Introduction
Lung cancer is the leading cause of cancer-related
death worldwide, 85% of which is non-small cell lung cancer (NSCLC)
(1), which usually invades the
regional or distant lymph nodes, brain and liver (2,3).
Metastasis is typically the most common risk of death in NSCLC,
which suggests that blocking the migration and invasion of tumor
cells might be a possible therapeutic strategy.
A previous study identified that migration and
invasion of tumor cells could be regulated by chemokines and their
receptors (4). Chemokines, which are
a family of small, structurally related heparin-binding proteins
classified as C, CXC, CC and CX3C sub-families (5,6), are
essential to the normal and pathologic trafficking of leukocytes.
The biological functions of chemokines are mediated via
G-protein-coupled receptors, which play an important role in
inflammation and leukocyte differentiation. It has been
demonstrated that various cell types may express different
chemokine receptors, such as C-X-C chemokine receptor type 2
(CXCR2) (7), C-X-C chemokine
receptor type 4 (CXCR4) (8), C-C
chemokine receptor type 6 (CCR6) (9)
and C-C chemokine receptor type 9 (CCR9) (10). High expression of CXCR2, CXCR4 and
CCR9 with their respective ligands C-X-C motif chemokine 8, C-X-C
motif chemokine 12 and CC motif chemokine ligand 25 (CCL25) was
associated with tumor metastasis and poor survival in NSCLC
(11-13).
We previously demonstrated that CCR9 and CCL25 interaction could
suppress apoptosis in a phosphoinositide 3-kinase/Akt-dependent
model in the NSCLC cell lines A549 and SK-MES-1(14). However, the exact molecular mechanism
of migration and invasion in NSCLC remains elusive.
The present study aimed to examine the potential
association between the CCL25/CCR9 axis and key molecules involved
in lung cancer cell migration, and to verify further the
association between the CCL25/CCR9 axis and key molecules examined
(VEGF-C, VEGF-D, MMP-1 and MMP-7) by CCR9 drug treatment
experiments. A Kaplan-Meier survival analysis was used to further
confirm the association between the CCL25/CCR9 axis and prognosis
of lung cancer.
Materials and methods
Study design
A total of 60 patients with NSCLC who underwent
surgery in The Huadong Hospital Affiliated with Fudan University
were enrolled in the present study between September 2012 and
December 2015. Tumor tissues and adjacent normal tissues were
collected and paraffin-embedded. Neither chemotherapy nor
immunotherapy was performed prior to surgery. A double pathological
diagnosis was performed prior to enrolling the patients in the
present study. The median age of these patients was 58 years, with
the age ranging from 31 to 76. Further patient information is
presented in Table I. The present
study protocol was approved by The Ethics Committee Review Board of
The Huadong Hospital Affiliated with Fudan University, and written
informed consent was obtained from every participant.
 | Table IAssociation between CCR9 and
clinicopathological characteristics. |
Table I
Association between CCR9 and
clinicopathological characteristics.
| Clinicopathological
characteristic | Number of
patients | CCR9 postivity [N,
(%)] | χ2 | P-value |
|---|
| Sex | | | | |
|
Male | 40 | 25 (62.50) | 0.330 | 0.566 |
|
Female | 20 | 14 (70.00) | | |
| Age (years) | | | | |
|
≤60 | 37 | 22 (59.46) | 1.302 | 0.254 |
|
>60 | 23 | 17 (73.91) | | |
| Histology | | | | |
|
SCC | 25 | 12 (48.00) | 5.444 | 0.020 |
|
AC | 35 | 27 (77.14) | | |
| Lymph node
metastasis | | | | |
|
No | 15 | 4 (26.67) | 12.918 | <0.001 |
|
Yes | 45 | 35 (77.78) | | |
| VEGF | | | | |
|
Positive | 31 | 25 (80.65) | 6.901 | 0.008 |
|
Negative | 29 | 14 (48.28) | | |
| MMP-1 | | | | |
|
Positive | 36 | 27 (75.00) | 3.956 | 0.047 |
|
Negative | 24 | 12 (50.00) | | |
| MMP-7 | | | | |
|
Positive | 33 | 29 (87.88) | 16.873 | <0.001 |
|
Negative | 27 | 10 (37.04) | | |
Immunohistochemistry (IHC) assay
Formalin-fixed (10%, for 8 h at room temperature),
paraffin-embedded tissues were sliced into 4-µm sections using a
microtome, then successively deparaffinized with xylene, rehydrated
in a descending alcohol series and rinsed with PBS followed by
high-pressure antigen retrieval in a microwave oven with citrate
buffer (100˚C for 5 min and then 24˚C for a further 3 min).
