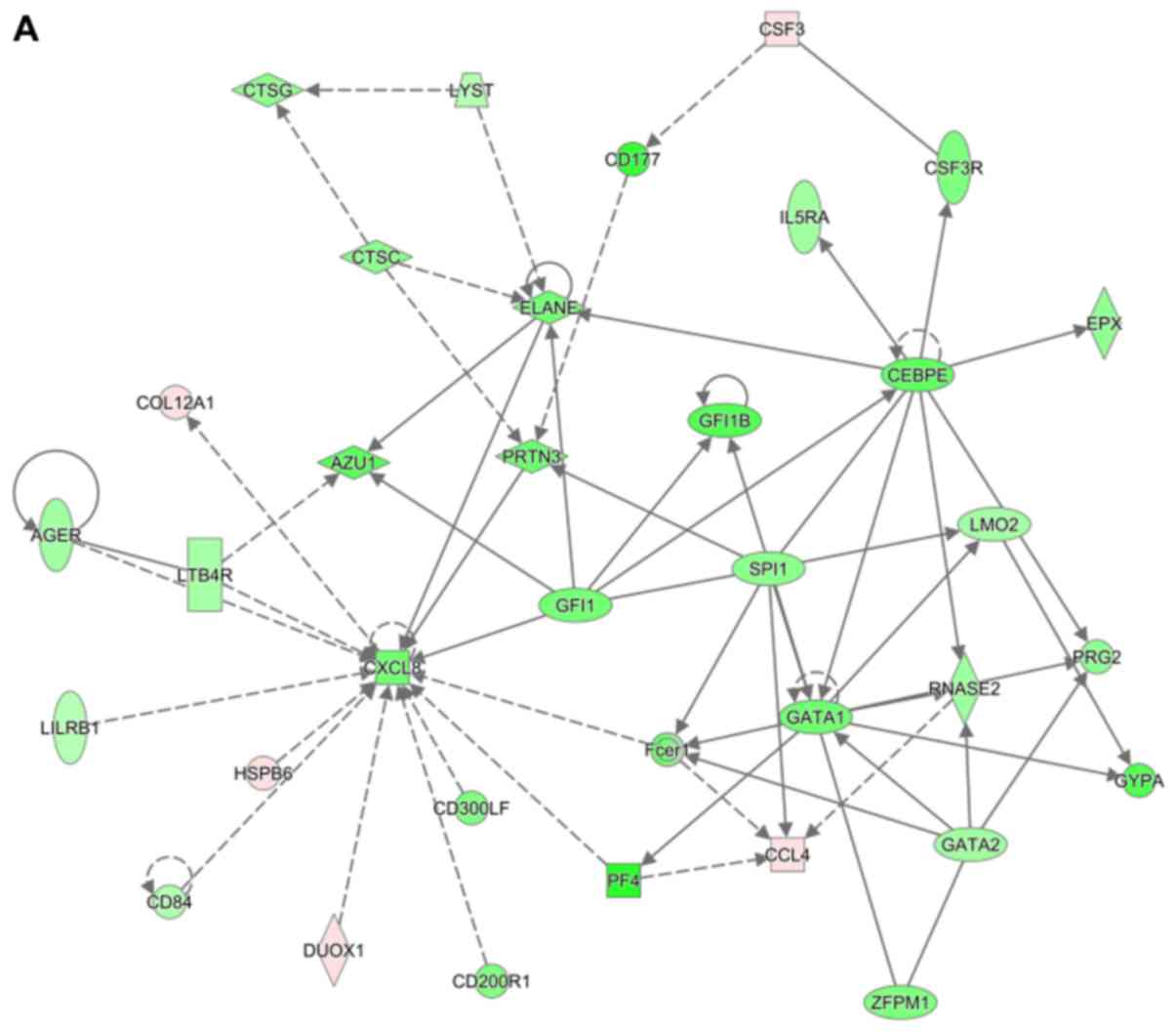 | Figure 7Top 5 ingenuity pathway analysis
networks in facet joint osteoarthritis. Upregulated genes are
labeled in red, while downregulated genes are labeled in green. The
saturation of color is correlated with the fold change of the gene
(high saturation means high fold change and low saturation means
low fold change). Solid lines indicate direct connections, while
dotted lines indicate indirect connections (circular arrows means
influence itself). Ellipses represent transcription regulators,
rhombuses represent enzymes, trapezoids represent transporters,
double circles represent a complex/group and circles represent
others. (A) CXCL8, ELANE, GFI1, SPI1, CEBPE, GATA1, (B) TAL1, BTG2,
BRCA1, (C) CCND1, (D) CHAF1A, histone, (E) TREM1 and TP63 were
located in the central positions of IPA networks. |
The expression levels of certain central genes
identified, including ELANE, BTG2, BRCA1, CHAF1A, TP63, CXCL8,
GFI1, CCND1, SPI1, CEBPE, GATA1, TAL1, MCM4 and TREM1, were further
examined by RT-qPCR. The results indicated that, consistent with
the sequencing results, most of these genes were dysregulated in
lesioned facet joint tissue samples collected from patients with
FJOA compared with those in the healthy facet joint tissue samples
(Fig. 8).
