Introduction
Implantation failure is the most frequent cause of pregnancy loss after in vitro fertilization (IVF) and embryo transfer (ET), and a substantial number of patients fail to achieve successful pregnancy, even after repeated IVF attempts (1). Recurrent implantation failure (RIF) refers to a clinical condition in which good-quality embryos repeatedly fail to implant following two or more IVF cycles (1). The precise definition, however, remains controversial, and different fertility centers practicing IVF may use different definitions for this condition (2,3). Despite various known causes of RIF, including embryonic or uterine factors, cytogenetic abnormalities, genetics, and immunologic factors, the etiology of RIF remains undetermined in most cases, and this presents a major challenge for both clinicians and researchers (2).
One potential molecular mechanism that may influence embryo implantation is regulation of microRNAs (miRNAs) (4). These are an abundant set of evolutionarily conserved small RNAs that control posttranscriptional gene expression by targeting messenger RNAs (mRNAs) for degradation, translational repression, or both (5,6). Functionally, miRNAs exert their activity by binding to the 3'-untranslated region (UTR) of their target mRNA, thereby repressing gene expression. A single miRNA can regulate multiple target mRNAs simultaneously, thereby acting as a master regulator of gene expression and cellular function. Therefore, miRNAs play pivotal roles in both physiological and pathological processes (5).
In a cell, miRNA biosynthesis involves the activity of several miRNA machinery genes in multiple steps. RNA polymerase II mediates transcription of primary miRNA transcripts (pri-miRNAs) in the nucleus. The endonuclease Drosha ribonuclease III (DROSHA) and its cofactor, DiGeorge syndrome critical region gene 8 (DGCR8), then cleave this long pri-miRNAs to form precursor-miRNAs (pre-miRNA). This pre-miRNA is exported from the nucleus to the cytoplasm by the combined activity of Ras-related nuclear protein (RAN) GTPase and exportin 5 (XPO5), and is then converted into mature miRNA by Dicer 1, ribonuclease III (DICER1) (7). These miRNA machinery genes are located in chromosome 5 (DROSHA), chromosome 14 (DICER1), chromosome 12 (RAN), chromosome 6 (XPO5), and chromosome 22 (DGCR8) (8-12).
The miRNA machinery proteins must function precisely to generate correct miRNAs, in order for these species to perform their numerous essential roles accurately. Abnormal expression of even a single miRNA can have a detrimental impact on the outcome of diverse cellular functions via modulation of target gene activity, and there is increasing evidence for deregulated miRNA expression in a variety of reproductive disorders, including endometriosis, leiomyoma, and implantation failure (4). In particular, Revel et al (13) reported altered miRNA expression in the secretory endometrium of RIF patients, as compared to fertile women, indicating that aberrantly expressed miRNAs may contribute to RIF development. In addition, a number of recent studies have shown that single nucleotide polymorphisms (SNPs) or mutations within the miRNA sequence can lead to altered miRNA expression and/or maturation, and that SNPs in specific miRNAs contribute to the genetic predisposition for pregnancy loss (14).
To date, however, few studies have investigated the potential relationship between specific SNPs in miRNA machinery genes and the risk of RIF. Here, we investigated whether polymorphisms in the miRNA machinery genes DROSHA, DICER1, RAN, XPO5, and DGCR8 are associated with increased risk of RIF in Korean women.
Materials and methods
Study participants
The study population consisted of women who experienced RIF (n=119) and normal controls (n=228). RIF was defined as the failure to achieve pregnancy after two completed fresh IVF-ET cycles, in subjects whose serum human chorionic gonadotrophin concentrations were <5 mIU/ml at 14 days after ET. Each transferred embryo was cleaved into more than 10 cells, and these were examined by the embryologist and judged to be of good quality by morphological assessment, according to the Gardner criteria (15), before transfer. Grade 1 embryos, with regular blastomeres and no cytoplasmic fragments, and grade 2 embryos, with regular blastomeres and minor cytoplasmic fragments, were considered to be good-quality embryos.
Both the male and female partners experiencing RIF were evaluated. Subjects who were diagnosed with RIF due to anatomic, chromosomal, hormonal, infectious, autoimmune, or thrombotic causes were excluded from the study group. Anatomical abnormalities were evaluated using several imaging modalities, including sonography, hysterosalpingogram, hysteroscopy, computerized tomography, and magnetic resonance imaging. Karyotyping was conducted using standard protocols. Hormonal causes for RIF, including hyperprolactinemia, luteal insufficiency, and thyroid disease, were excluded by measuring prolactin, thyroid-stimulating hormone (TSH), free T4, follicle-stimulating hormone (FSH), luteinizing hormone (LH), and progesterone levels in peripheral blood. Lupus anticoagulant and anti-cardiolipin antibodies were measured to identify the autoimmune diseases. Thrombotic disorders, defined as thrombophilia, were evaluated by protein C and protein S deficiencies, and by the presence of anti-α2 glycoprotein antibodies. Semen analysis, karyotyping, and hormonal assays, including measurements of estradiol, testosterone, FSH, and LH, were performed for male partners. Among the initial 152 patients who were enrolled for the study, 33 subjects who had Müllerian anomaly, hypothyroidism, chromosomal abnormality, or antiphospholipid syndrome were excluded from the patient group, resulting in 119 RIF patients for the study.
Women in the control group were recruited from CHA Bundang Medical Center. Enrollment criteria for controls included regular menstrual cycles, normal karyotype (46, XX), a history of at least one naturally conceived pregnancy, and no history of pregnancy loss. Data were collected identically for both groups. Furthermore, this study was approved by the Institutional Review Board of CHA Bundang Medical Center (reference no. PBC09-120) and all participants provided written informed consent. All study protocols followed the recommendations of the Declaration of Helsinki.
Analysis of miRNA biogenesis gene polymorphisms
Peripheral blood samples for genotyping were collected from all subjects, using an anticoagulant. Genomic DNA was extracted from blood samples using the G-DEX IIb Genomic DNA Extraction Kit-BLOOD (iNtRON Biotechnology, Seongnam, South Korea). Nucleotide sequence variations in miRNA processing genes were determined by polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) analysis. Restriction enzyme digestion was carried out using the following enzymes (New England BioLabs) and conditions: NlaIII (DROSHA, rs10719), BanI (DICER1, rs3742330), BslI (RAN, rs14035), with incubation at 55̊C for 16 h, and EcoRI (XPO5, rs11077) and NsiI (DGCR8, rs417309), with incubation at 37̊C for 16 h. Genotypes determined by RFLP analysis were confirmed by two independent investigators, and then checked again by sequencing 10% of the samples. For RAN rs14035 genotyping of the control group, and for XPO5 rs11077 genotyping of RIF patients, genotypes were confirmed three times to rule out possible errors attributed to violation of the Hardy-Weinberg equilibrium (HWE).
Hormone assays
Blood samples were collected by venipuncture on day 2 or 3 of the menstrual cycle for the measurement of FSH, LH, E2, TSH, and prolactin levels. Serum was prepared as previously described (16), and hormone levels were determined using either radioimmunoassays (E2, TSH, and prolactin; Beckman Coulter), or enzyme immunoassays (FSH and LH; Siemens AG), according to the manufacturer protocols.
Immune cell assays
Immune cell measurements were performed by flow cytometry and analyzed using CellQuest software (BD FACSCalibur; BD Biosciences), as previously described (17). Fluorescently labeled [fluorescein isothiocyanate, phycoerythrin (PE), peridinin chlorophyll protein, and allophycocyanin] monoclonal antibodies specific for CD3, CD4, CD8, CD19, and CD56 were obtained from BD Biosciences. Anti-NKG2A-PE antibodies were obtained from Immunotech (Beckman Coulter). These experiments were performed in the department of Diagnostic Laboratory Medicine, CHA Bundang Medical Center.
