Introduction
Human papillomaviruses (HPVs) are important sexually
transmitted human pathogens (1). The
most common high-risk HPV types, 16 and 18, are frequently detected
in gynecological and urological cancers (2). The low risk HPV types, 6 and 11, are
associated with anogenital warts and laryngeal papillomatosis
(3). HPV-related lesions require
continued production of the oncogenic E6 protein, which has targets
in both the nucleus and cytoplasm, and promotes cell proliferation
in human keratinocytes (4). Small
molecule inhibitors and small interfering RNAs (siRNAs) targeting
the E6 oncogene can be used to improve the therapeutic effects of
current therapies for treating HPV-related gynecological and
urological diseases (5).
RNA-guided, RNA degradation is a natural phenomenon
mediated by the bacterial type II CRISPR/CRISPR-associated protein
(Cas)13a system (6). This system can
be reprogrammed to induce mRNA and long non-coding RNA knockdown at
specific sites (7). The potential of
CRISPR-Cas13a to treat or diagnose human genetic diseases has yet
to be demonstrated. Although it has been revealed that the E6 gene
can be inactivated by CRISPR-Cas9 (8,9), to the
best of our knowledge, there are no reports regarding the study of
a designed CRISPR-Cas13a targeted against the E6 gene of HPV 16/18,
the pathogen behind gynecological and urological cancers. Both
CRISPR-Cas9 and siRNA may cause significant off-target effects
(7), while the targeting specificity
of CRISPR-Cas13a is considered to be excellent. Therefore,
CRISPR-Cas13a may be a new and useful molecular tool for targeting
the HPV E6 gene.
To test the potential application value of
CRISPR-Cas13a, the present study constructed a CRISPR-Cas13a system
that targets the sequence of the HPV16/18 E6 gene. The aim of the
present study was to fully investigate the feasibility of using
CRISPR-Cas13a to degrade E6 mRNA and resist the pathogenic effects
of HPVs. The study on the silencing effect of CRISPR-Cas13a on the
pathogenic genes of HPV may bring new means for the treatment of
HPV-related infectious diseases.
Materials and methods
Cell lines and cell culture
Human keratinocytes were isolated from the neonatal
foreskin by two-step digestion using trypsin and dispase II and
grown in RPMI-1640 media supplemented with 10% FBS (both from
Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C in an
atmosphere of 5% CO2. Informed patient consent was
obtained from the parents of the newborn and the patient protocols
were approved by the institutional review board of the Foshan
Maternal and Child Health Hospital (approval no. 201914258).
Construction of plasmids
To construct a reprogrammed CRISPR/Cas13a system
that targets the entire E6 protein of HPV 16/18, two different
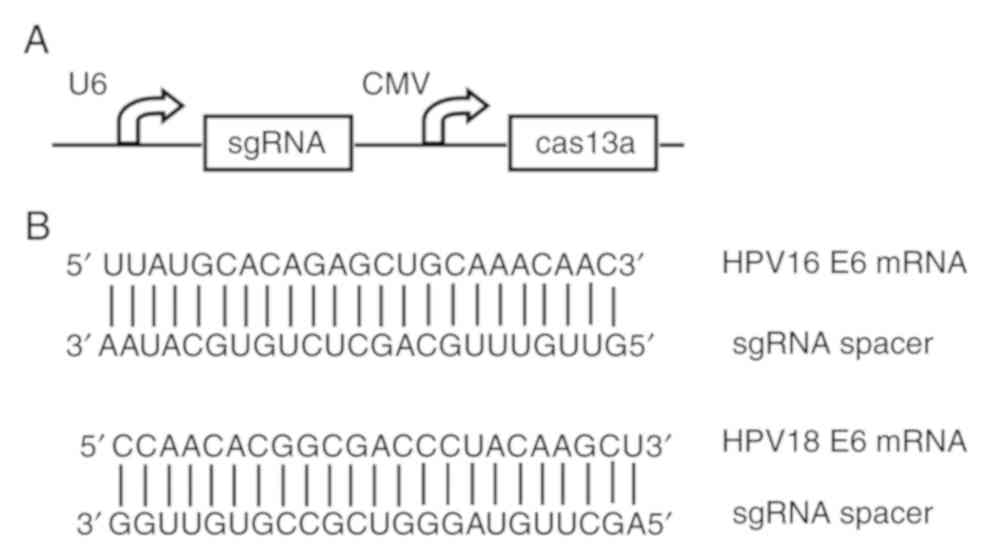
single guide RNA (sgRNA) sequences (Fig.