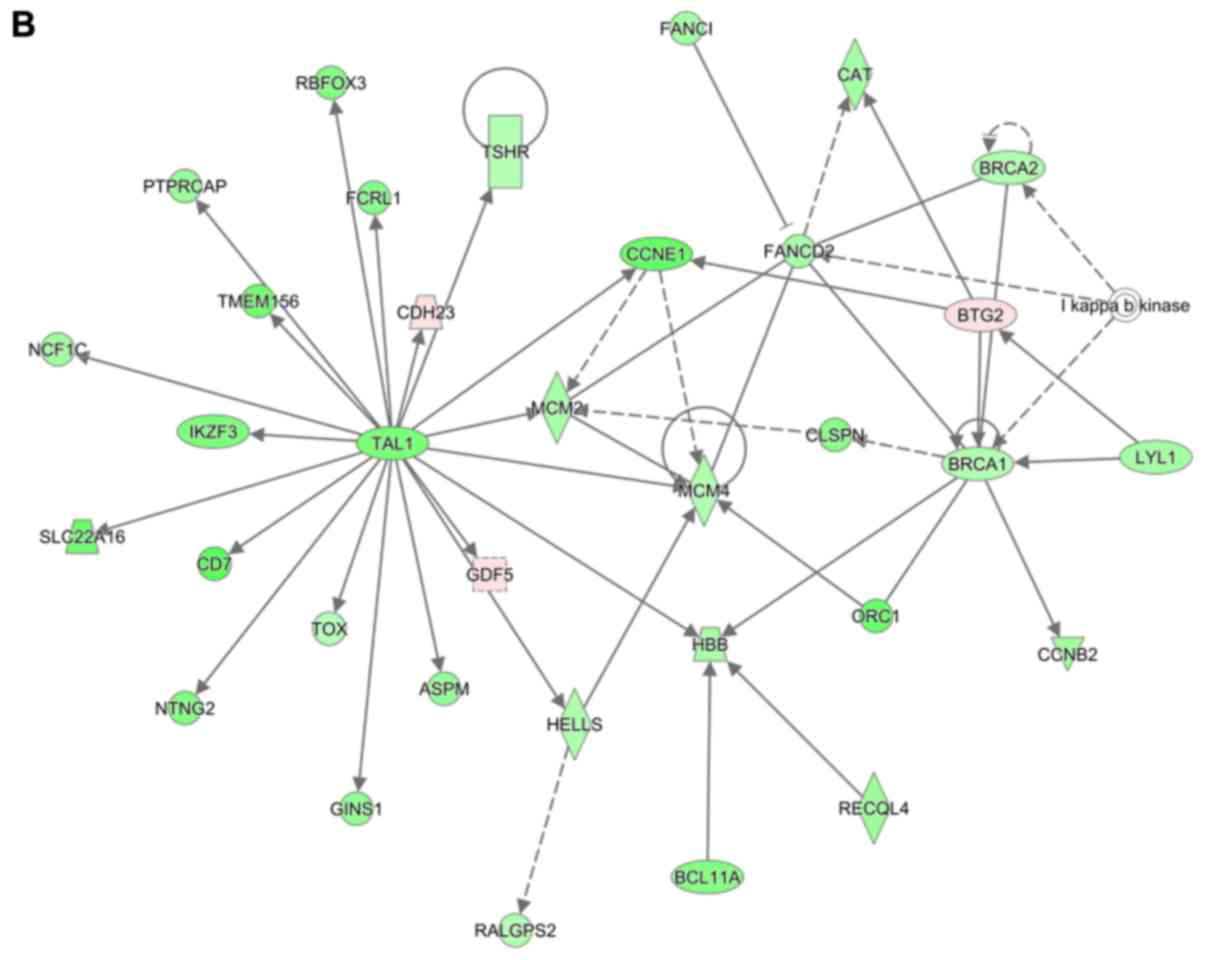 | Figure 8Validation of hub genes in ingenuity
pathway analysis networks. The relative mRNA abundance of ELANE,
BTG2, BRCA1, CHAF1A, TP63, CXCL8, GFI1, CCND1, SPI1, CEBPE, GATA1,
TAL1, MCM4 and TREM1 was determined by reverse transcription-qPCR
with normalization to GAPDH. Results are presented as the mean ±
standard deviation from 3 independent experiments. RNA-seq, RNA
sequencing; qPCR, quantitative PCR; ELANE, elastase, neutrophil
expressed; BTG2, BTG anti-proliferation factor 2; BRCA1, BRCA1 DNA
repair-associated; CHAF1A, chromatin assembly factor 1 subunit A;
TP63, tumor protein p63; CXCL8, C-X-C motif chemokine ligand 8;
GFI1, growth factor independent 1 transcriptional repressor; CCND1,
cyclin D1; SPI1, Spi-1 proto-oncogene; CEBPE, CCAAT enhancer
binding protein epsilon; GATA1, GATA binding protein 1; TAL1, TAL
bHLH transcription factor 1, erythroid differentiation factor;
MCM4, minichromosome maintenance complex component 4; TREM1,
triggering receptor expressed on myeloid cells 1. |
Discussion
In the present study, a high-resolution analysis of
transcriptional profiling was performed to characterize the changes
in expression in FJOA and IPA core analysis was used to interpret
gene expression profiles. IPA has been broadly used in the
interpretation of high-throughput data, including RNA sequencing,
small RNA sequencing, DNA sequencing, microarray, metabolomics,
proteomics and small-scale experiments (16). By using the built-in
literature-supported IPKB database, the global molecular network in
numerous physiological and pathological processes may be searched
and discovered. In the present study, by using IPA core analysis,
DEGs in FJOA were categorized into IPA canonical signaling pathways
and the top 25 enriched IPA canonical signaling pathways in FJOA
were identified.
These top enriched canonical signaling pathways were
mainly categorized into cellular stress and injury-associated
pathways, disease-specific pathways, cellular immune
response-associated pathways, cell cycle regulation-associated
pathways, intracellular and second messenger signaling,
pathogen-influenced signaling, biosynthesis-associated pathways,
and organismal growth and development-associated pathways. From the
aspect of the number of signaling pathways involved, cellular
immune response appeared to be most significant in FJOA, since
nearly half of the canonical signaling pathways (12 out of 25) were
cellular immune response-associated pathways. Inflammation and
immune responses are generally considered as key components of
osteoarthritis (17-19).