Statistical analysis
To compare baseline characteristics between RIF patients and controls, we used Chi-square tests to analyze categorical data, and the unpaired-samples Student's t-test to analyze continuous data. Also, we used analysis of variance (ANOVA) followed by Tukey's test to analyze continuous data. Genetic frequencies of each polymorphism in RIF patients and controls were compared using multivariate logistic regression. Allele frequencies were calculated to identify deviations from HWE using P=0.05 as a threshold. Odds ratios (ORs), adjusted odds ratios (AOR), and 95% confidence intervals (CIs) were used to measure the strength of the association between DICER1, DROSHA, RAN, XPO5, and DGCR8 polymorphisms and risk of RIF with GraphPad Prism 4.0 (GraphPad Software Inc.) and StatsDirect software, version 2.4.4 (StatsDirect Ltd.) (18). Associations between polymorphisms and RIF incidence were calculated using AORs and 95% CIs from logistic regression, adjusted for age. Two-tailed P-values <0.05 were considered statistically significant. A false discovery rate (FDR) correction was performed to adjust for multiple comparisons and associations with FDR-P-values <0.05 were considered statistically significant (19). Gene-gene interactions among SNP loci were analyzed using multifactor dimensionality reduction (MDR), with MDR, version 2.0 (www.epistasis.org) (20). We determined the best multilocus combinations based on MDR identification of the most significant models, using the optimal maximized cross-validation values. HAPSTAT, version 3.0 (www.bios.unc.edu/~lin/hapstat/), was used to estimate which allele combination frequencies, determined by MDR analysis, are predicted to have strong synergistic effects (21).
Results
Genotype frequencies of miRNA machinery gene polymorphisms
The demographic characteristics and clinical variables of RIF patients and control subjects are presented in Table I. The genotype and allele frequencies for the five miRNA machinery genes in RIF patients and unaffected controls are shown in Table II. All genes that were analyzed contained polymorphisms and the genotypic distributions in both groups were in HWE. We found that the DROSHA rs10719T>C and RAN rs14035C>T 3'UTR polymorphisms were negatively associated with the prevalence of RIF. Further, the frequencies of the TC genotype of DROSHA rs10719 and that of CT genotype of RAN rs14035 were lower in the RIF group than in control group (AOR=0.550, 95% CI, 0.339-0.893, P=0.016; AOR=0.590, 95% CI, 0.363-0.958, P=0.033, respectively). Similarly, the combined RAN rs14035 CT+TT and combined DROSHA rs10719 TC+CC genotypes were associated with decreased prevalence of RIF (AOR=0.616, 95% CI, 0.386-0.982, P=0.042; AOR=0.620, 95% CI, 0.394-0.976, P=0.039, respectively). For the DICER1 rs3742330A>G, XPO5 rs11077A>C, and DGCR8 rs417309G>A genotypes, there were no statistically significant differences between the RIF group and controls (Table SI).
 |
Table I
Baseline characteristics of controls and RIF patients.
|
Table I
Baseline characteristics of controls and RIF patients.
| Characteristic |
Controls (n=210), mean ± SD |
RIF patients (n=119), mean ± SD |
P-valuea |
| Age (years) |
34.06±5.66 |
34.24±3.35 |
0.760 |
| Platelet (103/µl) |
239.47±66.35 |
238.46±63.65 |
0.919 |
| PT (sec) |
11.61±3.32 |
11.42±0.62 |
0.001b |
| aPTT (sec) |
33.45±3.65 |
30.93±3.41 |
<0.001 |
| FSH (mIU/ml) |
8.14±2.86 |
8.76±4.50 |
0.662b |
| LH (mIU/ml) |
3.26±1.78 |
4.75±2.33 |
<0.001b |
| Estradiol-2 (pg/ml) |
26.17±14.83 |
34.20±23.25 |
0.001b |
| Body mass index (kg/m2) |
21.78±3.49 |
20.89±2.55 |
0.276b |
| Hematocrit (%) |
35.95±4.19 |
- |
- |
| Folate (nmol/l) |
- |
15.63±11.29 |
- |
| Homocysteine (umol/l) |
- |
6.56±1.34 |
- |
| BUN (mg/dl) |
- |
10.03±2.86 |
- |
| Creatinine (mg/dl) |
- |
0.78±0.10 |
- |
| Uric acid (mg/dl) |
- |
3.93±0.96 |
- |
| Total cholesterol (mg/dl) |
- |
192.93±42.77 |
- |
| White blood cell (103/µl) |
- |
7.22±2.85 |
- |
| TSH (uIU/ml) |
- |
2.26±1.44 |
- |
| Prolactin (ng/ml) |
- |
12.64±6.18 |
- |
| Hemoglobin (g/dl) |
- |
12.57±1.40 |
- |
| CD19+ B cell (%) |
- |
66.27±11.23 |
- |
| CD3+ pan T (%) |
- |
33.83±8.65 |
- |
| CD4+ helper T (%) |
- |
29.11±7.88 |
- |
| CD56+ NK cell (%) |
- |
11.01±4.44 |
- |
| CD8+ T (%) |
- |
20.00±9.49 |
- |
 |
Table II
Genotype frequency of miRNA machinery gene polymorphisms between RIF patients and control groups.
|
Table II
Genotype frequency of miRNA machinery gene polymorphisms between RIF patients and control groups.
