Introduction
Hepatocellular carcinoma (HCC) is the most common
primary liver cancer worldwide and represents the third leading
cause of cancer-related mortality (1). It is the fifth most common cancer in
men and the seventh most common in women, with an increasing
incidence, particularly in Western countries (1,2).
Hepatitis B virus infection, liver cirrhosis and the occurrence of
malignant liver neoplasia are considered the three gradations of
HCC. This process involves a series of gene mutations in
hepatocytes, and the accumulation of these mutations, which can
lead to tumor cell proliferation and invasion/metastasis, is
responsible for the formation of malignancy and metastases to the
surrounding liver parenchyma/distant organs (3). Clinically, ablation, liver
transplantation and surgical resection are used for the treatment
of HCC and enable a high rate of complete excision of this disease
(2). However, as HCC frequently
relapses or is diagnosed at an advanced stage, these treatments may
be ineffective. Furthermore, no adjuvant therapy has been
demonstrated to improve recurrence-free survival rate following
curative treatments (4).
In recent years, long non-coding RNAs (lncRNAs) have
gained increasing attention in several fields in molecular and
cellular medicine. As a product of transcription with little to no
protein-coding functions, lncRNAs are involved in multiple
biological functions, including intercellular signaling, protein
localization or functions, post-transcriptional mRNA processing,
control of gene transcription and regulation of gene expression
(5-7). In
addition to the investigation and recognition of lncRNAs, there is
increasing evidence that they have significant roles as tumor
oncogenes or/and suppressors (8-10).
Furthermore, several lncRNAs are important in the progression and
development of liver cancer (11,12).
The lncRNA gastric cancer associated transcript 3
(GACAT3), also known as AC130710), is located on human chromosome
2p24.3. It is upregulated in gastric cancer tissues and is
significantly associated with distal metastasis, tumor-node
metastasis stages and tumor size (13). In addition, GACAT3 promotes gastric
cancer progression by negatively regulating the expression of
microRNA (miR)-497, and the knockdown of GACAT3 significantly
inhibits the invasion, migration, colony formation and
proliferation of gastric cancer cells in vitro (14). However, the biological function of
GACAT3 in HCC, including its role in proliferation, apoptosis and
migration, remains to be elucidated. Therefore, the present study
investigated the correlation between the expression levels of
GACAT3 in HCC tissues and cell lines. Furthermore, the effects of
decreasing the expression levels of GACAT3 on the proliferation,
apoptosis and migration of liver cancer cells in vitro were
examined.
Materials and methods
Cell culture and HCC samples
The QSG-7701 normal human liver cell line and the
HepG2, HCCLM3, SK-Hep-1 and SMMC-7721 liver cancer cell lines were
purchased from the American Type Culture Collection (Rockville, MD,
USA). The Huh7 HCC cell line was purchased from the Chinese Academy
of Sciences Cell Bank (Shanghai, China). All cells were grown in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
and 100 U/ml penicillin/streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37˚C in a 5% CO2 incubator. HCC and
adjacent normal tissues were obtained with informed consent from
120 patients at the First Affiliated Hospital of Nanjing Medical
University (Nanjing, China) between January 2016 and December 2017.
The adjacent noncancerous liver tissues were obtained from >5 cm
away from the tumor sites, and verified simultaneously by two
pathologists separately. All specimens were placed in liquid
nitrogen immediately following resection. Patient charts were
obtained regarding age, sex, tumor size, α-fetoprotein levels,
hepatitis B surface antigen positivity, tumor differentiation and
Tumor, Node, Metastasis stage (American Joint Committee on Cancer;
Table I) (15). The study protocol was approved by the
Ethics Committee of the Yinzhou People's Hospital Affiliated to
Ningbo University School of Medicine (Ningbo, China).
 | Table IClinicopathological characteristics of
patients. |
Table I
Clinicopathological characteristics of
patients.
| Variable | Number |
|---|
| Sex | |
|
Male | 86 |
|
Female | 34 |
| Age (years) | |
|
≤50 | 48 |
|
>50 | 72 |
| Tumor size (cm) | |
|
≤3 | 36 |
|
>3 | 84 |
| α-fetoprotein
(ng/ml) | |
|
≤400 | 92 |
|
>400 | 28 |
| Hepatitis B surface
antigen | |
|
Positive | 70 |
|
Negative | 50 |
| Liver cirrhosis | |
|
Yes | 91 |
|
No | 29 |
| Barcelona clinic
Liver Cancer | |
|
0-B | 104 |
|
C | 16 |
| Tumor, node,
metastasis stage | |
|
I-II | 113 |
|
III-IV | 7 |
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA from the HCC tissues and the liver cancer
cell lines was extracted using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. The total RNA was eluted with RNase-free water and
the concentration was determined by ultraviolet spectrophotometry.