It was identified that numerous cell types in joints, including
synovial cells and articular chondrocytes, may express inflammatory
mediators (20). The modulation of
immune response was even regarded as a novel and effective
treatment of osteoarthritis (21).
On the other hand, the specific roles of inflammation and immune
responses in FJOA, a universal form of lumbar osteoarthritis, were
not well demonstrated. The IPA analysis, from the aspect of genetic
expression, demonstrated the importance of inflammation and immune
responses in FJOA. The observations of the present study were also
consistent with outcomes from a previous KEGG pathway analysis,
which indicated that Wnt signaling and NF-κ signaling pathways were
deeply involved in the process of facet joint degeneration
(12). Furthermore, it is worth
noting that it has been well elucidated that aging is closely
associated with dysregulated inflammation and immune responses
(22-24).
Since FJOA is also highly associated with aging, it is expected
that inflammation and immune responses may be critical causes of
FJOA.
Besides the integral analysis of IPA canonical
signaling pathways, the canonical signaling pathways with high
absolute values of z-scores were selected, DEGs in FJOA in these
signaling pathways were identified and the expression levels of
representative genes were validated by RT-qPCR. Leukocyte
extravasation signaling is the migration of leukocytes from blood
to tissue, a critical step during inflammation. Tec kinase
signaling is essential for the development and activation of B
cells and T cells. Therefore, although Tec kinase signaling is
grouped to intracellular and second messenger signaling due to its
intercellular localization, Tec kinase signaling is also highly
associated with inflammation and immune responses. Similarly,
osteoarthritis pathways are also associated with inflammation and
immune responses. The diagram of osteoarthritis pathways suggested
that osteoarthritis may be elicited and activated by inflammation.
Numerous pro-inflammatory and anti-inflammatory factors are also
critical in the signaling pathway. Therefore, a detailed study of
canonical signaling pathways with high absolute values of z-scores
will be explored in future to further clarify the importance of
inflammation and immune responses in FJOA. Further studies should
also clarify the specific roles of inflammatory mediators in FJOA
and the complex interactions between them. The results of the
present IPA core analysis of DEGs in FJOA may potentially aid the
discovery of inflammatory mediators as novel therapeutic strategies
to combat FJOA.
In the present study, the focus was on the
biological functions of DEGs in FJOA and the top enriched diseases
and functions in FJOA were identified and classified into different
categories. Detailed investigation of diseases and functions
indicated that cellular behavior-associated biological functions,
tissue development-associated diseases and functions, and
inflammation and immune response-associated diseases and functions
were most significantly enriched in FJOA. These results connected
the alterations of genes in FJOA with the pathological features of
the condition and indicated that cell development, tissue
remodeling, as well as inflammation and immune response, were
critical in FJOA. This was also consistent with a previous
observation that inflammation-associated signaling pathways were
critical in the pathological process of FJOA (12). Furthermore, by categorizing the DEGs
into diseases and tox functions, the connections between FJOA and
other diseases were also preliminarily revealed and diseases that
may be associated with FJOA were identified.
Besides the entire and systematical analysis of
significantly involved diseases and functions in FJOA, DEGs in
different disease and function categories were connected with one
another to build gene-gene interaction and functional networks
between differentially expressed genes. Consistent with analytic
outcomes from disease and functional categories, cell and tissue
growth diseases and functions also involved top diseases and
functions in IPA networks. Of the 25 IPA networks in FJOA, the top
5 IPA networks (with a score >30) were selected for further
study.
It was observed that CXCL8, ELANE, GFI1, SPI1, GATA1
(in IPA network 1), TAL1, BTG2 (in IPA network 2), TREM1 (in IPA
network 5) were located in the central positions of IPA networks
and connected with numerous other differentially expressed genes.