| Genotypes |
Controls, n (%) (n=210) |
RIF patients, n (%) (n=119) |
AOR (95% CI)a |
P-value |
| DICER rs3742330A>G |
|
|
|
|
| AA |
65 (31.0) |
40 (33.6) |
1.000 (reference) |
|
| AG |
114 (54.3) |
60 (50.4) |
0.856 (0.517-1.417) |
0.546 |
| GG |
31 (14.8) |
19 (16.0) |
0.991 (0.493-1.992) |
0.980 |
| Dominant (AA vs. AG+GG) |
|
|
0.890 (0.550-1.440) |
0.634 |
| Recessive (AA+AG vs. GG) |
|
|
0.752 (0.275-0.000) |
0.752 |
| HWE P |
0.096 |
0.656 |
|
|
| DROSHA rs10719T>C |
|
|
|
|
| TT |
95 (45.2) |
68 (57.1) |
1.000 (reference) |
|
| TC |
99 (47.1) |
39 (32.8) |
0.550 (0.339-0.893) |
0.016 |
| CC |
16 (7.6) |
12 (10.1) |
1.062 (0.472-2.390) |
0.884 |
| Dominant (TT vs. TC+CC) |
|
|
0.620 (0.394-0.976) |
0.039 |
| Recessive (TT+TC vs. CC) |
|
|
1.365 (0.623-2.994) |
0.437 |
| HWE P |
0.154 |
0.085 |
|
|
| RAN3'UTR rs14035C>T |
|
|
|
|
| CC |
112 (53.3) |
77 (64.7) |
1.000 (reference) |
|
| CT |
89 (42.4) |
37 (31.1) |
0.590 (0.363-0.958) |
0.033 |
| TT |
9 (4.3) |
5 (4.2) |
0.759 (0.239-2.417) |
0.641 |
| Dominant (CC vs. CT+TT) |
|
|
0.616 (0.386-0.982) |
0.042 |
| Recessive (CC+CT vs. TT) |
|
|
0.976 (0.319-2.984) |
0.965 |
| HWE P |
0.092 |
0.835 |
|
|
| XPO5 rs11077A>C |
|
|
|
|
| AA |
176 (83.8) |
99 (83.2) |
1.000 (reference) |
|
| AC |
32 (15.2) |
18 (15.1) |
0.983 (0.523-1.846) |
0.957 |
| CC |
2 (1.0) |
2 (1.7) |
1.529 (0.202-11.601) |
0.681 |
| Dominant (AA vs. AC+CC) |
|
|
1.036 (0.564-1.903) |
0.910 |
| Recessive (AA+AC vs. CC) |
|
|
1.726 (0.236-12.618) |
0.591 |
| HWE P |
0.687 |
0.283 |
|
|
| DGCR8 rs417309G>A |
|
|
|
|
| GG |
197 (93.8) |
110 (92.4) |
1.000 (reference) |
|
| GA |
13 (6.2) |
9 (7.6) |
1.237 (0.513-2.988) |
0.636 |
| AA |
0 (0.0) |
0 (0.0) |
NA |
NA |
| Dominant (GG vs. GA+AA) |
|
|
1.237 (0.513-2.988) |
0.636 |
| Recessive (GG+GA vs. AA) |
|
|
NA |
NA |
| HWE P |
0.643 |
0.668 |
|
|
Allele combination analysis
An allele-allele combination-based analysis of the five polymorphisms of the miRNA machinery genes (DICER1, DROSHA, RAN, XPO5, DGCR8) was conducted to evaluate whether polymorphic site interactions exert synergistic effects on RIF risk (Table III). Among the models of five polymorphic loci, one allele combination (A-C-T-A-G) showed synergistic effects on reduced RIF risk (OR=0.208, 95% CI, 0.070-0.618, FDR-P=0.022), whereas two other allele combinations (G-T-C-C-G and G-C-T-A-G) were associated with increased risk of RIF (OR=28.290, 95% CI, 1.634-490.000, FDR-P=0.021; OR=17.510, 95% CI, 0.972-315.500, FDR-P=0.048, respectively), when the reference combination was A-T-C-A-G. However, it was difficult to make a conclusion with five allele combinations of G-T-C-C-G and G-C-T-A-G due to the wide range of CIs. Three-allele combination analysis indicated that DICER1/DROSHA/RAN A-C-T (OR=0.255, 95% CI, 0.103-0.633, FDR-P=0.008), as well as the DICER1/DROSHA/XPO5 A-T-C, A-C-A, and G-T-A genotypes were associated with decreased RIF risk (OR=0.345, 95% CI, 0.144-0.829, FDR-P=0.024; OR=0.457, 95% CI, 0.281-0.744, FDR-P=0.005; and OR=0.553, 95% CI, 0.369-0.827, FDR-P=0.009, respectively). Among the models with two loci, the DICER1/DROSHA A-C genotype was associated with a reduced relative risk of RIF (OR=0.549, 95% CI, 0.345-0.875, FDR-P=0.033). These associations also remained statistically significant after controlling for FDR. In addition, we performed combined genotype analysis with the miRNA machinery gene polymorphisms (Table IV) and found that the DROSHA TC and RAN CT combination decreased the risk of RIF. The combination of these two genotypes showed the lowest AOR with statistical significance in RIF risk (AOR=0.314, 95% CI, 0.147-0.673, FDR-P=0.023).
 |
Table III
Allele combination frequencies for the miRNA machinery genes polymorphisms in RIF patients and controls.
|
Table III
Allele combination frequencies for the miRNA machinery genes polymorphisms in RIF patients and controls.
| Allele combinations |
Controls, n (%) (2n=420) |
RIF patients, n (%) (2n=238) |
OR (95% CI) |
P-value |
FDR P-value |
| DICER1 A>G/DROSHA T>C/RAN 3'UTR C>T/XPO5 A>C/DGCR8 G>A |
| A-T-C-A-G |
112 (26.8) |
83 (34.8) |
1.000 (reference) |
|
|
| A-T-C-C-G |
21 (5.0) |
5 (2.1) |
0.321 (0.116-0.888) |
0.022 |
0.095 |
| A-C-T-A-G |
26 (6.3) |
4 (1.6) |
0.208 (0.070-0.618) |
0.002 |
0.022 |
| G-T-C-A-G |
81 (19.2) |
37 (15.7) |
0.616 (0.381-0.998) |
0.048 |
0.168 |
| G-T-C-C-G |
0 (0.0) |
10 (4.3) |
28.290 (1.634-490.000) |
<0.001 |
0.021 |
| G-T-T-A-G |
49 (11.6) |
18 (7.7) |
0.496 (0.269-0.913) |
0.023 |
0.095 |
| G-C-T-A-G |
0 (0.0) |
6 (2.4) |
17.510 (0.972-315.500) |
0.007 |
0.048 |
| DICER1 A>G/DROSHA T>C/RAN 3'UTR C>T/XPO5 A>C |
| A-T-C-A |
104 (24.8) |
84 (35.3) |
1.000 (reference) |
|
|
| A-T-C-C |
24 (5.7) |
7 (3.1) |
0.361 (0.148-0.879) |
0.021 |
0.062 |
| A-C-T-A |
30 (7.1) |
4 (1.7) |
0.165 (0.056-0.487) |
<0.001 |
0.007 |
| G-T-C-A |
91 (21.6) |
40 (16.8) |
0.544 (0.340-0.871) |
0.011 |
0.047 |
| G-T-C-C |
0 (0.0) |
10 (4.1) |
25.970 (1.499-450.000) |
<0.001 |
0.007 |
| G-T-T-A |
46 (11.0) |
19 (8.0) |
0.511 (0.279-0.938) |
0.029 |
0.062 |
| G-C-T-A |
1 (0.2) |
7 (3.2) |
8.667 (1.045-71.880) |
0.026 |
0.062 |
| DICER1 A>G/DROSHA T>C/RAN 3'UTR C>T/DGCR8 G>A |
| A-T-C-G |
133 (31.6) |
88 (37.0) |
1.000 (reference) |
|
|
| A-C-T-G |
30 (7.1) |
6 (2.5) |
0.302 (0.121-0.756) |
0.008 |
0.072 |
| G-C-T-G |
0 (0.0) |
5 (2.2) |
16.590 (0.906-304.100) |
0.011 |
0.072 |
| DICER1 A>G/RAN 3'UTR C>T/XPO5 A>C/DGCR8 G>A |
| A-C-A-G |
167 (39.7) |
107 (45.2) |
1.000 (reference) |
|
|
| G-C-C-G |
5 (1.3) |
11 (4.8) |
3.434 (1.160-10.160) |
0.019 |
0.262 |
| DICER1 A>G/DROSHA T>C/RAN 3'UTR C>T |
| A-T-C |
130 (30.9) |
90 (38.0) |
1.000 (reference) |
|
|
| A-C-T |
34 (8.1) |
6 (2.7) |
0.255 (0.103-0.633) |
0.002 |
0.008 |
| G-C-T |
0 (0.0) |
7 (2.9) |
21.630 (1.219-383.800) |
0.002 |
0.008 |
| DICER1 A>G/DROSHA T>C/XPO5 A>C |
| A-T-A |
127 (30.2) |
99 (41.5) |
1.000 (reference) |
|
|
| A-T-C |
26 (6.2) |
7 (2.8) |
0.345 (0.144-0.829) |
0.014 |
0.024 |
| A-C-A |
87 (20.6) |
31 (13.0) |
0.457 (0.281-0.744) |
0.002 |
0.005 |
| G-T-A |
137 (32.5) |
59 (24.9) |
0.553 (0.369-0.827) |
0.004 |
0.009 |
| G-T-C |
0 (0.0) |
10 (4.3) |
26.910 (1.557-465.100) |
<0.001 |
0.005 |
| DICER1 A>G/DROSHA T>C/DGCR8 G>A |
| A-T-G |
152 (36.1) |
103 (43.2) |
1.000 (reference) |
|
|
| A-C-G |
85 (20.2) |
34 (14.5) |
0.590 (0.369-0.944) |
0.027 |
0.190 |
| DICER1 A>G/RAN 3'UTR C>T/XPO5 A>C |
| A-C-A |
167 (39.7) |
109 (45.7) |
1.000 (reference) |
|
|
| A-T-C |
8 (1.8) |
0 (0.1) |
0.090 (0.005-1.576) |
0.025 |
0.178 |
| DICER1 A>G/XPO5 A>C/DGCR8 G>A |
| A-A-G |
212 (50.4) |
127 (53.5) |
1.000 (reference) |
|
|
| G-C-G |
5 (1.2) |
11 (4.6) |
3.672 (1.247-10.810) |
0.012 |
0.085 |
| DICER1 A>G/DROSHA T>C |
| A-T |
154 (36.8) |
106 (44.4) |
1.000 (reference) |
|
|
| A-C |
90 (21.3) |
34 (14.5) |
0.549 (0.345-0.875) |
0.011 |
0.033 |
| DROSHA T>C/RAN 3’UTR C>T |
| T-C |
222 (52.9) |
141 (59.4) |
1.000 (reference) |
|
|
| C-T |
40 (9.6) |
13 (5.6) |
0.512 (0.264-0.991) |
0.044 |
0.131 |
| RAN 3'UTR C>T/XPO5 A>C |
| C-A |
286 (68.0) |
169 (71.0) |
1.000 (reference) |
|
|
| T-C |
9 (2.1) |
0 (0.0) |
0.089 (0.005-1.539) |
0.030 |
0.089 |
 |
Table IV
Combined genotype analysis for the miRNA machinery gene polymorphisms in RIF patients and controls.