RNA was reverse transcribed to cDNA and RT-qPCR was performed using
Synergy Brands (SYBR)-green PCR Master mix (Takara Biotechnology
Co., Ltd., Dalian, China) in a Fast Real-time PCR 7500 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling steps were as follows: 5 min at 95˚C, followed by 40
cycles at 95˚C for 15 sec and 60˚C for 30 sec. The specific primers
for GACAT3 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
used were from Invitrogen; Thermo Fisher Scientific, Inc., and as
described previously (16). The
primer sequence details were as follows: GACAT3 (forward,
5'-CTTCCGGAGCAGGTCTGAGT-3' and reverse,
5'-CTTTCCCTGCAGAGACCAGT-3'), GAPDH (forward,
5'-GTCAACGGATTTGGTCTGTATT-3' and reverse,
5'-AGTCTTCTGGGTGGCAGTGAT-3'). Relative quantification was analyzed
via the comparative threshold cycle (2-∆∆Cq) method
(17) and the results were
internally normalized to the expression of GAPDH.
Gene silencing of GACAT3 in liver
cancer cell lines
To downregulate the expression of GACAT3, a small
interfering RNA (siRNA) sequence targeting GACAT3
(5'-AUCAGGGCUUGUGGAAUGGGAAG-3'; GeneChem Co., Ltd., Shanghai,
China) was inserted into the lentivirus vector pLL3.7
(Sigma-Aldrich; Merck KGaA) containing green fluorescent protein,
and designated as si-GACAT3. A hairpin siRNA with no human sequence
homology was used as the negative control (si-NC;
5'-UCACAACCUCCUAGAAAGAGUAGA-3'; Shanghai GeneChem Co., Ltd.). All
constructs were verified by sequencing. Recombinant lentivirus was
established from human 293T cells (American Type Culture
Collection) by co-transfection of pdelta-8.91 and pVSVG, together
with si-GACAT3 or si-NC using polybrene (1:1,000; Sigma-Aldrich;
Merck KGaA) and FBS-free MEM. After 12 h, the FBS-free MEM was
changed with DMEM containing 10% FBS. After 48 h the cells were
observed under fluorescence microscopy (magnification x40), to
identify GFP, then DMEM containing 10% FBS and 0.2% puromycin was
added to select the transfected cells for 7 days. The cells were
cultured in DMEM containing 10% FBS. The knockdown efficiency of
si-GACAT3 was determined by RT-qPCR.
Determination of cell viability with a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
The liver cancer cells were seeded into 96-well
plates and treatments were performed following adherence. Following
incubation periods of 24, 48, 72, or 96 h, 20 µl of a 5 mg/ml MTT
solution was added to each well and the plate was incubated at 37˚C
for 4 h. Thereafter, the medium was aspirated and the wells washed
with phosphate-buffered saline. Following drying for ~2 h, 200 µl
of dimethyl sulfoxide was added to each well. The formazan crystals
were dissolved by placing the microtiter plate on a shaker, and the
absorbance was determined spectrophotometrically at 570 nm using a
reference wavelength of 630 nm on an ELX800 UV universal microplate
reader (Bio-Tek Instruments, Inc., Winooski, VT, USA).
Determination of apoptosis by flow
cytometric analysis
The HepG2 and HCCLM3 cells transfected with
si-GACAT3 or si-NC for 48 h were subjected to analysis of apoptosis
using Annexin V/propidium iodide (PI), as described previously
(18). Liver cell apoptosis was
examined using an Annexin V-FITC apoptosis detection kit
(Sigma-Aldrich; Merck KGaA) according to the manufacturer's
recommendations. Annexin V-positive cells were considered early
apoptotic cells, and PI-positive cells were considered late
apoptotic/necrotic cells. All liver cell nuclei were counterstained
with Hoechst 33342 dye.
Western blot analyses
Following transfection of the HepG2 and HCCLM3 cells
with si-GACAT3 or si-NC for 48 h, total protein was extracted with
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Jiangsu, China), followed by quantification using a
bicinchoninic acid protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Equal quantities of protein (20 g) from each
sample were separated by sodium dodecyl sulfate polyacrylamide gel
electrophoresis on 8-12% gels and transferred onto a nitrocellulose
membrane (Amersham Biosciences, Piscataway, NJ, USA). Membranes
were blocked using 5% skimmed milk for 2 h at room temperature,
washed with TBST, and incubated primary antibodies against Bax
(cat. no. sc-20067; diluted 1:1,000), Bcl-2 (cat. no. sc-509;
diluted 1:500), E-cadherin (cat. no. sc-71008; diluted 1:1,000),
N-cadherin (cat. no. sc-7939; diluted 1:1,000; all Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), TGF-β1 (cat. no. ab155264;
diluted 1:500; Abcam), β-catenin (cat. no. 9582; diluted 1:500),
β-actin (cat. no. 4970; diluted 1:2,000) and GAPDH (cat. no. 8884;
diluted 1:1,000; all Cell Signaling Technology, Inc., Danvers, MA,
USA) overnight at 4˚C. The membranes were washed with TBST three
times, incubated with horseradish peroxidase-conjugated goat
anti-rabbit (cat. no. sc-2004; diluted 1:1,000; Santa Cruz
Biotechnology, Inc.) and goat anti-mouse (cat. no. ab97040;
1:5,000; Abcam) secondary antibodies for 2 h at room temperature
and again washed with TBST. The protein bands were detected using a
chemiluminescent detection system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). GAPDH or β-actin was used as the loading
control.