Subsequently, sections were treated with 3%
H2O2 for 10 min at room temperature to block
endogenous peroxidase. The sections were washed with PBS three
times for 10 min each and then blocked with 10% normal goat serum
(Beijing Solarbio Science & Technology Co., Ltd.) for 30 min at
room temperature. The tissue sections were subsequently incubated
with anti-CCR9 (1:200; cat. no. ab1662; Abcam), anti-vascular
endothelial growth factor (VEGF; 1:200; cat. no. AF0312; Beyotime
Institute of Biotechnology) anti-matrix metalloproteinase (MMP)-1
(1:500; cat. no. ab137332; Abcam) or anti-MMP-7 (1:500; cat. no.
ab205525; Abcam) antibodies overnight at 4˚C. The slides were then
incubated with horseradish peroxidase (HRP)-conjugated goat
anti-mouse antibody (1:50; cat. no. A0216, Beyotime Institute of
Biotechnology) or HRP-conjugated goat-anti-rabbit antibody (1:50;
cat. no. A0208; Beyotime Institute of Biotechnology) at room
temperature for 60 min. Finally, the sections were washed with PBS,
then stained with diaminobenzidine and hematoxylin (Beyotime
Institute of Biotechnology) at room temperature for 5 min. The
sections were observed under a light microscope (magnification,
x200; BX-43; Olympus Corporation). A semi-quantitative scoring
system (15) was applied to assess
the expression of CCR9, VEGF, MMP-1 and MMP-7 in the specimens
based on the average intensity and density of positively stained
cells by IHC. Staining intensity was graded as follows: i) 0, no
staining; ii) 1, weak staining; iii) 2, moderate staining; and iv)
3, strong staining. The density of positively stained cells was
scored according to the following criteria: i) 0, ≤5%; ii) 1,
5-25%; iii) 2, 26-50%; and iv) 3, >50%. The total IHC score was
calculated by multiplying the intensity score by the density score.
A total IHC score >0 was defined as positive expression and a
total IHC score=0 was defined as negative expression.
Cell culture
The NSCLC cell lines SK-MES-1 [squamous cell
carcinoma (SCC)] and A549 [adenocarcinoma (AC)] were purchased from
The Kunming Institute of Zoology, Chinese Academy of Sciences.
Cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, lnc.) containing 10% FBS (Gemini Bio Products), 100
U/ml penicillin and 100 µg/ml streptomycin in 5% CO2 at
37̊C. Before migration and invasion studies, lung cancer cells were
cultured for 24 h in RPMI-1640 with 2% FBS.
Reverse transcription-quantitative PCR
(RT-qPCR) primer design
The human mRNA sequences for VEGF-C, VEGF-D, MMP-1,
MMP-7 and GAPDH were obtained from The National Institutes of
Health National Center for Biotechnology Information (Genbank
Database Accession nos. NM_005429.4, NM_004469.4, NM_001145938.2,
NM_002423.5 and BC004109, respectively). These sequences were used
to design primers for RT-qPCR analysis. The primer sequences were
as follows: VEGF-C forward, 5'-AGCACGAGCTACCTCAGCAAGAC-3' and
reverse, 5'-TTTAGACATGCATCGGCAGGAA-3'; VEGF-D forward,
5'-AGCTGCCTGATGTCAACTGCTTA-3' and reverse,
5'-GTTCATTACTGGAGCCCTGCAC-3'; MMP-1 forward
5'-AAGAATGATGGGAGGCAAGT-3' and reverse, 5'-GGTTTCAGCATCTGGTTTCC-3';
MMP-7 forward 5'-GAGTGAGCTACAGTGGGAACA-3' and reverse,
5'-CTATGACGCGGGAGTTTAACAT-3' and GAPDH forward,
5'-GACGGCAAGATGCACATCAC-3' and reverse,
5'-GAGATGTAGCACGGGATCATGG-3'.
RNA isolation and RT-qPCR
analysis
Total RNA was isolated from lung cancer cells using
the RNA Simple Total RNA kit (Tiangen Biotech Co., Ltd.). Cells
were untreated, CCL25-treated or CCL25-treated and stained with
anti-CCR9 antibody, cDNA was generated using the Quantscript RT kit
(Tiangen Biotech Co., Ltd.) according to the manufacturer's
protocols. Real-time RT-qPCR was performed using a 2X Taq PCR
MasterMix (Tiangen Biotech Co., Ltd.) in a ABI7500 instrument
(Applied Biosystems; Thermo Fischer Scientific, Inc.) using the
following thermocycling conditions: Initial denaturation for 3 min
at 94˚C, followed by 30 cycles of 94˚C for 30 sec, 55˚C for 30 sec
and 72˚C for 1 min, prior to final extension at 72˚C for 5 min. The
expression of GAPDH was measured in each sample as an endogenous
control. The ratio of mRNA expression was calculated for each group
using the 2-ΔΔCq method (16).
Western blot analysis
The A549 and SK-MES-1 cell lines were treated with
100 ng/ml CCL25 (cat. no. 300-45; PeproTech, Inc.) or CCL25 (100
ng/ml) with 5 µg/ml anti-CCR9 antibody (cat. no. ab1662; Abcam) at
37̊C and 5% CO2 for 48 h. Untreated cells were used as
controls. Total protein from untreated and treated cells was washed
with cold PBS and isolated in modified radio-immunoprecipitation
assay buffer (50 mM Tris-Cl at pH 7.5; 150 mM NaCl; 1% NP-40; 1.0%
sodium deoxycholate; 1% PMSF and 0.1% SDS) and quantified using the
bicinchoninic acid method (Beyotime Institute of Biotechnology).