|
Table IV
Combined genotype analysis for the miRNA machinery gene polymorphisms in RIF patients and controls.
| Genotype combinations |
Controls, n (%) (n=210) |
RIF patients, n (%) (n=119) |
AOR (95% CI)a |
P-value |
FDR P-value |
| DICER1 A>G/DROSHA T>C |
| AA/TT |
26 (12.4) |
24 (20.2) |
1.000 (reference) |
|
|
| AA/TC |
31 (14.8) |
13 (10.9) |
0.457 (0.194-1.081) |
0.075 |
0.299 |
| AG/TC |
56 (26.7) |
19 (16.0) |
0.358 (0.163-0.786) |
0.011 |
0.084 |
| DICER1 A>G/RAN 3'UTR C>T |
| AA/CC |
38 (18.1) |
28 (23.5) |
1.000 (reference) |
|
|
| AG/CT |
51 (24.3) |
19 (16.0) |
0.495 (0.240-1.021) |
0.057 |
0.454 |
| DROSHA T>C/RAN3'UTR C>T |
| TT/CC |
54 (25.7) |
45 (37.8) |
1.000 (reference) |
|
|
| TC/CT |
44 (21.0) |
12 (10.1) |
0.314 (0.147-0.673) |
0.003 |
0.023 |
| DROSHA T>C/XPO5 A>C |
| TT/AA |
79 (37.6) |
55 (46.2) |
1.000 (reference) |
|
|
| TC/AA |
83 (39.5) |
33 (27.7) |
0.581 (0.341-0.991) |
0.046 |
0.277 |
| DROSHA T>C/DGCR8 G>A |
| TT/GG |
90 (42.9) |
62 (52.1) |
1.000 (reference) |
|
|
| TC/GG |
93 (44.3) |
37 (31.1) |
0.577 (0.350-0.952) |
0.031 |
0.157 |
| RAN 3'UTR C>T/XPO5 A>C |
| CC/AA |
94 (44.8) |
61 (51.3) |
1.000 (reference) |
|
|
| CT/AC |
14 (6.7) |
4 (3.4) |
0.333 (0.096-1.162) |
0.085 |
0.437 |
| RAN 3'UTR C>T/DGCR8 G>A |
| CC/GG |
108 (51.4) |
72 (60.5) |
1.000 (reference) |
|
|
| CT/GG |
81 (38.6) |
33 (27.7) |
0.597 (0.360-0.992) |
0.046 |
0.232 |
Clinical variables and miRNA machinery gene polymorphisms
We next stratified the mean values for our clinical variables according to miRNA machinery gene polymorphism status using one-way ANOVA (Table SII). From this analysis, we found that the mean proportions of CD8+ T cells and the mean total cholesterol levels were significantly different in subjects with the RAN genotypes (P=0.041 and P=0.048, respectively). Specifically, the proportion of the CD8+ T cells was lower in those with the RAN C allele [mean ± standard deviation (SD), 28.50±7.58) than in subjects with the T allele (mean ± SD, 31.39±8.55, P=0.041; Fig. 1). Further, patients with the RAN C allele contained a significantly higher level of total cholesterol (mean ± SD, 196.68±45.01) than those with the T allele (mean ± SD, 176.33±23.89, P=0.048; Fig. 2). In subjects with the various DICER1 genotypes, the mean proportions of CD19+ B cells were significantly different; CD19+ B cells were higher in those with the GG vs. the AA genotype (P=0.012, Fig. S1]. Further, the mean activated partial thromboplastin times (aPTT) and blood urea nitrogen (BUN) levels were significantly different (P=0.009 and P=0.037) in those with distinct XPO5 genotypes and alleles (Figs. 3 and S2). That is, aPTT time was higher in those with the AC vs. the AA genotype and in those with the C vs. the A allele, and BUN levels were higher in those with the AA vs. the CC genotype (Fig. 3A and B). Moreover, the optimal sample size was determined based on statistical power analysis (Tables SIII and SIV).
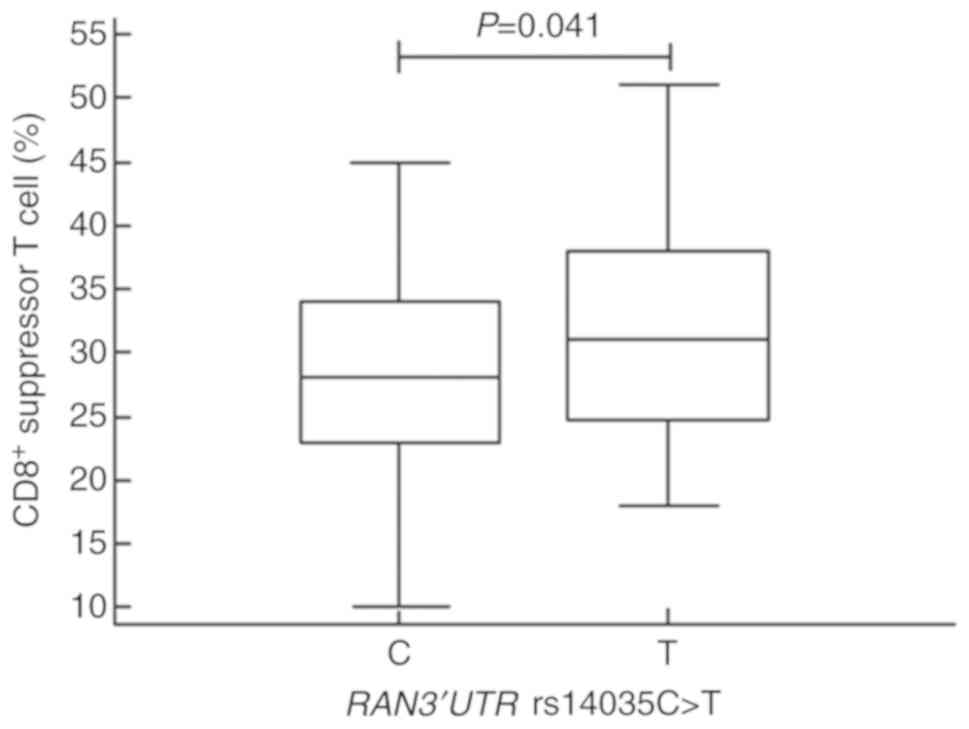 |
Figure 1
Proportions of CD8+ T cells in subjects with the RAN 3'UTR C and T alleles. Different proportions of CD8+ T cells (P=0.041) were detected in subjects with the RAN 3'UTR C (mean ± SD, 28.50±7.58) and T alleles (mean ± SD, 31.39±8.55). UTR, untranslated region; RAN, Ras-related nuclear protein.