Transwell migration assay
The migration ability of liver cancer cells was
assessed using a Transwell assay with 8 µm porous membrane inserts
(Corning, Inc., Corning, NY, USA). The siRNA-transfected liver
cancer cells were treated with a trypsin/EDTA solution, and then
washed with PBS and suspended in serum-free medium. The cells
(5x104) in 200 µl serum-free medium were inoculated into
the upper chamber and 500 µl of medium containing 20% fetal bovine
serum were added into the lower chamber. Following incubation at
37˚C for 48 h, cells that migrated to the bottom surface of the
Transwell chamber were fixed with 100% methanol at room temperature
for 10 min. The cells were then stained with 0.5% crystal violet
for 10 min and washed three times with phosphate-buffered saline.
The cells on the top surface of the Transwell chamber were removed
with a cotton swab. The migration capacity was quantified by
counting the number of migratory cells in five fields of each
Transwell using an inverted microscope (x200 magnification; Olympus
Corporation, Tokyo, Japan). Each experiment was repeated three
times.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism5 software (GraphPad Software, Inc., La Jolla, CA, USA). The
results are presented as the mean ± standard error of the mean.
Statistical analyses were performed with Student's t-test or
one-way ANOVA followed by Tukey's post hoc test. For all
experiments, P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression of GACAT3 is increased
aberrantly in HCC tissues
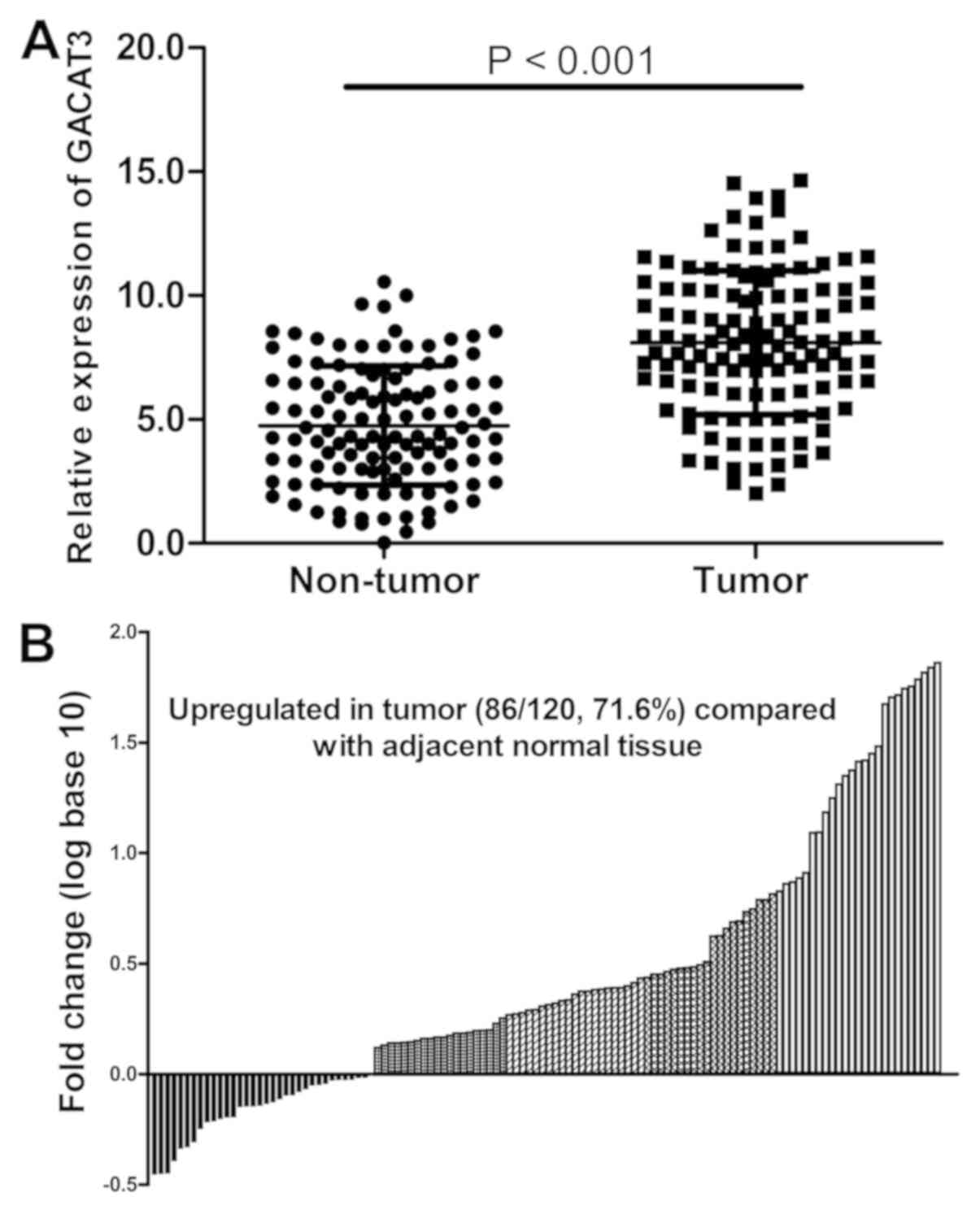
The mRNA expression level of GACAT3 was measured in
tissues from 120 patients with HCC. As indicated in Fig. 1A and B, the mRNA expression of GACAT3 was
significantly upregulated in HCC tissues compared with that in
adjacent normal tissues (P<0.001). These results indicate that
GACAT3 may serve a vital role in the development of HCC.