The expression levels of these hub genes were validated by RT-qPCR
and consistent with RNA-seq and it was confirmed that their
expression levels in lesioned facet joint tissue samples collected
from patients with FJOA were significantly different from their
expression levels in healthy facet joint tissue samples. CXCL8, the
hub gene in IPA network 1, was also a significantly differentially
expressed gene in the NF-κ signaling pathway, an immune and
inflammation-associated signaling pathway, as previously
demonstrated and validated (12).
Likewise, BTG2, the hub gene in IPA network 2, was identified and
examined, as it is a critical a mechanosensitive and
inflammation-related gene in osteoarthritis (25). The present study suggests that these
hub genes may not only exert their biological functions in
activated signaling pathways but also affect other genes and
signaling pathways.
Besides CXCL8 and BTG2, certain other hub genes
identified in the present study have also been previously studied
in osteoarthritis. For instance, quantitative analysis of ELANE, a
gene coding for neutrophil elastase, a proteinase secreted by
neutrophils and macrophages during inflammation, indicated that the
abundance of ELANE mRNA in peripheral blood CD14+ cells from
patients with osteoarthritis was lower than that in healthy
subjects or in patients with rheumatoid arthritis (26). This was consistent with the present
study with regard to the RNA deep sequencing and RT-qPCR results.
SPI1, a gene coding for an ETS-domain transcription factor that
activates gene expression during myeloid and B-lymphoid cell
development, and GATA1, a gene coding for a member of the GATA
family of transcription factors, were identified as candidate
transcription factors for osteoarthritis (27,28).
BRCA1, a gene encoding a tumor suppressor nuclear phosphoprotein
that has a role in maintaining genomic stability, was indicated to
have a relatively lower abundance in patients with FJOA in the
present study; this was in line with the result of a previous study
on patients with knee/hip osteoarthritis (29). BRCA1 was also identified as a crucial
transcription factor that regulates numerous DEGs located
downstream with a role in the development of osteoarthritis
(30). Similar to the observations
of the present study on FJOA, TREM1, a gene encoding an Ig
superfamily receptor that amplifies neutrophil and
monocyte-mediated inflammatory responses, was also indicated to be
upregulated in patients with osteoarthritis by using microarray
gene expression profiling, western blot and immunohistochemical
analyses (31,32). A recent study further demonstrated
that knockdown of upregulated TREM1 in a mouse model of
osteoarthritis inhibited the production of matrix
metallopeptidase-13, promote the synthesis of collagen type II,
suppress the metabolic imbalance of extracellular matrix and
decrease the activity of the NF-κ signaling pathway (33). The participation of other hub genes
identified, including GFI1, CEBPE, TAL1, CHAF1A and TP63, in
osteoarthritis, let alone in FJOA, has not been previously
determined, to the best of our knowledge. The present study
revealed the involvement of these hub genes, indicated their
central roles in FJOA and thus proposed potential therapeutic
targets for FJOA. The presence of these hub genes in patients with
different degrees of FJOA and the biological roles of these hub
genes should be further investigated in future studies.
In conclusion, the present study performed an IPA
core analysis and a systemic investigation of the transcriptomic
signature of FJOA. Diseases, biological functions and tox functions
significantly enriched by the DEGs in FJOA, as well as the
gene-gene interactions of those DEGs were identified. The present
study provides implications for understanding the mechanisms of
FJOA and the identification of novel therapeutic approaches for
this condition.
Supplementary Material
Full list of canonical signaling
pathways in facet joint osteoarthritis.
Acknowledgements
Not applicable.
Funding
This work was supported by The National Natural
Science Foundation of China (grant no. 81771319) and Nantong
Science and Technology Project (grant no. HS2018002).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CC, YS and ZC conceived and designed the study. CC,
SC, WL, HJ, JF and YS performed the experiments. CC analyzed the
data. YS and ZC provided reagents/materials/analysis tools. CC and
ZC wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All procedures were ethically approved by The Human
Ethics Committee of the Second Affiliated Hospital of Nantong
University (Nantong, China) and documents of informed consent were
signed by patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lewinnek GE and Warfield CA: Facet joint
degeneration as a cause of low back pain. Clin Orthop Relat Res.