Proteins were loaded (100 µg per lane) and resolved by SDS-PAGE on
10% gels using Tris-glycine-SDS buffer (25 mM Tris; 192 mM glycine
and 0.1% SDS; pH 8.8). Proteins were transferred to polyvinylidene
fluoride membranes (EMD Millipore) using a wet transfer apparatus
(Bio-Rad Laboratories, Inc.). Membranes were incubated in blocking
buffer (5% non-fat milk dissolved in TBS + 0.1% Tween-20) for 1 h
at room temperature. After blocking, membranes were incubated with
anti-VEGF-C (1:500; cat. no. ab83905; Abcam), anti-VEGF-D (1:800;
cat. no. ab155288; Abcam), anti-MMP-1 (1:1,000; cat. no. ab137332;
Abcam), anti-MMP-7 (1:1,000; cat. no. ab205525; Abcam) and
anti-GAPDH (1:2,000; cat. no. ab8245; Abcam) antibodies overnight
at 4̊C. Membranes were then washed in wash buffer (TBS + 0.1%
Tween-20) three times for 10 min each time. After washing,
membranes were incubated with the secondary antibodies for 2 h at
37̊C. The secondary antibodies were horseradish
peroxidase-conjugated goat anti-rabbit antibody (1:8,000; cat. no.
ab205718; Abcam) or goat-anti-mouse antibody (1:8,000; cat. no.
ab205719; Abcam). Following incubation, membranes were washed five
times with washing buffer for 6 min each time. The immunoreactive
bands were detected using the ECL Plus reagent (Beyotime Institute
of Biotechnology). ImageJ software (version 1.5.2; https://imagej.nih.gov/ij) was used to quantify the
data from three independent experiments. Bands from untreated cells
were given a value of 1.0 and the protein levels of treated groups
were normalized to this value.
Migration and invasion assays
Migration and invasion studies were performed using
Corning Biocoat™ Matrigel® invasion chambers
with 8-µm pore size (Corning, Inc.). Serum-free RPMI-1640 was added
to the bottom and top chambers and allowed to hydrate the membrane
for 2 h at 37̊C with 5% CO2. Subsequently,
1x104 cells were seeded to the top chamber and 100 ng/ml
CCL25 (PeproTech, Inc.) was added into the bottom chamber as a
chemo-attractant. To determine if the migration and invasion of
lung cancer cells was mediated specifically by CCL25/CCR9
interaction, cells pre-incubated with 5.0 µg/ml anti-CCR9 antibody
(Abcam) were added to the top chamber and allowed to migrate or
invade in the presence of CCL25 at 37̊C with 5% CO2
overnight. After incubation, non-migrating cells in the upper
surface of the membrane were removed. Cells at the bottom surface
of the insert were fixed with 100% methanol for 10 min at room
temperature, stained for 2 min with crystal violet (Beijing
Solarbio Science & Technology Co., Ltd.) at room temperature
and washed twice with deionized water. Migrated or invaded cells
were counted using a light microscope (magnification, x200). All
experiments were repeated three times for validation.
Bioinformatics analysis
The Kaplan-Meier Plotter online tool (http://kmplot.com/analysis/index.php?p=service&cancer=lung)
was used to investigate the association between CCR9 and CCL25 mRNA
expression and clinical prognosis for patients with NSCLC (n=1,926)
(17). In this database (http://kmplot.com), multiple independent
transcriptomic datasets, generated using caBIG (http://cabig.cancer.gov), GEO (http://www.ncbi.nlm.nih.gov/geo/) and TCGA (http://cancergenome.nih.gov), are merged and the gene
expression data and survival information were analysed, where the
hazard ratio (HR), 95% confidence interval (CI) and log-rank
P-value was determined and displayed. P<0.05 was considered to
indicate a statistically significant difference. The mutational
level of CCR9 and CCL25 was determined using the cBioPortal for
Cancer Genomics website (http://www.cbioportal.org/).
Statistical analysis
All values are presented as the mean ± SD.
Associations between CCR9 expression and clinicopathological
parameters were assessed using the χ2 test as well as
the Fisher's exact test. Statistically significant differences
between groups were analyzed by one-way ANOVA with Tukey's post hoc
test. Data were analyzed using SPSS 20.0 software (IBM Corp.) and
GraphPad Prism 7 (GraphPad Software, Inc.). P<0.05 was used to
indicate a statistically significant difference.
Results
CCR9, VEGF, MMP-1 and MMP-7 expression
in NSCLC tissue
CCR9, VEGF, MMP-1 and MMP-7 were located on the cell
membrane and/or the cytoplasm in the cancer tissues (Fig. 1). The positive rates of CCR9, VEGF,
MMP-1 and MMP-7 were significantly higher in the lung cancer
tissues compared with the adjacent normal tissues (Table I).