|
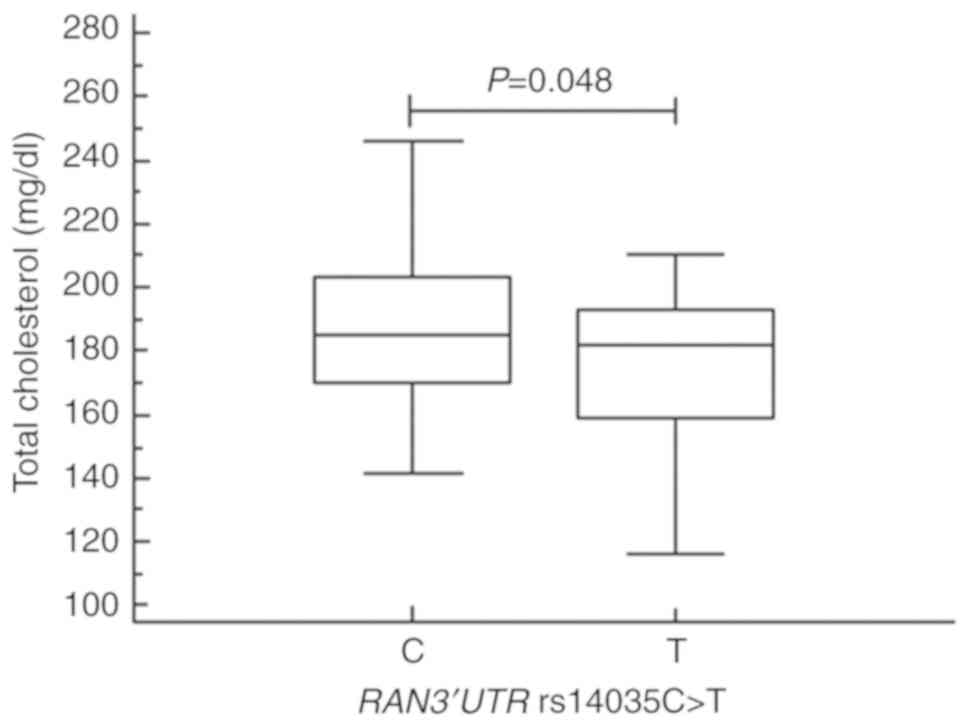 |
Figure 2
Total cholesterol levels in subjects with the RAN 3'UTR C and T alleles. Total cholesterol levels were significantly different (P=0.048) in subjects with the RAN 3'UTR C (mean ± SD, 196.68±45.01) and T alleles (mean ± SD, 176.33±23.89). UTR, untranslated region; RAN, Ras-related nuclear protein.
|
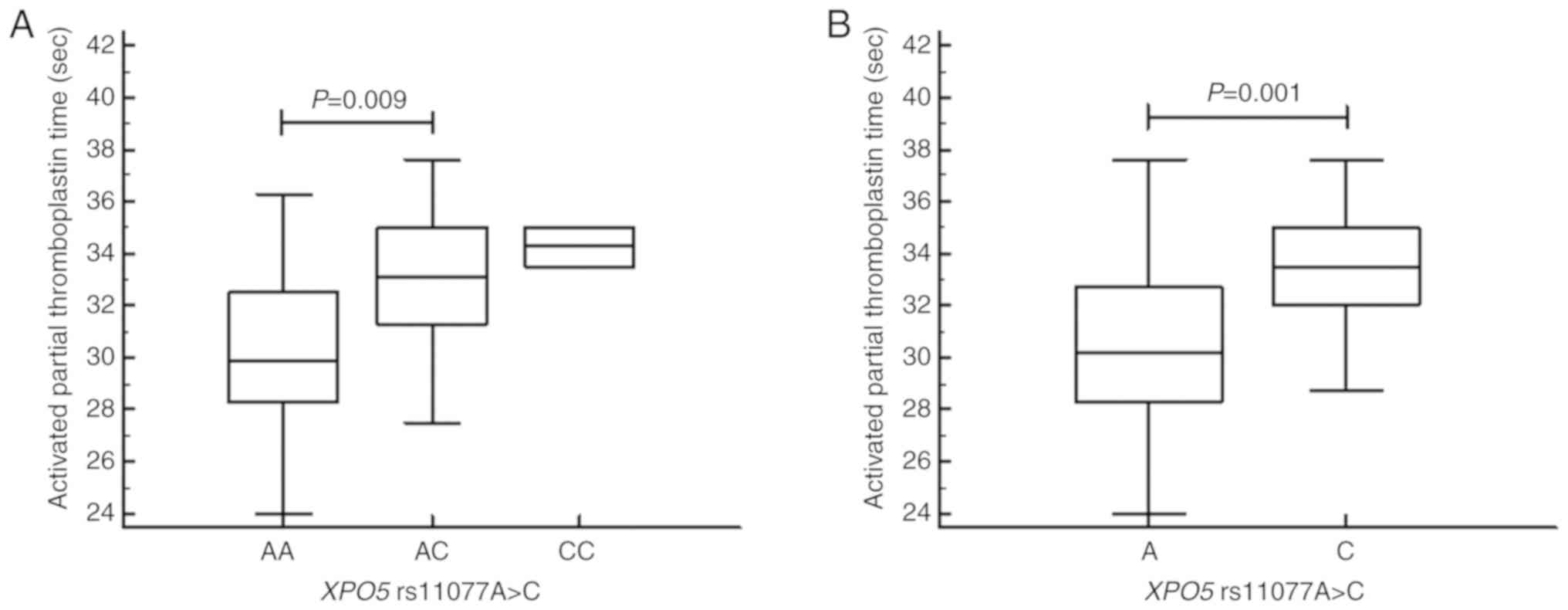 |
Figure 3
Activated partial thromboplastin time in subjects with XPO5 A and C alleles. (A) Activated partial thromboplastin time was significantly different (P=0.009) in subjects with the XPO5AA (mean ± SD, 30.25±3.07) and AC (mean ± SD, 33.26±3.84) genotypes. (B) Activated partial thromboplastin time was significantly different (P=0.001) in subjects with the XPO5A (mean ± SD, 30.56±3.26) and C alleles (mean ± SD, 33.53±3.30). XPO5, exportin 5.