mRNA expression of GACAT3 in liver
cancer cell lines and the effect of GACAT3 knockdown on the
viability in HepG2 and HCCLM3 cells
To assess the biological function of GACAT3 in HCC,
the mRNA expression level of GACAT3 was examined in the QSG-7701
normal human liver cell line and the HepG2, HCCLM3, SK-Hep-1,
SMMC-7721 and Huh7 liver cancer cell lines. The results from
RT-qPCR analysis showed that the mRNA levels of GACAT3 were
significantly upregulated in the liver cancer cell lines compared
with that in the QSG-7701 cells (Fig.
2A).
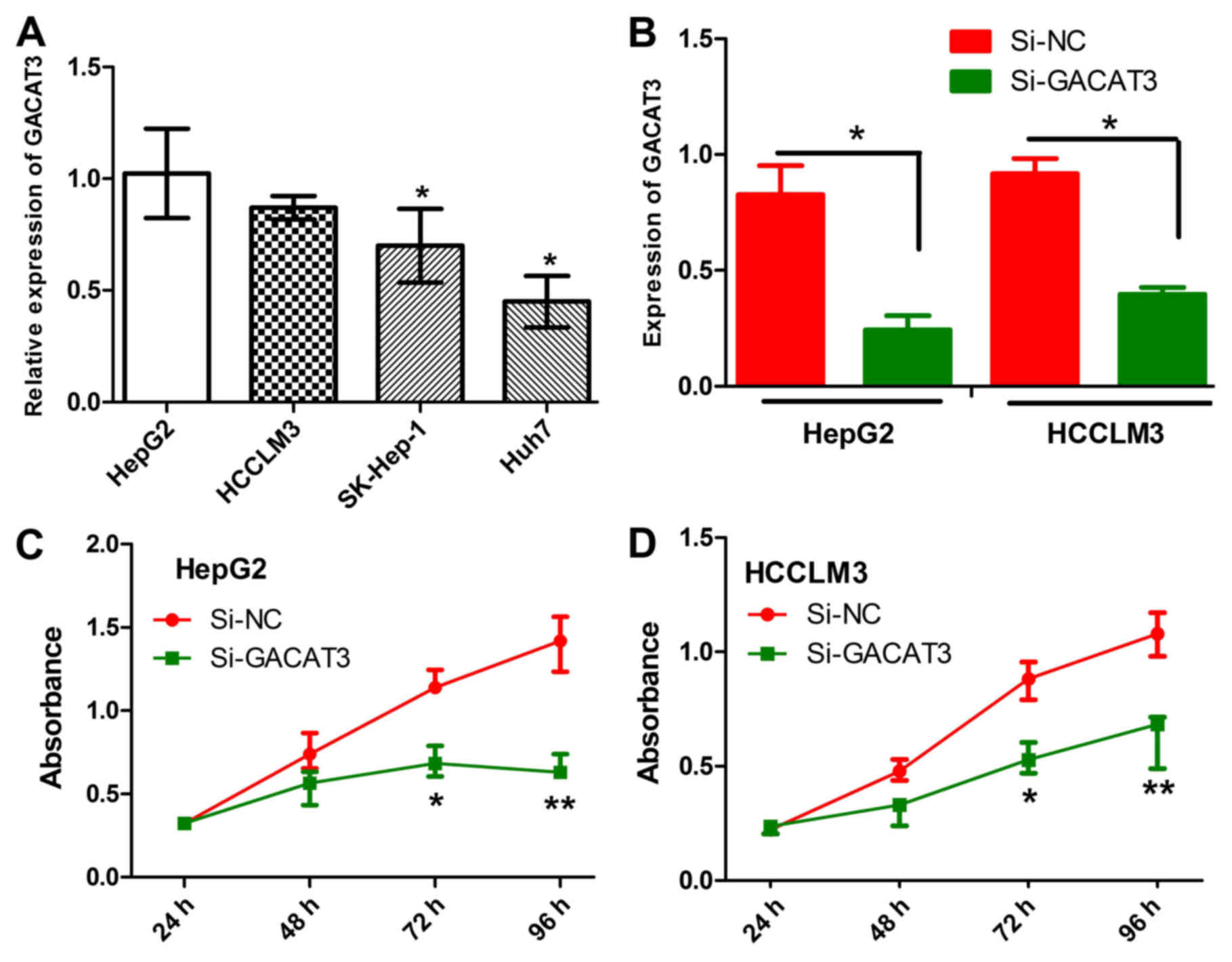 | Figure 2Effects of GACAT3 knockdown on liver
cancer cell lines. (A) mRNA expression levels of GACAT3 in HepG2,
HCCLM3, SK-Hep-1, SMMC-7721, Huh7 and QSG-7701 cells were detected
by the RT-qPCR analysis. (B) Relative mRNA expression of GACAT3 in
HepG2 and HCCLM3 cells transfected with si-GACAT3 mRNA or si-NC was
detected by RT-qPCR analysis. Viability of (C) HepG2 and (D) HCCLM3
cells transfected with si-GACAT3 or si-NC was determined using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay.