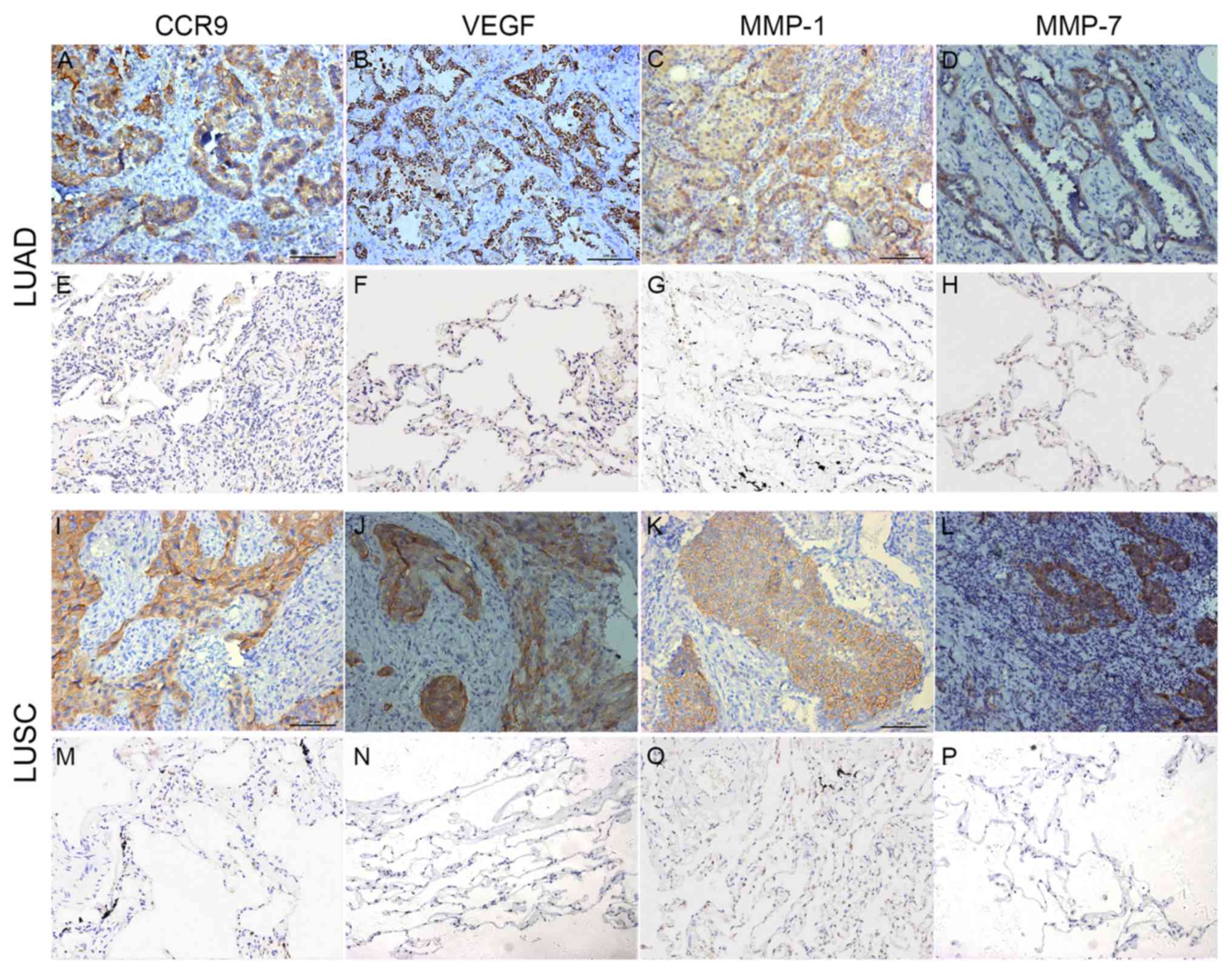 | Figure 1CCR9, VEGF, MMP-1 and MMP-7
expression in the non-small cell lung cancer and adjacent normal
tissues. Positive (A) CCR9, (B) VEGF, (C) MMP-1 and (D) MMP-7
expression in LUAD tissue. Negative (E) CCR9, (F) VEGF, (G) MMP-1
and (H) MMP-7 expression in lung adjacent normal tissue. Positive
(I) CCR9, (J) VEGF, (K) MMP-1 and (L) MMP-7 expression in LUSC
tissue. Negative (M) CCR9, (N) VEGF, (O) MMP-1 and (P) MMP-7
expression in lung adjacent normal tissue. Magnification x200.
CCR9, CC chemokine receptor 9; VEGF, vascular endothelial growth
factor; MMP-1, matrix metalloproteinase-1; MMP-7, matrix
metalloproteinase-7; LUAD, lung adenocarcinoma; LUSC, lung squamous
cell carcinoma. |
Association between CCR9 expression
and clinical parameters in NSCLC
The CCR9 positive rate was higher in AC than SCC
tissues. Additionally, CCR9 positivity was also significantly
higher in the presence of lymph node metastasis. Moreover, CCR9
positive rates were significantly higher in VEGF, MMP-1 or
MMP-7-positive tissue, compared with VEGF, MMP-1 or MMP-7-negative
tissue, respectively. However, no significant difference in CCR9
expression was observed when patients were stratified by sex or age
(Table II).
 | Table IIExpression of CCR9, VEGF, MMP-1 and
MMP-7 in tumor tissues and adjacent normal tissues in patients with
non-small cell lung cancer. |
Table II
Expression of CCR9, VEGF, MMP-1 and
MMP-7 in tumor tissues and adjacent normal tissues in patients with
non-small cell lung cancer.