|
Discussion
In this study, we evaluated SNPs in five genes, DROSHA, DICER1, RAN, XPO5, and DGCR8, and their possible association with RIF. At present, the role that genetic variation in miRNA machinery genes may play in the development of RIF is not fully understood, although modifications in these genes significantly affect cellular transformation in various diseases (22-25) and carcinogenesis in several cancers (26-29). To the best of our knowledge, this is the first report to evaluate the association between miRNA machinery gene polymorphisms and RIF. We found that the DROSHA rs10719T>C and RAN rs14035C>T 3'UTR polymorphisms are significantly associated with decreased prevalence of RIF in Korean women. Further, the DROSHA rs10719 TC and RAN rs14035 CT genotype frequencies were found to be lower in the RIF group than in the control group. This observation suggests that each of these confer protective effects against RIF occurrence. We also examined potential interactive effects of different genetic polymorphisms on RIF using genotype-based MDR analysis, which can detect gene-gene interactions, regardless of chromosomal location (20). An allele-allele combination-based analysis of the five polymorphisms indicated that there is a gene-gene interaction between the DROSHA rs10719 and the RAN rs14035 polymorphisms. In particular, five-allele combination analysis showed that the DICER1/DROSHA/RAN/XPO5/DGCR8 A-C-T-A-G combination is associated with decreased RIF risk, when the reference combination is A-T-C-A-G, which indicates that both the DROSHA and RAN polymorphisms exert synergistic effects on reducing RIF risk. Four, three, and two-allele combination analysis revealed similar results. In our genotype combination analysis, the DROSHA TC genotype showed a highly significant negative association with RIF prevalence when combined with the RAN CT genotype, which implies that defective DROSHA function, arising due to genetic polymorphisms, might be intensified by RAN polymorphisms.
DROSHA is a nuclear RNase III enzyme responsible for cleaving pri-miRNAs into pre-miRNAs, and thus is essential for miRNA biogenesis and the maturation. DROSHA also recognizes and directly cleaves many protein-coding messenger RNAs with secondary stem-loop structures (7). In addition to DROSHA, miRNA biogenesis requires the RNase III enzyme, DICER1. Numerous studies have identified roles for DROSHA and DICER1 in various diseases (22-25) and in a variety of cancers (27-31). For example, Kim et al (22) found that the DICER rs3742330 A>G and DROSHA rs10719 T>C polymorphisms are more common in ischemic stroke patients. Additionally, Saeki et al (23) reported an association between the development of Graves' disease and the DICER rs1057035 TT and DROSHA rs644236 CC genotypes. Zhang et al (24) further showed that the DROSHA rs10719 T>C polymorphism promotes the development of primary hypertension by disrupting the interaction between miR-27b and DROSHA, and both the DROSHA rs10719 TC and CC genotypes were found to be associated with increased susceptibility to preeclampsia (25).
In reproductive disease, however, the expression of DROSHA and its role remain unclear. Zhang et al (32) analyzed Drosha expression patterns in the mouse uterus during early pregnancy and concluded that this gene is involved in stromal decidualization and may participate in the process of embryo implantation in mice. Drosha expression in the endometrium was gradually up-regulated around the implantation window, reached a peak on the sixth day of pregnancy, and was reduced thereafter. However, few reports have evaluated the clinical significance of the DROSHA, DICER, and RAN SNPs in human reproduction. A recent study by Jung et al (33) revealed that the RAN rs14035 CC genotype and the combined DICER rs3742330/DROSHA rs10719 GG/TC+CC, DICER rs3742330/RAN rs14035 GG/CC, and DICER rs3742330/XPO5 rs11077 GG/AC+CC genotypes were significantly associated with an increased risk of recurrent pregnancy loss (RPL) in Korean women. Moreover, they showed that the RAN rs14035 CT genotype and combined DICER rs3742330/RAN rs14035 AA+AG/CT+TT, DROSHA rs10719/RAN rs14035 TC+CC/CT+TT, and RAN rs14035/XPO5 rs11077 CT+TT/AA genotypes were associated with a reduced risk of RPL. Fu et al (34) further reported that the DICER rs3742330/DROSHA rs10719 GG/TC+TT combinations were associated with an increased risk of RPL in the Chinese population. Additionally, down-regulation of DROSHA has been associated with higher histological grades in endometrial cancer (28) and decreased median survival of ovarian cancer patients (29), suggesting a role for DROSHA in reproductive cancers, as well.
Here, we found that the combined RAN rs14035 CT+TT genotype was associated with decreased RIF risk, and that RAN rs14035 CT heterozygotes have a significantly decreased relative risk of RIF, as compared to wild type homozygotes. RAN, a member of the Ras superfamily of GTPases, is essential for the transport of pre-miRNAs from the nucleus to the cytoplasm through the nuclear pore complex (35). RAN also plays key roles in assembly of the mitotic spindle and nuclear envelope during mitosis. When RAN-GTP is depleted, pre-miRNA export is greatly reduced, indicating that disruption of the RAN pathway perturbs the miRNA transport processes (36). Therefore, it is possible that RAN mutations mediate pathologic changes in miRNA transport and expression. RAN is also a well-known downstream modulator of osteopontin-induced malignant transformation of cells, which mediates cancer cell invasion and metastasis (37). Overexpression of RAN stimulates cancer cell differentiation, invasion, and ultimately metastasis in colorectal cancer (38). Consistent with these data, Yuen et al (39) demonstrated that knockdown of RAN suppressed adhesion, migration, and invasion of cancer cells in several breast and lung cancer cell lines.
To our knowledge, there are no published studies describing the effects of these SNPs specifically on RIF. Critically, we note that the pathophysiologies of RIF and RPL are distinct, although their respective etiologies have not been clearly elucidated. RPL is a disease that is distinct from infertility, whereas RIF occurs in a specific group of infertile patients undergoing IVF (40). In our study, both the DROSHA rs10719 TC and RAN rs14035 CT genotypes were associated with the prevalence of RIF. However, Jung et al (33) did not find an association between the DROSHA rs10719 polymorphism and RPL risk. Furthermore, four miRNA polymorphisms of the same patient and control groups in our previous study were not associated with the prevalence of RIF in genotype analysis but significantly associated with the RIF risk in allele combination analysis and interaction analysis (41). The results of combined genotype analyses for RPL and RIF were also different, which further implies distinct etiologies for these conditions. Interestingly, however, the RAN rs14035 CT genotype was related to a reduced risk of both RPL and RIF, suggesting that these conditions may also share some underlying mechanistic features, as well.
During early implantation, the endometrium undergoes dynamic cellular processes, including DNA synthesis and angiogenesis, which are required for cell proliferation and decidualization, in order to become receptive to the embryo (42). We hypothesize that dysregulation of miRNA machinery genes may locally affect angiogenesis and decidualization in the endometrium during implantation and pregnancy maintenance in early pregnancy. DROSHA may be particularly important for implantation, as this enzyme is essential for posttranscriptional miRNA processing. In both the individual and combined genotyping analysis of our study, the DROSHA rs10719 TC genotype was inversely associated with the RIF prevalence. In the combination analysis, DROSHA TC genotype combined with the RAN CT genotype showed the most significant negative association with RIF development. However, it remains unclear how the polymorphisms in these genes may influence one another during implantation.