All data are expressed as the mean ± SEM (*P<0.05,
**P<0.01). GACAT3, gastric cancer associated
transcript 3; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; si, small interfering RNA; NC, negative
control. |
HepG2 and HCCLM3 cells, with the highest mRNA levels
of GACAT3, were selected for functional investigation. GACAT3 was
silenced in these cells by transfecting them with the si-GACAT3
lentivirus. An MTT assay was used to determine their viability. The
efficiencies of GACAT3 knockdown were verified by the RT-qPCR
assay. The results showed that the mRNA expression of GACAT3 was
significantly decreased in the HepG2 and HCCLM3 cells transfected
with the si-GACAT3 lentivirus, compared with that in the control
cells (Fig. 2B). The MTT assay
indicated that the knockdown of GACAT3 significantly decreased the
viability of HepG2 and HCCLM3 cells (Fig. 2C and D, P<0.05 and P<0.01). These results
demonstrated that the expression of GACAT3 was high in liver cancer
cell lines and its knockdown significantly inhibited liver cancer
cell viability.
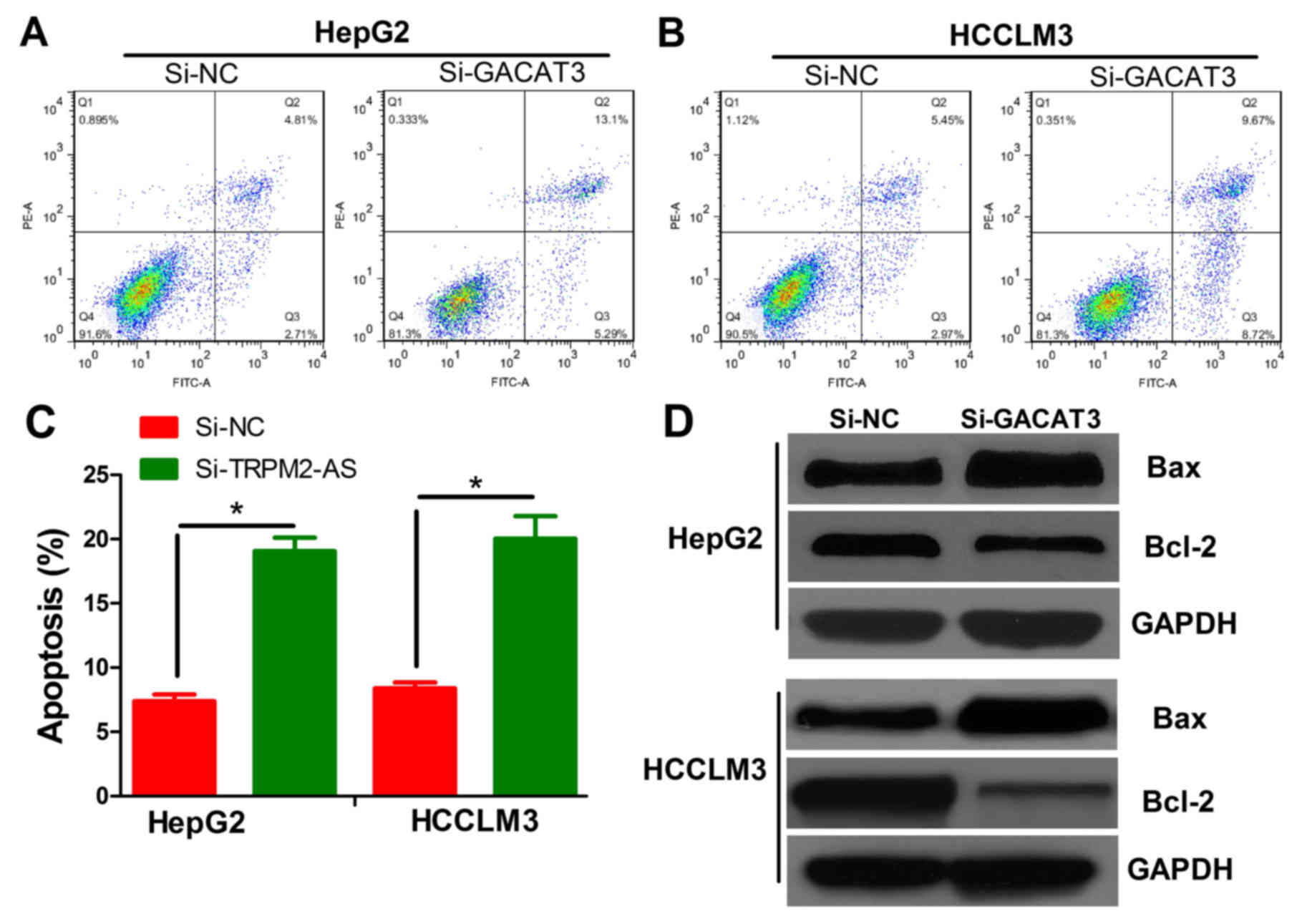
Effects of GACAT3 knockdown on
apoptosis and apoptosis-related proteins of HepG2 and HCCLM3
cells
To examine whether GACAT3 knockdown is involved in
the apoptosis of liver cancer cell lines, the HepG2 and HCCLM3
cells were transfected with si-GACAT3 or si-NC lentivirus. These
cells were then stained with Annexin-V/PI, followed by the
detection of apoptosis using flow cytometry. The results showed
that the knockdown of GACAT3 in both cell lines significantly
promoted apoptosis (Fig. 3A-C
P<0.05). Two apoptosis-related proteins (Bax and Bcl-2) were
measured by western blot analysis to confirm the effects of GACAT3
on apoptosis. Bax was upregulated and Bcl-2 was downregulated in
the HepG2 and HCCLM3 cells transfected with si-GACAT3 lentivirus,
compared with levels in the siNC-transfected cells (Fig. 3D).
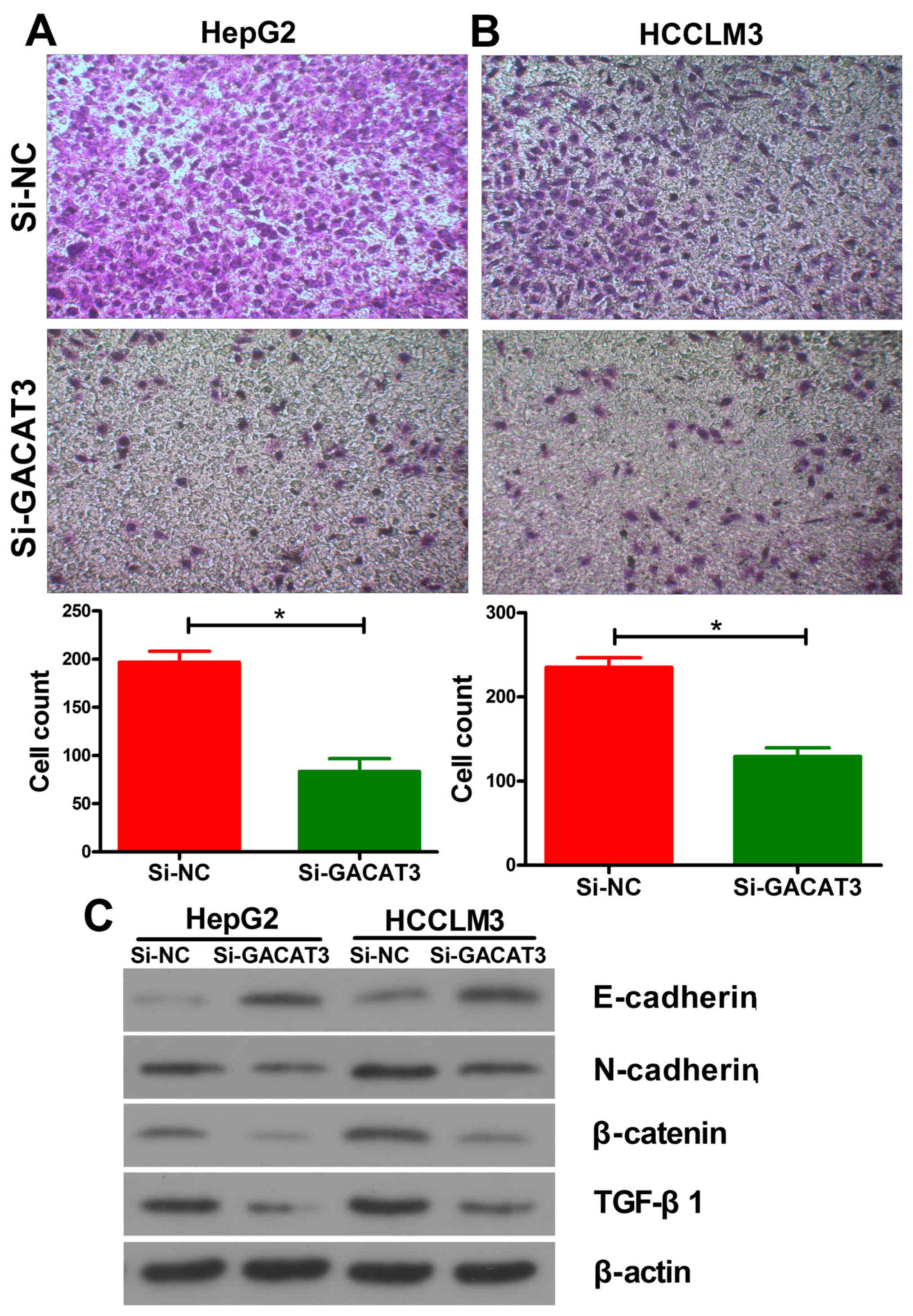
Effects of GACAT3 on cell migration in
liver cancer cell lines
To evaluate the effect of GACAT3 on the development