| Parameter | Tumour tissue, n
(%) | Adjacent normal
tissue, n (%) | χ2 | P-value |
|---|
| CCR9 | 39 (65.00) | 15 (25.00) | 19.394 | <0.0001 |
| VEGF | 31 (51.67) | 10 (16.67) | 16.338 | <0.0001 |
| MMP-1 | 36 (60.00) | 11 (18.33) | 21.860 | <0.0001 |
| MMP-7 | 33 (55.00) | 6 (10.00) | 27.693 | <0.0001 |
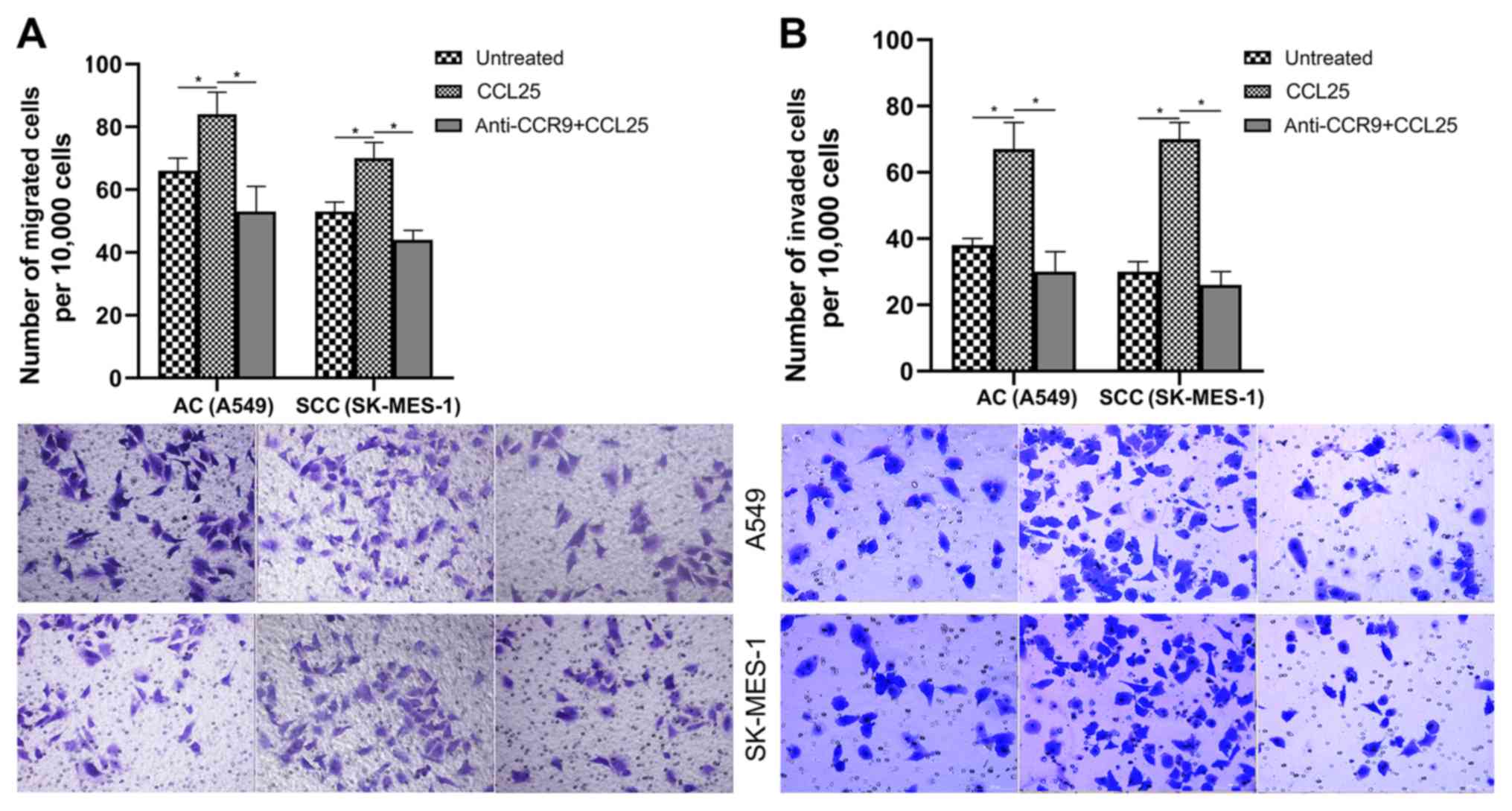
CCL25 induces the migration and
invasion of NSCLC cells
A549 and SK-MES-1 cells lines demonstrated increased
proliferative ability compared with the control (P<0.05) when
treated with 100 ng/ml recombinant CCL25. Both A549 and SK-MES-1
cells were highly responsive in both migration and invasion assays
(Fig. 2). Notably, the
chemo-attractant effect of CCL25 was specific to CCR9 as
demonstrated by the anti-CCR9 antibody blocking the
chemo-attracttant effect (Fig. 2;
P<0.05).