1B) were designed using the CHOPCHOP v3 online
tool (https://chopchop.cbu.uib.no) and
inserted into the sgRNA expression cassettes of the pCRISPR-CG01
vector (cat. no. PVT10921; Guangzhou FulenGen, Co., Ltd.)
containing the U6 promoter to drive the transcription of each
sgRNA, as well as the cytomegalovirus (CMV) promoter to drive the
expression of the Cas13a protein. The same vector, however,
expressing sgRNAs that lack the complete complementary region, was
also constructed and used as the negative control.
To construct the plasmids pcDNA3.1-HPV16-E6 and
pcDNA3.1-HPV18-E6, the coding sequences for HPV16 E6 and HPV18 E6,
were chemically synthesized and inserted into pcDNA3.1 (+),
digested with HindIII and EcoRI, respectively.
Cell transfections
For stable transfection experiments, human
keratinocytes stably expressing the HPV16 E6 or HPV18 E6 gene were
obtained by selecting cells using the G418 drug after transfection
with the related plasmids. For transient transfection experiments,
the cells (5x105) were cultured in the 12-well plates
until they reached 70% confluency. They were then transfected with
1.25 µg Cas13a- and sgRNAs-expressing plasmids using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the supplier's protocols. Cells were
used for further experimentation after 24-72 h following
transfection.
Western blot analysis
Cells were washed in ice-cold PBS and lysed in
modified RIPA buffer (Amresco, LLC). The protein concentration was
calculated using the bicinchoninic acid protein assays. A total of
40 µg protein extract per lane were separated by 10%
SDS-polyacrylamide gels and transferred onto PVDF membranes (EMD
Millipore). Membranes were blocked overnight at 4˚C in 5% non-fat
milk in TBS and incubated overnight with the mouse anti-HPV16 E6 +
HPV18 E6 antibodies (cat. no. ab70; 1:10,000; Abcam) at 4˚C.
Subsequently, the membranes were incubated with horseradish
peroxidase-conjugated anti-mouse secondary antibodies at 1:10,000
dilution (cat. no. NA931; GE Healthcare Life Sciences) and the
immunoblots were developed using Super-Signal chemiluminescence
reagents (Pierce; Thermo Fisher Scientific, Inc.).
Cell proliferation assay
Cell proliferation was examined using CCK-8 assays
according to the manufacturer's protocol (Beyotime Institute of
Biotechnology). After 24, 48 or 72 h post-transfection, 10 µl CCK-8
solution was added to each well of the 96-well plates and the cells
were maintained for 4 h at 37˚C. Absorbance values were determined
at a wavelength of 450 nm using an ELISA microplate reader (Bio-Rad
Laboratories, Inc.). The assay was repeated at least three times
independently.
Cell apoptosis assay
Morphological assessment of apoptotic cells was
performed using a Hoechst-33258 staining kit (Beyotime Institute of
Biotechnology) according to the supplier's protocols. Briefly, the
cells were fixed in 4% paraformaldehyde for 10 min at room
temperature, and were washed twice with PBS. The cells were then
stained using 0.5 ml Hoechst-33258 for 5 min at room temperature,
and were subsequently observed using a fluorescence microscope
(magnification, x100; Nikon Corporation) at 350 nm.
Fluorescence images were captured from six randomly
chosen fields per well and each experiment was repeated
three times).
Cell apoptosis was examined by analyzing the
activity of Caspase-3 using the enzyme-linked immunosorbent assay
kit (R&D Systems, Inc.) according to the manufacturer's
instructions. The optical density (OD) values were measured using
an ELISA microplate reader (Bio-Rad Laboratories, Inc.).
Statistical analyses
Data are expressed as the mean ± SD. Statistical
significance was determined using Student's t-tests or ANOVAs with
post hoc Tukey's tests. P<0.05 was considered to indicate a
statistically significant difference. All the related statistical
tests were performed using SPSS version 17.0 software (SPSS,
Inc.).