of HCC, the present study observed its effect on the migration in
HepG2 and HCCLM3 cells using a Transwell assay. The results showed
that GACAT3 knockdown significantly suppressed the migration
ability of the HepG2 (Fig. 4A) and
HCCLM3 (Fig. 4B) cells. These
results suggested that the inhibition of GACAT3 decreased the
migration ability of HCC cells. In order to analyze the mechanism
of inhibition, the expression of EMT-related proteins were
detected. As is shown in Fig. 4C,
the expression levels of N-cadherin, β-catenin and TGF-β1 were
decreased and that of E-cadherin was increased following the
inhibition of GACAT3 in both liver cancer cell lines.
Discussion
lncRNAs perform crucial and complicated roles in
cancer progression and carcinogenesis at the chromatin
organizational, translational and post-translational levels
(19). Accumulated evidence has
shown that numerous lncRNAs are closely associated with the
progression and development of HCC. For example, lncRNA UCA1
downregulated miR-216 and activated the fibroblast growth
factor-1/extracellular signal-regulated kinase pathway, inhibiting
the proliferation of HCC cells in vitro (20). lncRNA linc-USP16 regulated the
phosphatase of tensin homolog and negatively regulated the
invasion/migration of HCC cells (21). In vitro, the upregulation of
the lncRNA HULC inhibited the expression of p18 and promoted HCC
cell proliferation (22). However,
numerous novel lncRNAs remain unknown.
Increasing evidence indicates that lncRNA GACAT3
serves complex roles in various aspects of cancer progression and
carcinogenesis. Zhong et al (23) found that GACAT3 predicted poor
prognosis and promoted cell proliferation in breast cancer through
the regulation of miR-497/CCND2. Zhou et al (24) reported that GACAT3 promoted
colorectal cancer cell proliferation, invasion and migration
through miR-149. Yang et al (25) demonstrated that a high expression of
GACAT3 inhibited the invasion and metastasis of non-small cell lung
cancer, enhancing the effect of radiotherapy. In addition, GACAT3