A critical role for miRNAs in the differentiation of human T cells is becoming increasing evident (43). For example, it has been shown that miRNAs regulate thymic development of both CD4+ and CD8+ T cells and are further involved in the differentiation of mature T cells (44). DICER1 is also involved in the control of CD8+ T cell activation, proliferation, migration, and accumulation (44,45). Experiments in mice with a conditional deletion of Dicer1 in mature CD8+ T cells revealed a dramatically reduced CD8+ effector T cell response in mutant animals, suggesting an essential function for miRNAs in the regulation of T cells (45). In our study, we found that subjects with the RAN rs14035 T allele had a significantly higher proportion of CD8+ T cells than those with the RAN rs14035 C allele (P=0.041). Notably, this RAN polymorphism was associated with decreased risk of RIF in our population. Several observational studies have suggested that T cells and NK cells may be involved in RIF (46,47). For example, Yang et al (46) found that women with RIF have increased T cell activation in peripheral blood lymphocytes. Our results also suggest a role for T cells in RIF, although we found that a RAN polymorphism associated with a higher proportion of CD8+ T cells is associated with decreased risk of RIF. In contrast, Yin et al (47) detected no significant difference in the proportions of peripheral CD8+ T cells in women with RIF and in healthy controls. A potential explanation for these conflicting results is that RIF has various unknown etiologies, and the role of immunologic factors in implantation failure is both complex and poorly understood (48). Thus, although our data suggest that CD8+ T cells are involved in implantation failure, the precise role that these cells play in RIF remains unclear. Conversely, we found no significant association between NK cells and the other miRNA machinery gene polymorphisms. In addition, the question of whether other miRNA machinery genes may affect the differentiation of T cells and the risk of RIF has not yet been elucidated and will require further investigation.
There are several limitations to this study. First, the association between miRNA machinery gene polymorphisms and RIF risk found in our investigation is not a causal relationship. Although our results suggest potential roles of these SNPs in the development of RIF, it is difficult to definitively conclude that miRNA machinery genes affect RIF. Therefore, further research will be needed to explore this possibility. Second, the mechanism underlying the role of these miRNA machinery gene polymorphisms and the development of RIF has not been demonstrated. Functional studies of which polymorphisms affect miRNA biogenesis and promote the RIF pathogenesis were not conducted, as they were beyond the scope of this study. Third, we recruited women with normal pregnancy history for the control group, rather than women with successful pregnancy after IVF-ET. Thus, with the current comparison, we cannot rule out the possibility that these SNPs are associated with infertility rather than with RIF, specifically. Lastly, miRNA expression in local tissue, such as endometrial tissue, in individuals with different SNP genotypes, was not examined. Altered expression of miRNAs could change endometrial expression of specific genes critical for embryo implantation (49,50). Lastly, the sample size of our RIF group was not big enough to provide sufficient statistical power for demonstrating association between these miRNA SNPs and RIF. Thus, future studies with larger sample sizes will be required to confirm these results.
In conclusion, we have identified an association between the DROSHA rs10719 and RAN rs14035 3'UTR polymorphisms and the risk of RIF in Korean women. Our data demonstrate that the frequencies of the TC genotype of DROSHA rs10719 and that of the CT genotype of RAN rs14035 are lower in RIF patients than in controls, suggesting that these alleles may be associated with reduced risk of RIF. The underlying mechanisms by which these alleles affect RIF remain unknown and will be investigated in future studies. Regardless, our data suggest that these miRNA machinery gene polymorphisms could function as potential diagnostic markers to predict the RIF occurrence.
Supplementary Material
Figure S1. CD19+ B cell proportion is significantly different (P=0.012) among DICERAA (mean ± SD, 12.17±5.48), AG (mean ± SD, 9.69±3.00) and GG (mean ± SD, 13.62±5.01). DICER, dicer 1, ribonuclease III.
Figure S2. Blood urea nitrogen levels are significantly different (P=0.037) among XPO5AA (mean ± SD, 10.10±2.89), AC (mean ± SD, 10.62±2.12) and CC (mean ± SD, 5.10±0.71). XPO5, exportin 5.
Table SI. Genotype frequency of miRNA machinery gene polymorphisms between subgroups of RIF patients and controls.
Table SII. Clinical variables in recurrent implantation failure patients stratified by the miRNA machinery gene polymorphisms status.
Table SIII. Statistical power to detect various genetic associations in the present case-control study.
Table SIV. Statistical power to detect various genetic associations in the present case-control study.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Research Foundation of Korea Grants funded by the Korean Government (grant no. 2018R1D1A1B07044096), and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant. no. HI18C19990200).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
YL, EHA and NKK conceived and designed the experiments. YL, EHA, CSR, HJA, SHC, JHK and YRK performed the experiments. YL, EHA, CSR, JOK, YRK, WSL and NKK analyzed the data. YL, EHA and NKK wrote the paper. YL, EHA and NKK edited the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Institutional Review Board of CHA Bundang Medical Center reviewed and approved the current study on February 23, 2010 (reference no. PBC09-120), and written informed consent was obtained from all participants.
Patient consent for publication
All patients provided written informed consent and agreed to the publication of the present study.
Competing interestsin
The authors declare that they have no competing interests.
References
|
1
|
Polanski LT, Baumgarten MN, Quenby S, Brosens J, Campbell BK and Raine-Fenning NJ: What exactly do we mean by ‘recurrent implantation failure’? A systematic review and opinion. Reprod Biomed Online. 28:409–423. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Simon A and Laufer N: Repeated implantation failure: Clinical approach. Fertil Steril. 97:1039–1043. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Coughlan C, Ledger W, Wang Q, Liu F, Demirol A, Gurgan T, Cutting R, Ong K, Sallam H and Li TC: Recurrent implantation failure: Definition and management. Reprod Biomed Online. 28:14–38. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Santamaria X and Taylor H: MicroRNA and gynecological reproductive diseases. Fertil Steril. 101:1545–1551. 2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM: Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Olena AF and Patton JG: Genomic organization of microRNAs. J Cell Physiol. 222:540–545. 2010.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ and Littman DR: Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 24:1951–1960. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Harden ME and Munger K: Perturbation of DROSHA and DICER expression by human papilloma virus 16 oncoproteins. Virology. 507:192–198. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Roberts TC: The microRNA machinery. Adv Exp Med Biol. 887:15–30. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Al-Zahrani J, Al-Dosari N, Abudheim N, Alshidi TA, Colak D, Al-Habit O, Al-Odaib A, Sakati N, Meyer B, Ozand PT and Kaya N: Chromosome 12q24.31-q24.33 deletion causes multiple dysmorphic features and developmental delay: First mosaic patient and overview of the phenotype related to 12q24qter defects. Mol Cytogenet. 4(9)2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bueno MT, Martinez-Rios C, la Puente Gregorio A, Ahyad RA, Villani A, Druker H, van Engelen K, Gallinger B, Aronoff L, Grant R, et al: Pediatric imaging in DICER1 syndrome. Pediatr Radiol. 47:1292–1301. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Patrao AS, Dias F, Teixeira AL, Mauricio J and Medeiros R: XPO5 genetic polymorphisms in cancer risk and prognosis. Pharmacogenomics. 19:799–808. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Revel A, Achache H, Stevens J, Smith Y and Reich R: MicroRNAs are associated with human embryo implantation defects. Hum Reprod. 26:2830–2840. 