Results
Design and construction of the
reprogrammed CRISPR-Cas13a system
To determine whether CRISPR-Cas13a could silence the
HPV16/18 E6 gene, a single plasmid was constructed to express the
Cas13a and guide RNAs that can be used to make cuts at designated
sites (Fig. 1A). The targets of the
sgRNAs were designed to select sequences of the E6 genes of
HPV16/18 (Fig. 1B). A CRISPR-Cas13a
system that expresses sgRNAs lacking the complementary regions was
used as the negative control.
Promotion of cell growth by the
HPV16/18 E6 gene
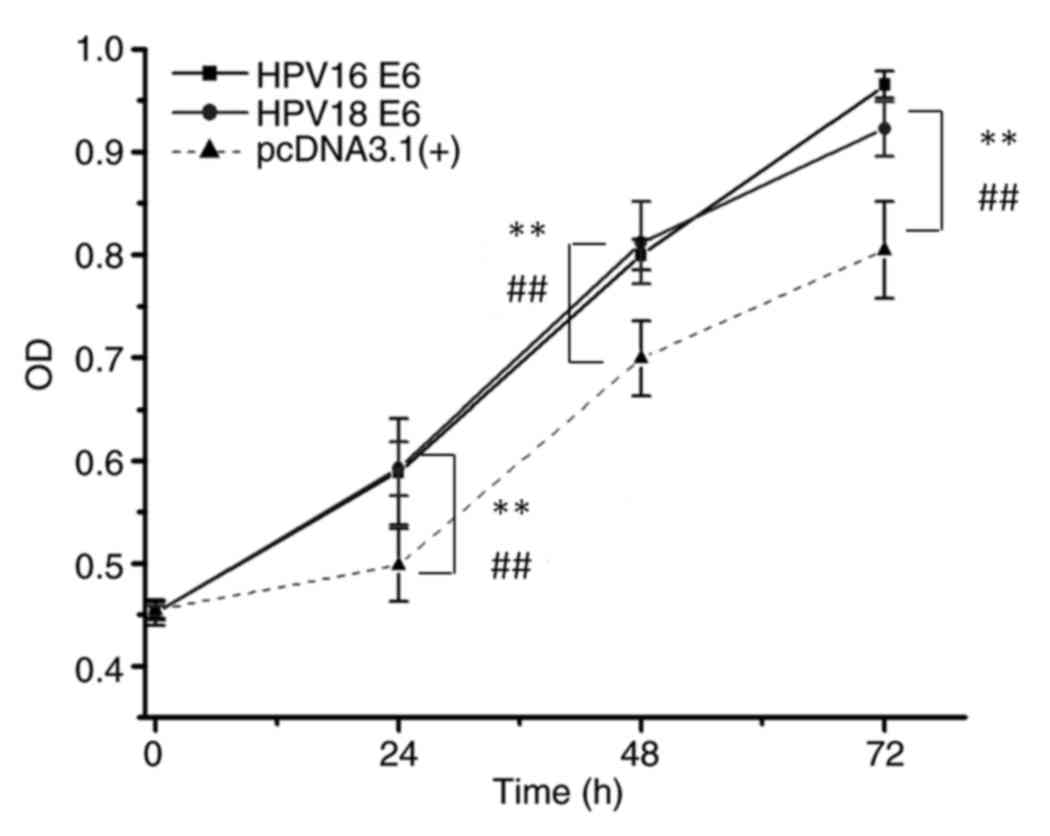
Plasmids encoding HPV16 E6 and HPV18 E6 (Fig. S1) were first constructed, following
which protein overexpression was verified by western blotting. The
potential alteration of cellular phenotypes after HPV16/18 E6
overexpression was examined by stable transfection of the
corresponding plasmids. As shown in Fig.
2, consistent with results from a previous study (4), the E6 gene of HPV16 or HPV18
significantly promoted cell growth (P<0.01), as indicated by OD
values when compared with the OD values of normal keratinocytes
transfected with the empty pcDNA3.1 (+) vector. The transformed
keratinocytes were then used as test models for analyzing the
effects of the reprogrammed CRISPR-Cas13a system on the HPV16/18 E6
genes in the subsequent experiments.