CCL25 modulates VEGF-C, VEGF-D, MMP-1
and MMP-7 expression in NSCLC cells
At the transcriptional and protein level, the
expressions of VEGF-C, VEGF-D, MMP-1 and MMP-7 were upregulated in
the A549 and SK-MES-1 cell lines after CCL25 stimulation (Figs. 3 and 4; P<0.05 in all cases). Furthermore,
CCL25-induced upregulation was reversed by anti-CCR9 antibody
treatment (Figs. 3 and 4; P<0.05 in all cases). However, MMP-7
protein expression was not modified by CCL25 or anti-CCR9 antibody
in the SK-MES-1 cells (P>0.05).
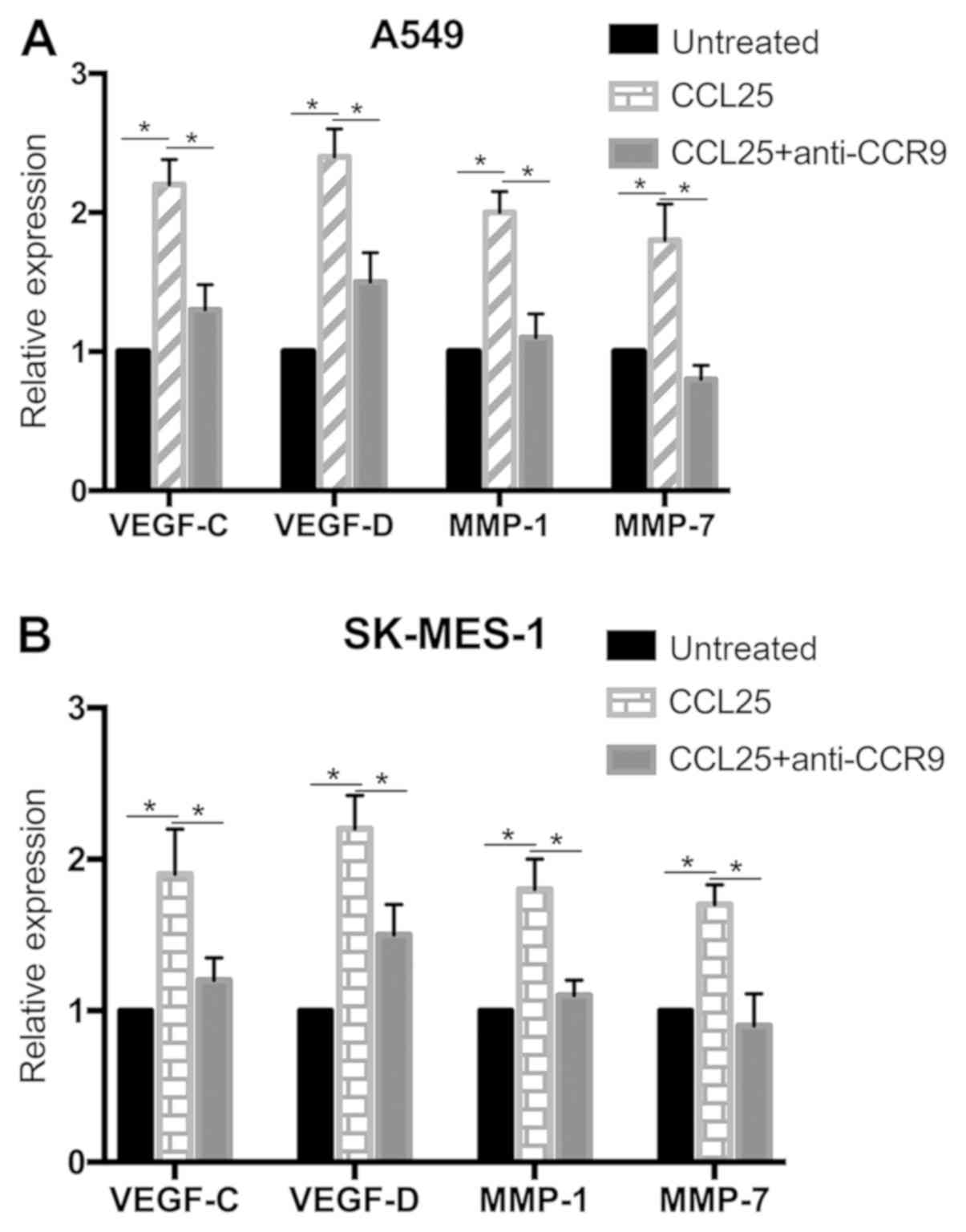 | Figure 3mRNA expression of VEGF-C, VEGF-D,
MMP-1 and MMP-7 in non-small cell lung cancer cell lines. (A)
Expression of VEGF-C, VEGF-D, MMP-1 and MMP-7 in A549 cells
significantly increased after CCL25 treatment, and the addition of
an anti-CCR9 antibody inhibited CCL25-mediated upregulation. (B)
Expression of VEGF-C, VEGF-D, MMP-1 and MMP-7 in SK-MES-1 cells
increased after CCL25 treatment, and the anti-CCR9 antibody
abrogated this CCL25-mediated increase. *P<0.05.