216–222. 1986.PubMed/NCBI
|
|
2
|
Ko S, Vaccaro AR, Lee S, Lee J and Chang
H: The prevalence of lumbar spine facet joint osteoarthritis and
its association with low back pain in selected Korean populations.
Clin Orthop Surg. 6:385–391. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Netzer C, Distel P, Wolfram U, Deyhle H,
Jost GF, Schären S and Geurts J: Comparative analysis of bone
structural parameters reveals subchondral cortical plate resorption
and increased trabecular bone remodeling in human facet joint
osteoarthritis. Int J Mol Sci. 19(pii: E845)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kalichman L, Li L, Kim DH, Guermazi A,
Berkin V, O'Donnell CJ, Hoffmann U, Cole R and Hunter DJ: Facet
joint osteoarthritis and low back pain in the community-based
population. Spine (Phila Pa 1976). 33:2560–2565. 2008.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kim JS, Ahmadinia K, Li X, Hamilton JL,
Andrews S, Haralampus CA, Xiao G, Sohn HM, You JW, Seo YS, et al:
Development of an experimental animal model for lower back pain by
percutaneous injury-induced lumbar facet joint osteoarthritis. J
Cell Physiol. 230:2837–2847. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kim DS, Lee SJ, Park SY, Yoo HJ, Kim SH,
Kim KJ and Cho HJ: Differentially expressed genes in rat dorsal
root ganglia following peripheral nerve injury. Neuroreport.
12:3401–3405. 2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Netzer C, Urech K, Hügle T, Benz RM,
Geurts J and Schären S: Characterization of subchondral bone
histopathology of facet joint osteoarthritis in lumbar spinal
stenosis. J Orthop Res. 34:1475–1480. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bleil J, Maier R, Hempfing A, Schlichting
U, Appel H, Sieper J and Syrbe U: Histomorphologic and
histomorphometric characteristics of zygapophyseal joint remodeling
in ankylosing spondylitis. Arthritis Rheumatol. 66:1745–1754.
2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Appel H, Maier R, Loddenkemper C, Kayser
R, Meier O, Hempfing A and Sieper J: Immunohistochemical analysis
of osteoblasts in zygapophyseal joints of patients with ankylosing
spondylitis reveal repair mechanisms similar to osteoarthritis. J
Rheumatol. 37:823–828. 2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Eisenstein SM and Parry CR: The lumbar
facet arthrosis syndrome. Clinical presentation and articular
surface changes. J Bone Joint Surg Br. 69:3–7. 1987.PubMed/NCBI
|
|
11
|
Kim JS, Ali MH, Wydra F, Li X, Hamilton
JL, An HS, Cs-Szabo G, Andrews S, Moric M, Xiao G, et al:
Characterization of degenerative human facet joints and facet joint
capsular tissues. Osteoarthritis Cartilage. 23:2242–2251.
2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen C, Bao GF, Xu G, Sun Y and Cui ZM:
Altered Wnt and NF-κ signaling in facet joint osteoarthritis:
Insights from RNA deep sequencing. Tohoku J Exp Med. 245:69–77.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yu J, Gu X and Yi S: Ingenuity pathway
analysis of gene expression profiles in distal nerve stump
following nerve injury: Insights into wallerian degeneration. Front
Cell Neurosci. 10(274)2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yi S, Zhang H, Gong L, Wu J, Zha G, Zhou
S, Gu X and Yu B: Deep sequencing and bioinformatic analysis of
lesioned sciatic nerves after crush injury. PLoS One.
10(e0143491)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ghosh S, Dutta S, Thorne G, Boston A,
Barfield A, Banerjee N, Walker R and Banerjee HN: Core canonical
pathways involved in developing human glioblastoma multiforme
(GBM). Int J Sci Res Sci Eng Technol. 3:458–465. 2017.PubMed/NCBI
|
|
17
|
Raman S, FitzGerald U and Murphy JM:
Interplay of inflammatory mediators with epigenetics and cartilage
modifications in osteoarthritis. Front Bioeng Biotechnol.