Inhibition of HPV16/18 E6 expression
by the reprogrammed CRISPR-Cas13a
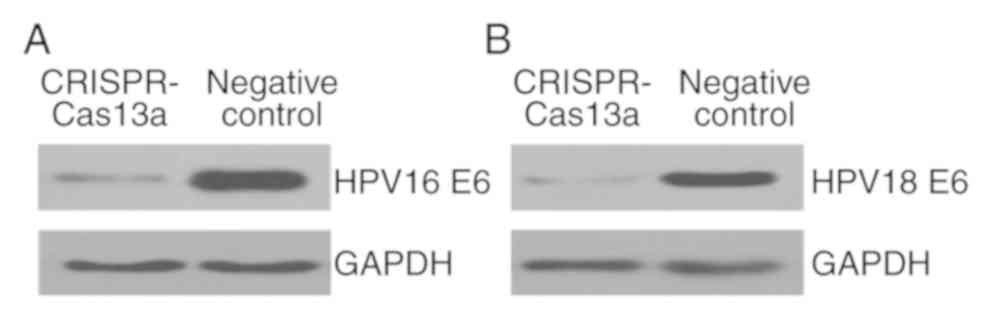
We then investigated whether the endogenous
HPV-16/18 E6 genes can be inactivated in the transformed
keratinocytes using the reprogrammed CRISPR-Cas13a system. Plasmids
expressing the specific CRISPR-Cas13a or the negative control were
transiently transfected into keratinocytes. The expression levels
of E6 proteins were then monitored at 48-h post-transfection using
western blot analyses. Interestingly, expression of the single
CRISPR-Cas13a system resulted in a marked loss of both the HPV16 E6
protein (Fig. 3A) and the HPV18 E6
protein (Fig. 3B). It was therefore
concluded that the reprogrammed CRISPR-Cas13a system could
effectively silence the expression of the HPV16/18 E6 genes.
Inhibition of proliferation of the
E6-transformed keratinocytes by the reprogrammed CRISPR-Cas13a
E6 expression is known to be essential for
HPV-transformed cell growth and survival (4). Therefore, whether the proliferation of
E6-transformed keratinocytes can be affected by the reprogrammed
CRISPR-Cas13a was examined.
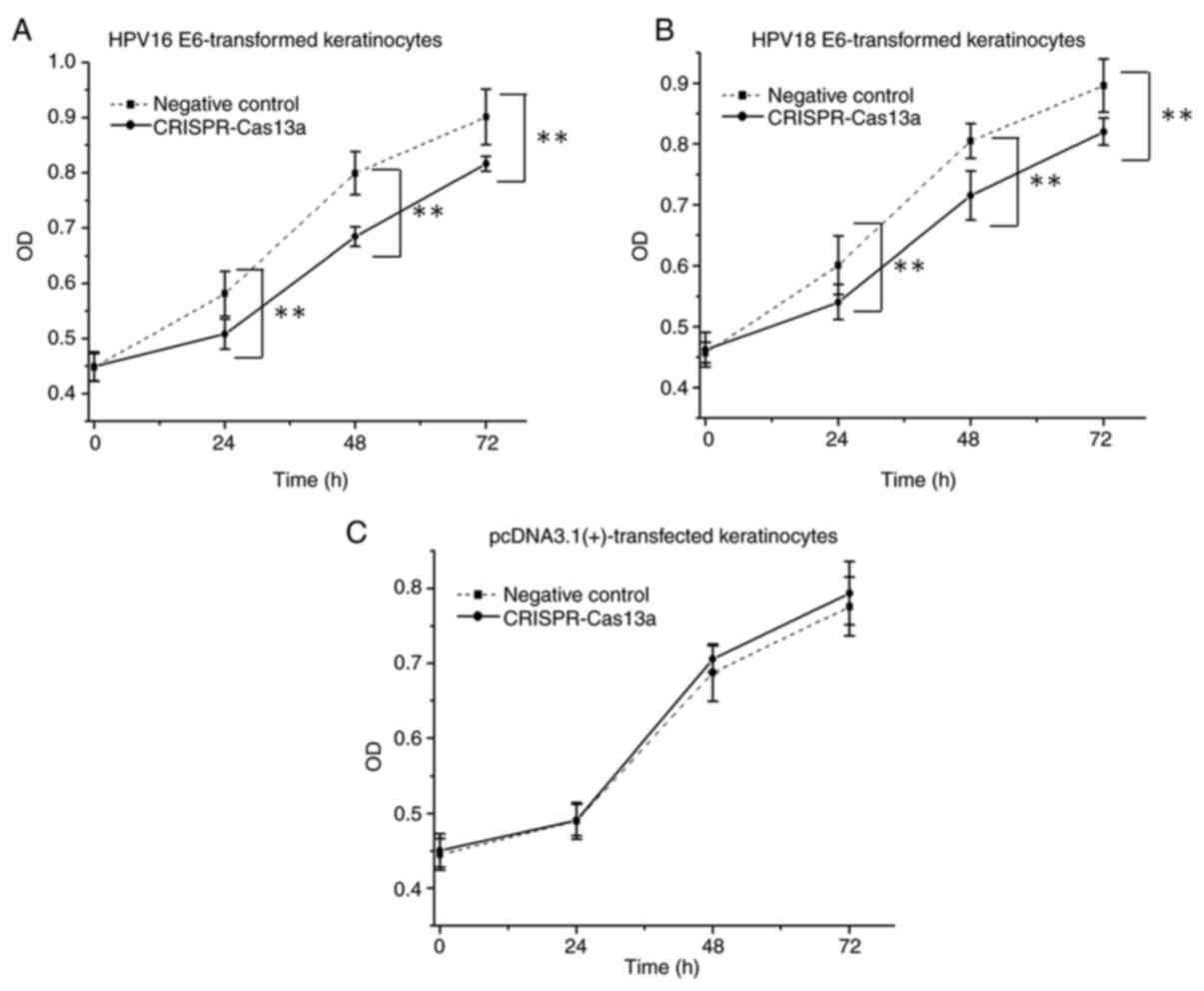
The keratinocytes transformed with the HPV16/18 E6
genes were transiently transfected with the reprogrammed