promotes gastric cancer progression and cell proliferation by
negatively regulating the expression of miR-497 and the
interleukin-6/STAT3 signaling pathway, respectively (14,13).
However, there has been no investigation on the effects of GACAT3
in HCC.
In the present study, it was demonstrated that the
expression of GACAT3 was elevated in HCC tissues and cell lines.
The results indicated that GACAT3 may be an important factor that
can affect the pathological characteristics of HCC. Furthermore,
experiments were performed at the tissue and cell level to confirm
the biological function and mechanism of proliferation and
metastasis of GACAT3 in HCC in vitro. The results revealed
that the knockdown of GACAT3 in liver cancer cells significantly
inhibited cell proliferation and migration by promoting apoptosis
and influencing EMT. These results indicate that GACAT3 may be an
oncogene in liver cancer. Therefore, GACAT3 may be a therapeutic
target for the treatment of liver cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MC and DL were involved in the collection of HCC
tissues. CJ and LC were mainly involved in cell culture and western
blot analyses. LD was involved in designing the experiment,
processing the main content of this experiment, and drafting the
manuscript and revising it critically for important intellectual
content. KZ was involved in the conception and design of the
present study. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Human HCC tissues were obtained with written and
signed informed consent under a general waiver for the appropriate
secondary use of human material and were obtained from Yinzhou
People's Hospital. The study protocol was approved by the Ethics
Committee of Yinzhou People's Hospital Affiliated to Ningbo
University School of Medicine (Ningbo, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
European Association For The Study Of The
Liver and European Organisation For Research And Treatment Of
Cancer: EASL-EORTC clinical practice guidelines: Management of
hepatocellular carcinoma. J Hepatol 56: 908-943, 2012.