2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Hu Y, Liu CM, Qi L, He TZ, Shi-Guo L, Hao CJ, Cui Y, Zhang N, Xia HF and Ma X: Two common SNPs in pri-miR-125a alter the mature miRNA expression and associate with recurrent pregnancy loss in a Han-Chinese population. RNA Biol. 8:861–872. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gardner DK, Surrey E, Minjarez D, Leitz A, Stevens J and Schoolcraft WB: Single blastocyst transfer: A prospective randomized trial. Fertil Steril. 81:551–555. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Choi DH, Kim EK, Kim KH, Lee KA, Kang DW, Kim HY, Bridges P and Ko C: Expression pattern of endothelin system components and localization of smooth muscle cells in the human pre-ovulatory follicle. Hum Reprod. 26:1171–1180. 2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rosen HR, Doherty DG, Madrigal-Estebas L, O'Farrelly C and Golden-Mason L: Pretransplantation CD56(+) innate lymphocyte populations associated with severity of hepatitis C virus recurrence. Liver Transpl. 14:31–40. 2008.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ryu CS, Sakong JH, Ahn EH, Kim JO, Ko D, Kim JH, Lee WS and Kim NK: Association study of the three functional polymorphisms (TAS2R46G>A, OR4C16G>A, and OR4X1A>T) with recurrent pregnancy loss. Genes Genomics. 41:61–70. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Benjamini Y, Drai D, Elmer G, Kafkafi N and Golani I: Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 125:279–284. 2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hahn LW, Ritchie MD and Moore JH: Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 19:376–382. 2003.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jeon YJ, Choi YS, Rah H, Kim SY, Choi DH, Cha SH, Shin JE, Shim SH, Lee WS and Kim NK: Association study of microRNA polymorphisms with risk of idiopathic recurrent spontaneous abortion in Korean women. Gene. 494:168–173. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kim JO, Bae J, Kim J, Oh SH, An HJ, Han IB, Oh D, Kim OJ and Kim NK: Association of microRNA biogenesis genes polymorphisms with ischemic stroke susceptibility and post-stroke mortality. J Stroke. 20:110–121. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Saeki M, Watanabe M, Inoue N, Tokiyoshi E, Takuse Y, Arakawa Y, Hidaka Y and Iwatani Y: DICER and DROSHA gene expression and polymorphisms in autoimmune thyroid diseases. Autoimmunity. 49:514–522. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhang Y, Cao AL and Dong C: Rs10719 Polymorphism located within DROSHA 3'-untranslated region is responsible for development of primary hypertension by disrupting binding with microRNA-27b. Med Sci Monit. 23:911–918. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Rezaei M, Eskandari F, Mohammadpour-Gharehbagh A, Teimoori B, Yaghmaei M, Mokhtari M and Salimi S: The Drosha rs10719 T>C polymorphism is associated with preeclampsia susceptibility. Clin Exp Hypertens. 40:440–445. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Huang JT, Wang J, Srivastava V, Sen S and Liu SM: MicroRNA machinery genes as novel biomarkers for cancer. Front Oncol. 4(113)2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Lin S and Gregory RI: MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 15:321–333. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Torres A, Torres K, Paszkowski T, Jodłowska-Jędrych B, Radomański T, Książek A and Maciejewski R: Major regulators of microRNAs biogenesis Dicer and Drosha are down-regulated in endometrial cancer. Tumour Biol. 32:769–776. 2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al: Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 359:2641–2650. 2008.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Cho SH, Ko JJ, Kim JO, Jeon YJ, Yoo JK, Oh J, Oh D, Kim JW and Kim NK: 3'-UTR polymorphisms in the MiRNA machinery genes DROSHA, DICER1, RAN, and XPO5 are associated with colorectal cancer risk in a Korean population. PLoS One. 10(e0131125)2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, et al: Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 212:368–377. 2007.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Zhang C, Long X, Ding Y, Chen X, He J, Liu S, Geng Y, Wang Y and Liu X: Expression of DROSHA in the uterus of mice in early pregnancy and its potential significance during embryo implantation. Reprod Sci. 23:154–162. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jung YW, Jeon YJ, Rah H, Kim JH, Shin JE, Choi DH, Cha SH and Kim NK: Genetic variants in microRNA machinery genes are associated [corrected] with idiopathic recurrent pregnancy loss risk. PLoS One. 9(e95803)2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Fu M, Xu WM, Qin TZ and Xu KH: The association between the polymorphisms of miRNA biogenesis related genes (DICER,DROSHA and RAN) and unexplained recurrent spontaneous abortion in Chinese women. Sichuan Da Xue Xue Bao Yi Xue Ban. 48:880–885. 2017.(In Chinese). PubMed/NCBI
|
|
35
|
Khuperkar D, Helen M, Magre I and Joseph J: Inter-cellular transport of ran GTPase. PLoS One. 10(e0125506)2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Aoki K and Niki H: Release of condensin from mitotic chromosomes requires the Ran-GTP gradient in the reorganized nucleus. Biol Open. 6:1614–1628. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kurisetty VV, Johnston PG, Johnston N, Erwin P, Crowe P, Fernig DG, Campbell FC, Anderson IP, Rudland PS and El-Tanani MK: RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene. 27:7139–7149. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Fan H, Lu Y, Qin H, Zhou Y, Gu Y, Zhou J, Wang X and Fan D: High Ran level is correlated with poor prognosis in patients with colorectal cancer. Int J Clin Oncol. 18:856–863. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yuen HF, Chan KK, Platt-Higgins A, Dakir el-H, Matchett KB, Haggag YA, Jithesh PV, Habib T, Faheem A, Dean FA, et al: Ran GTPase promotes cancer progression via Met receptor-rmediated downstream signaling. Oncotarget. 7:75854–75864. 2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Practice Committee of American Society for Reproductive Medicine: Definitions of infertility and recurrent pregnancy loss: A committee opinion. Fertil Steril 99: 63, 2013.
|
|
41
|
Rashid NA, Lalitkumar S, Lalitkumar PG and Gemzell-Danielsson K: Endometrial receptivity and human embryo implantation. Am J Reprod Immunol. 66 (Suppl 1):S23–S30. 2011.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Sheppard HM, Verdon D, Brooks AE, Feisst V, Ho YY, Lorenz N, Fan V, Birch NP, Didsbury A and Dunbar PR: MicroRNA regulation in human CD8+ T cell subsets-cytokine exposure alone drives miR-146a expression. J Transl Med. 12(292)2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Baumann FM, Yuzefpolskiy Y, Sarkar S and Kalia V: Dicer regulates the balance of short-lived effector and long-lived memory CD8 T cell lineages. PLoS One. 11(e0162674)2016.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Zhang N and Bevan MJ: Dicer controls CD8+ T-cell activation, migration, and survival. Proc Natl Acad Sci USA. 107:21629–21634. 2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chernyshov VP, Sudoma IO, Dons'koi BV, Kostyuchyk AA and Masliy YV: Elevated NK cell cytotoxicity, CD158a expression in NK cells and activated T lymphocytes in peripheral blood of women with IVF failures. Am J Reprod Immunol. 64:58–67. 2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yang KM, Ntrivalas E, Cho HJ, Kim NY, Beaman K, Gilman-Sachs A and Kwak-Kim J: Women with multiple implantation failures and recurrent pregnancy losses have increased peripheral blood T cell activation. Am J Reprod Immunol. 63:370–378. 2010.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yin B, Zeng Y, Wu T, Yu S, Xu J, Liu S, Diao L, Zhao Z, Liang D and Li Y: Functional properties of peripheral CD8+ T cells in patients with repeated implantation failure. Am J Reprod Immunol. 78:2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Alecsandru D and Garcia-Velasco JA: Why natural killer cells are not enough: A further understanding of killer immunoglobulin-like receptor and human leukocyte antigen. Fertil Steril. 107:1273–1278. 2017.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Pan Q and Chegini N: MicroRNA signature and regulatory functions in the endometrium during normal and disease states. Semin Reprod Med. 26:479–493. 2008.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Galliano D and Pellicer A: MicroRNA and implantation. Fertil Steril. 101:1531–1544. 2014.PubMed/NCBI View Article : Google Scholar
|

