CRISPR-Cas13a or the negative control in 96-well plates. In CCK-8
assays, it was demonstrated that the proliferative ability of
keratinocytes was decreased (by 14±1.8% on average; P<0.01) when
cells were treated with the reprogrammed CRISPR-Cas13a (Fig. 4A and B). Additionally, the same effects were not
observed in the negative control keratinocytes (P>0.05) that
were stably transfected with the empty pcDNA3.1 (+) vector
(Fig. 4C), indicating that the
reprogrammed CRISPR-Cas13a does not have off-target effects.
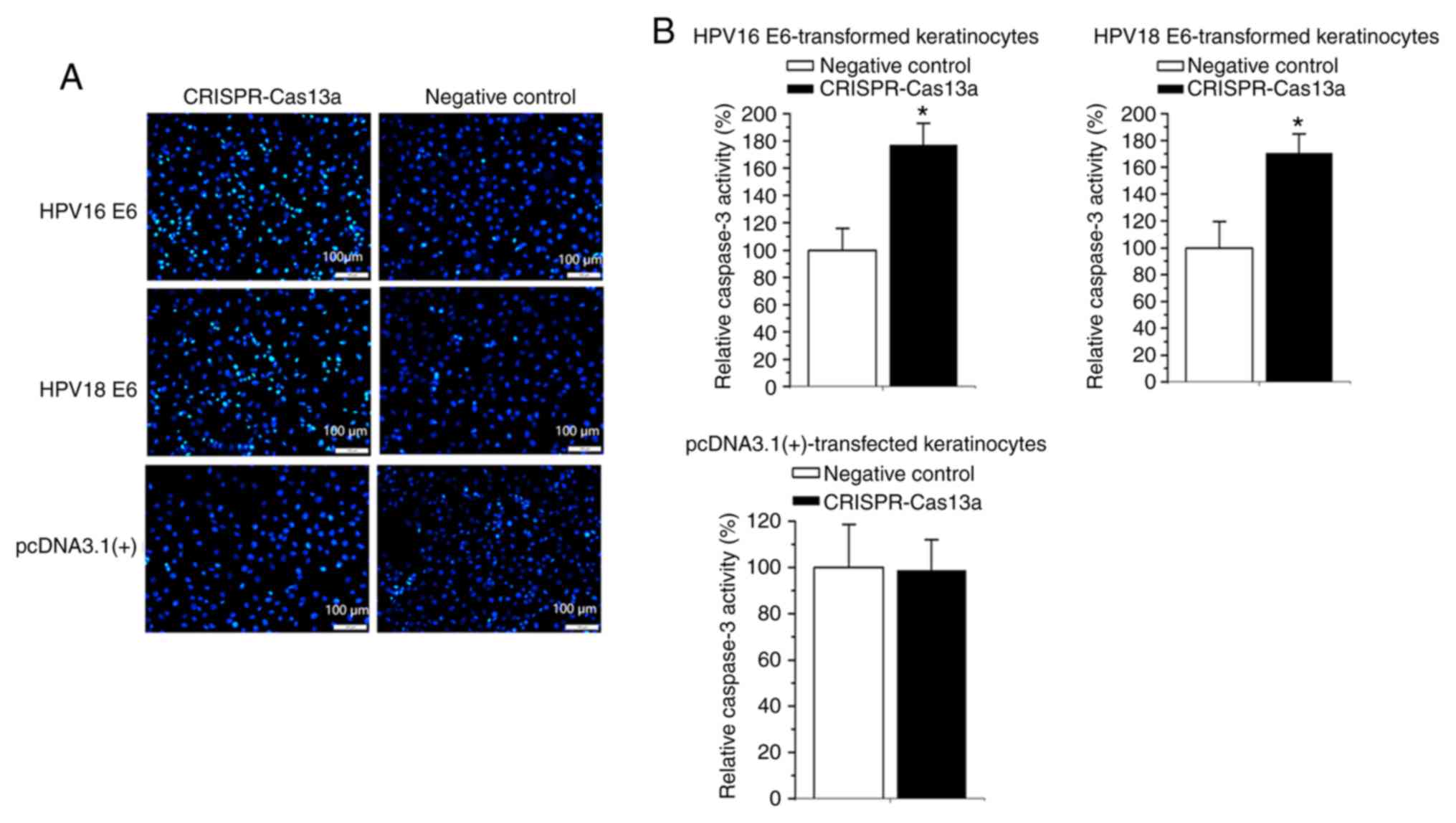
Induction of apoptosis of the
E6-transformed keratinocytes by the reprogrammed CRISPR-Cas13a
Finally, whether the apoptosis of E6-transformed
keratinocytes can be affected by the reprogrammed CRISPR-Cas13a was
also examined. Forty-eight hours after transfection of the
reprogrammed CRISPR-Cas13a or negative control, changes in the rate
of apoptosis of the E6-transformed keratinocytes were determined
using Hoechst-33258 staining and ELISAs. Cells transfected with the
reprogrammed CRISPR-Cas13a exhibited strong blue fluorescence in
the nucleolus after staining with Hoechst 33258, revealing the
apoptotic characteristics of the cells (Fig. 5A). An increase in caspase 3 levels
(80.2±3.2% on average; P<0.05) was also observed in both the
HPV16/18 E6-transformed keratinocytes transfected with the
reprogrammed CRISPR-Cas13a (Fig.
5B). The reprogrammed CRISPR-Cas13a did not induce apoptosis
(P>0.05) in keratinocytes stably transfected with the empty
plasmid (Fig. 5A and B).
Discussion
The current treatments for gynecological and
urological tumors usually include surgical removal, chemotherapy
and radiotherapy (10). Although
there are multiple approaches for treating these diseases, the rate
of recurrence and fatality after successful clearance is still very
high (10). Further improvements in
treatment outcomes may evolve from combinations of conventional
therapy with novel molecular agents (5). The expression of the HPV E6 protein is
positively associated with uncontrolled proliferation of
keratinocytes during the development of gynecological and
urological tumors (5). With this in
mind, the E6 gene is an ideal target for molecular agents targeting
HPV 16/18. In previous studies, anti-sense oligodeoxynucleotides
(11), ribozymes (12) and siRNA (13) have already been constructed to
inhibit the expression levels of E6 mRNA/protein. However, those
approaches have only achieved limited success due to the low
accessibility of most sites on the HPV16/18 E6 mRNA (11). Therefore, treatments based on new
molecular agents should be developed.