CCR9, CC chemokine receptor 9; CCL25, CC motif chemokine ligand 25;
VEGF, vascular endothelial growth factor; MMP-1, matrix
metalloproteinase-1; MMP-7, matrix metalloproteinase-7. |
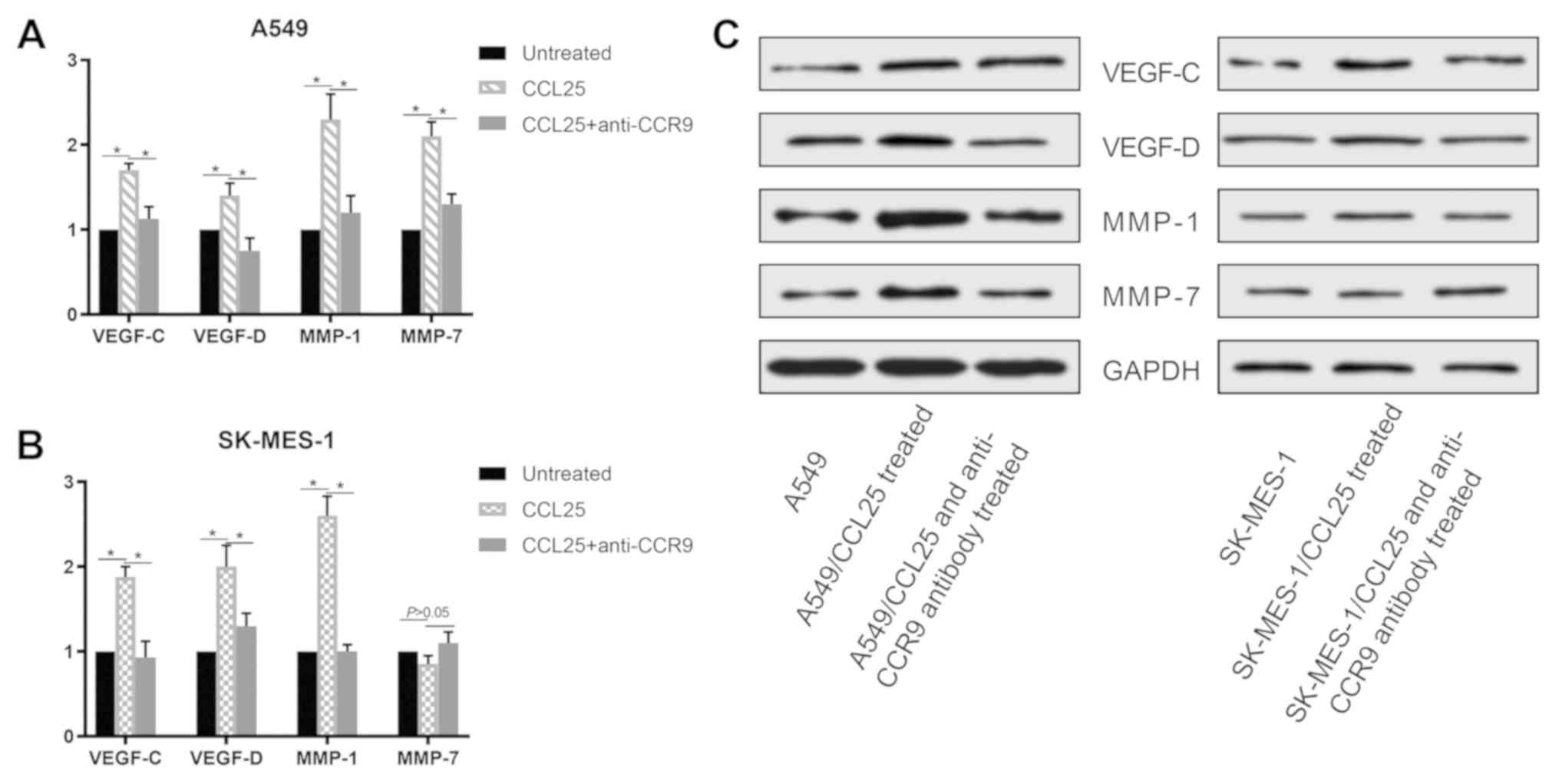 | Figure 4Protein expression of VEGF-C, VEGF-D,
MMP-1 and MMP-7 in non-small cell lung cancer cell lines. (A)
Expression of VEGF-C, VEGF-D, MMP-1 and MMP-7 in A549 cells
increased following CCL25 treatment and the addition of an
anti-CCR9 antibody abrogated this upregulation. (B) Expression of
VEGF-C, VEGF-D and MMP-1 in SK-MES-1 cells increased following
CCL25 treatment. Addition of anti-CCR9 could abrogate this
increase. (C) Protein expression of VEGF-C, VEGF-D, MMP-1 and MMP-7
was examined in A549 and SK-MES-1 cells. *P<0.05.