6(22)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Goldring MB and Goldring SR:
Osteoarthritis. J Cell Physiol. 213:626–634. 2007.PubMed/NCBI
|
|
19
|
Ray A and Ray BK: An
inflammation-responsive transcription factor in the pathophysiology
of osteoarthritis. Biorheology. 45:399–409. 2008.PubMed/NCBI
|
|
20
|
Shen J, Abu-Amer Y, O'Keefe RJ and
McAlinden A: Inflammation and epigenetic regulation in
osteoarthritis. Connect Tissue Res. 58:49–63. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Fahy N, Farrell E, Ritter T, Ryan AE and
Murphy JM: Immune modulation to improve tissue engineering outcomes
for cartilage repair in the osteoarthritic joint. Tissue Eng Part B
Rev. 21:55–66. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wu D and Meydani SN: Age-associated
changes in immune and inflammatory responses: Impact of vitamin E
intervention. J Leukoc Biol. 84:900–914. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Meydani SN and Wu D: Nutrition and
age-associated inflammation: Implications for disease prevention.
JPEN J Parenter Enteral Nutr. 32:626–629. 2008.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Han SN and Meydani SN: Antioxidants,
cytokines, and influenza infection in aged mice and elderly humans.
J Infect Dis. 182 (Suppl 1)(S74-S80)2000.PubMed/NCBI View
Article : Google Scholar
|
|
25
|
Guo Y, Song Y, Zhang Y and Yang L:
Identification of Btg2 As A mechanosensitive gene by functional
screening integrative analyses. Mol Cell Biomech. 16 (Suppl
2)(S119)2019. View Article : Google Scholar
|
|
26
|
Trzybulska D, Olewicz-Gawlik A, Graniczna
K, Kisiel K, Moskal M, Cieślak D, Sikora J and Hrycaj P:
Quantitative analysis of elastase and cathepsin G mRNA levels in
peripheral blood CD14(+) cells from patients with rheumatoid
arthritis. Cell Immunol. 292:40–44. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Dong S, Xia T, Wang L, Zhao Q and Tian J:
Investigation of candidate genes for osteoarthritis based on gene
expression profiles. Acta Orthop Traumatol Turc. 50:686–690.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang K, Zhao L, Liu X, Hao Z, Zhou Y, Yang
C and Li H: Differential co-expression analysis of rheumatoid
arthritis with microarray data. Mol Med Rep. 10:2421–2426.
2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Pellicelli M, Picard C, Wang D, Lavigne P
and Moreau A: E2F1 and TFDP1 Regulate PITX1 expression in normal
and osteoarthritic articular chondrocytes. PLoS One.
11(e0165951)2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Fei Q, Lin J, Meng H, Wang B, Yang Y, Wang
Q, Su N, Li J and Li D: Identification of upstream regulators for
synovial expression signature genes in osteoarthritis. Joint Bone
Spine. 83:545–551. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Lambert C, Dubuc JE, Montell E, Vergés J,
Munaut C, Noël A and Henrotin Y: Gene expression pattern of cells
from inflamed and normal areas of osteoarthritis synovial membrane.
Arthritis Rheumatol. 66:960–968. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Collins CE, La DT Yang HT, Massin F, Gibot
S, Faure G and Stohl W: Elevated synovial expression of triggering
receptor expressed on myeloid cells 1 in patients with septic
arthritis or rheumatoid arthritis. Ann Rheum Dis. 68:1768–1774.
2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tang J and Dong Q: Knockdown of TREM-1
suppresses IL-1β-induced chondrocyte injury via inhibiting the NF-κ
pathway. Biochem Biophys Res Commun. 482:1240–1245. 2017.PubMed/NCBI View Article : Google Scholar
|