The recent advancements in CRISPR engineering have
resulted in the development of more targeted and potentially safer
gene therapies for HPV-related diseases (14,15).
Previous studies have indicated that systemic delivery of
CRISPR/Cas9 targeting HPV oncogenes is effective at eliminating
established tumors (16-19).
However, Cas9 may cause many unexpected off-target effects. The
challenge of this technology is to identify Cas nucleases that
specifically target functional genes. CRISPR-Cas13a is a newly
discovered class of nucleases with a high targeting specificity
(6), but to the best of our
knowledge it has never been used to target HPV genes.
In the present study, the possibility of using the
reprogrammed CRISPR-Cas13a system as a novel molecular agent
against the E6 genes of high-risk HPV types (HPV16/18) was
investigated. The constructed CRISPR/Cas13a system can break E6
mRNA strands by co-expressing a single Cas13a protein with a sgRNA.
The present data showed that sgRNAs directed to the mRNA regions
could inactivate both HPV-16 E6 and HPV-18 E6. These results
highlight the potential for the reprogrammed CRISPR-Cas13a to be
used for the treatment of related viruses across various species.
From examining the cellular phenotypes, it was found that the
reprogrammed CRISPR-Cas13a inhibited the cell proliferation and
promoted cell apoptosis in the E6-transformed keratinocytes. These
results demonstrated the efficacy of CRISPR-Cas13a in regulating
the deregulated phenotypes induced by HPV 16/18. Because the E6 HPV
genes share no sequence homology to any human genes, knockdown of
this gene will not induce death of healthy cells (16).
Despite the potential application of this research,
it is still necessary to continue to test the ability of
CRISPR-Cas13a to target HPV genes in animal models. Genetic vectors
that deliver Cas13a genes to the target tissues are also yet to be
developed.
In conclusion, the present study developed a potent
CRISPR-Cas13a system that effectively suppressed cell growth and
induced apoptosis in HPV E6-transformed cells, while causing
minimal effects in E6-negative human cells. The reprogrammed
CRISPR-Cas13a may be further used to inactivate the functions of
the HPV E6 gene in future studies.
Supplementary Material
Figure S1. Detection of E6 protein
expression levels in keratinocytes transfected with the
overexpression vectors. Western blot analysis was performed to
detect the expression levels of (A) HPV16 E6 or (B) HPV18 E6 in
keratinocytes transfected with either the overexpression vector or
the negative control. HPV, human papillomavirus; overexpression
vector, pcDNA3.1.HPV 16/18 E6; negative control, pcDNA3.1.
Acknowledgements
Not applicable.
Funding
This study was funded by Foshan Science and
Technology Innovation Project (grant no. 1920001000300) and Fujian
Provincial Natural Fund (grant no. 2018J01223).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding authors on reasonable
request.
Authors' contributions
JFW and HHC planned and managed the project. CJL,
LWG, GQL, MJG, HLW and QQY were involved in performing the
experiments and collecting the data. All authors performed a final
review of the manuscript and all authors approved the submission of
this manuscript.
Ethics and approval
Informed patient consent was obtained by the parents
of the newborn and the patient protocols were approved by the
institutional review board of the Foshan Maternal and Child Health
Hospital (approval no. 201914258).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Remschmidt C, Fesenfeld M, Kaufmann AM and
Deleré Y: Sexual behavior and factors associated with young age at
first intercourse and HPVvaccine uptake among young women in
Germany: Implications for HPV vaccination policies. BMC Public
Health. 14(1248)2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ghosh I, Mittal S, Banerjee D, Singh P,
Dasgupta S, Chatterjee S, Biswas J, Panda C and Basu P: Study of
accuracy of colposcopy in VIA and HPV detection-based cervical
cancer screening program. Aust N Z J Obstet Gynaecol. 54:570–575.