|
|
3
|
Pollutri D, Gramantieri L, Bolondi L and
Fornari F: TP53/MicroRNA interplay in hepatocellular carcinoma. Int
J Mol Sci. 17(12)2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bruix J, Takayama T, Mazzaferro V, Chau
GY, Yang J, Kudo M, Cai J, Poon RT, Han KH, Tak WY, et al: Adjuvant
sorafenib for hepatocellular carcinoma after resection or ablation
(STORM): A phase 3, randomised, double-blind, placebo-controlled
trial. Lancet Oncol. 16:1344–1354. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Akhade VS, Pal D and Kanduri C: Long
noncoding RNA: Genome organization and mechanism of action. Adv Exp
Med Biol. 1008:47–74. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Carpenter S and Fitzgerald KA: Cytokines
and long noncoding RNAs. Cold Spring Harb Perspect Biol.
10(a028589)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Kwok ZH and Tay Y: Long noncoding RNAs:
Lincs between human health and disease. Biochem Soc Trans.
45:805–812. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wu X, Tudoran OM, Calin GA and Ivan M: The
many faces of long noncoding RNAs in cancer. Antioxid Redox Signal.
20:922–935. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Falcon T, Freitas M, Mello AC, Coutinho L,
Alvares-da-Silva MR and Matte U: Analysis of the cancer genome
atlas data reveals novel putative ncRNAs targets in hepatocellular
carcinoma. Biomed Res Int. 26(2864120)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Xie Z, Zhou F, Yang Y, Li L, Lei Y, Lin X,
Li H, Pan X, Chen J, Wang G, et al: Lnc-PCDH9-13:1 is a
hypersensitive and specific biomarker for early hepatocellular
carcinoma. EBioMedicine. 33:57–67. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Shen W, Yuan Y, Zhao M, Li J, Xu J, Lou G,
Zheng J, Bu S, Guo J and Xi Y: Novel long non-coding RNA GACAT3
promotes gastric cancer cell proliferation through the IL-6/STAT3
signaling pathway. Tumour Biol. 37:14895–14902. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Feng L, Zhu Y, Zhang Y and Rao M: LncRNA
GACAT3 promotes gastric cancer progression by negatively regulating
miR-497 expression. Biomed Pharmacother. 97:136–142.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
National Health and Family Planning
Commission of the People's Republic of China: Diagnosis, treatment
and treatment of hepatocellular carcinoma (V2017). J Clin Hepatol
33: 114-126, 2017.
|
|
16
|
Xu C, Shao Y, Xia T, Yang Y, Dai J, Luo L,
Zhang X, Sun W, Song H, Xiao B and Guo J: lncRNA-AC130710 targeting
by miR-129-5p is upregulated in gastric cancer and associates with
poor prognosis. Tumour Biol. 35:9701–9706. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gui Y and Zheng XL: Epidermal growth
factor induction of phenotype-dependent cell cycle arrest in
vascular smooth muscle cells is through the mitogen-activated
protein kinase pathway. J Biol Chem. 278:53017–53025.
2003.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yang G, Lu X and Yuan L: LncRNA: A link
between RNA and cancer. Biochim Biophys Acta. 1839:1097–1109.
2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang F, Ying HQ, He BS, Pan YQ, Deng QW,
Sun HL, Chen J, Liu X and Wang SK: Upregulated lncRNA-UCA1
contributes to progression of hepatocellular carcinoma through
inhibition of miR-216b and activation of FGFR1/ERK signaling
pathway. Oncotarget. 6:7899–7917. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Sui J, Yang X, Qi W, Guo K, Gao Z, Wang L
and Sun D: Long non-coding RNA linc-USP16 functions as a tumour
suppressor in hepatocellular carcinoma by regulating PTEN
expression. Cell Physiol Biochem. 44:1188–1198. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhong H, Yang J, Zhang B, Wang X, Pei L,
Zhang L, Lin Z, Wang Y and Wang C: LncRNA GACAT3 predicts poor
prognosis and promotes cell proliferation in breast cancer through
regulation of miR-497/CCND2. Cancer Biomark. 22:787–797.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhou W, Wang L, Miao Y and Xing R: Novel
long noncoding RNA GACAT3 promotes colorectal cancer cell
proliferation, invasion, and migration through miR-149. Onco
Targets Ther. 11:1543–1552. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yang X, Zhang W, Cheng SQ and Yang RL:
High expression of lncRNA GACAT3 inhibits invasion and metastasis
of non-small cell lung cancer to enhance the effect of
radiotherapy. Eur Rev Med Pharmacol Sci. 22:1315–1322.
2018.PubMed/NCBI View Article : Google Scholar
|