2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Muñoz N, Kjaer SK, Sigurdsson K, Iversen
OE, Hernandez-Avila M, Wheeler CM, Perez G, Brown DR, Koutsky LA,
Tay EH, et al: Impact of human papillomavirus (HPV)-6/11/16/18
vaccine on all HPV-associated genital diseases in young women. J
Natl Cancer Inst. 102:325–339. 2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
McKee CH, Onder Z, Ashok A, Cardoso R and
Moroianu J: Characterization of the transport signals that mediate
the nucleocytoplasmic traffic of low risk HPV11 E7. Virology.
443:113–122. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Stern PL, van der Burg SH, Hampson IN,
Broker TR, Fiander A, Lacey CJ, Kitchener HC and Einstein MH:
Therapy of human papillomavirus-related disease. Vaccine. 30 (Suppl
5):F71–F82. 2012.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gootenberg JS, Abudayyeh OO, Lee JW,
Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer
NM, Freije CA, et al: Nucleic acid detection with
CRISPR-Cas13a/C2c2. Science. 356:438–442. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Abudayyeh OO, Gootenberg JS,
Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT,
Kellner MJ, Regev A, et al: RNA targeting with CRISPR-Cas13.
Nature. 550:280–284. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhen S, Hua L, Takahashi Y, Narita S, Liu
YH and Li Y: In vitro and in vivo growth suppression of human
papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9.
Biochem Biophys Res Commun. 450:1422–1426. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kennedy EM, Kornepati AV, Goldstein M,
Bogerd HP, Poling BC, Whisnant AW, Kastan MB and Cullen BR:
Inactivation of the human papillomavirus E6 or E7 gene in cervical
carcinoma cells by using a bacterial CRISPR/Cas RNA-guided
endonuclease. J Virol. 88:11965–11972. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Paner GP, Stadler WM, Hansel DE, Montironi
R, Lin DW and Amin MB: Updates in the eighth edition of the
tumor-node-metastasis staging classification for urologic cancers.
Eur Urol. 73:560–569. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Bharti AC, Singh T, Bhat A, Pande D and
Jadli M: Therapeutic strategies for human papillomavirus infection
and associated cancers. Front Biosci (Elite Ed). 10:15–73.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Liu DZ, Jin YX, Hou H, Huang YZ, Yang GC
and Xu Q: Preparation and identification of activity of
anti-HPV-6b/11E1 universal ribozyme-Rz1198 in vitro. Asian J
Androl. 1:195–201. 1999.PubMed/NCBI
|
|
13
|
Chen XZ, Zhu KJ, Xu Y, Tang XY, Cai XZ,
Zhang X and Cheng H: RNA interference silences the human
papillomavirus 6b/11 early gene E7 in vitro and in vivo. Clin Exp
Dermatol. 35:509–515. 2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
De Buhr H and Lebbink RJ: Harnessing
CRISPR to combat human viral infections. Curr Opin Immunol.
54:123–129. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Hsu DS, Kornepati AVR, Glover W, Kennedy
EM and Cullen BR: Targeting HPV16 DNA using CRISPR/Cas inhibits
anal cancer growth in vivo. Future Virol. 13:475–482.
2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhu D, Shen H, Tan S, Hu Z, Wang L, Yu L,
Tian X, Ding W, Ren C, Gao C, et al: Nanoparticles based on poly
(β-Amino Ester) and HPV16-targeting CRISPR/shRNA as potential drugs
for HPV16-related cervical malignancy. Mol Ther. 26:2443–2455.
2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yoshiba T, Saga Y, Urabe M, Uchibori R,
Matsubara S, Fujiwara H and Mizukami H: CRISPR/Cas9-mediated
cervical cancer treatment targeting human papillomavirus E6. Oncol
Lett. 17:2197–2206. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu Y, Cai Z and Zhang X: Reprogrammed
CRISPR-Cas9 targeting the conserved regions of HPV6/11 E7 genes
inhibits proliferation and induces apoptosis in E7-transformed
keratinocytes. Asian J Androl. 18:475–479. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hu Z, Yu L, Zhu D, Ding W, Wang X, Zhang
C, Wang L, Jiang X, Shen H, He D, et al: Disruption of HPV16-E7 by
CRISPR/Cas system induces apoptosis and growth inhibition in HPV16
positive human cervical cancer cells. Biomed Res Int.
2014(612823)2014.PubMed/NCBI View Article : Google Scholar
|