CCR9, CC chemokine receptor 9; CCL25, CC motif chemokine ligand 25;
VEGF, vascular endothelial growth factor; MMP-1, matrix
metalloproteinase-1; MMP-7, matrix metalloproteinase-7. |
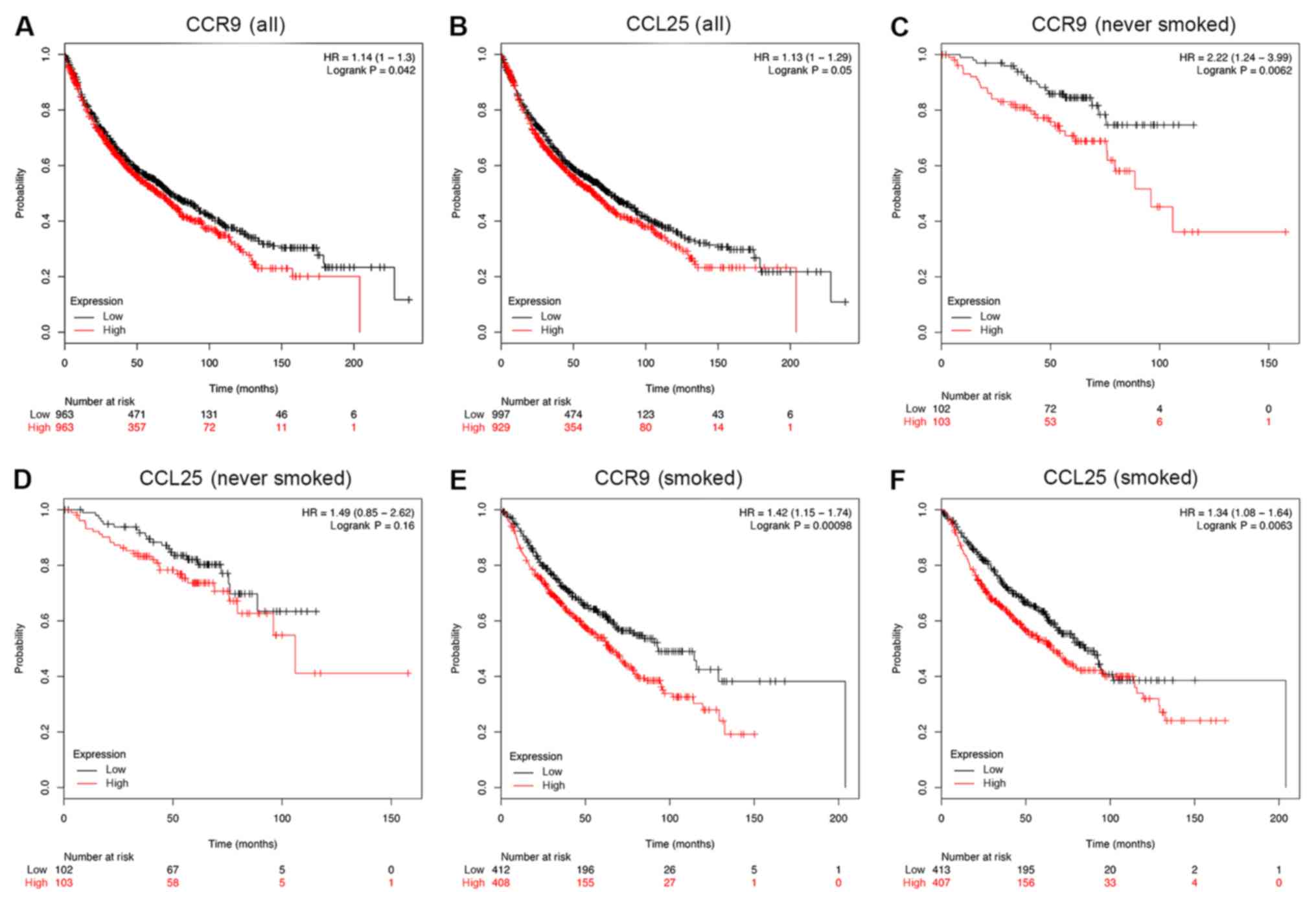
Survival analysis of CCR9 and CCL25 in
patients with NSCLC
The association between CCR9 and CCL25 positive
rates, and clinical prognosis in patients with NSCLC (n=1,926) was
examined using a Kaplan-Meier analysis. Patients with lower CCR9
expression demonstrated better overall survival (OS), compared with
those with higher CCR9 expression (HR=1.14; P=0.042; Fig. 5A). In addition, patients with lower
CCL25 expression had better OS, compared with those with higher
CCL25 expression (HR=1.13; P=0.05; Fig.
5B). Although the results presented in Fig. 5A are only marginally statistically
significant, the actual OS benefits of low CCR9 and CCL25
expression in months are substantial. Indeed, in the case of CCR9,
the OS benefit for high expression, compared with low expression,
was as follows: i) 50 months, 357 compared with 471; ii) 100
months, 72 compared with 131; iii) 150 months, 11 compared with 46;
and iv) 200 months, 1 compared with 6. In the case of CCL25, the OS
benefit for high expression, compared with low expression, was as
follows: i) 50 months, 354 compared with 474; ii) 100 months, 80
compared with 123; iii) 150 months, 14 compared with 43; and iv)
200 months, 1 compared with 6. Moreover, in a sub-group analysis,
low levels of CCR9 mRNA were significantly associated with an
improved OS for patients who never smoked (Fig. 5C; HR=2.22; P<0.01). However, there
was no similar association between the expression of CCL25 mRNA and
OS among patients who never smoked (Fig.
5D; HR=1.49; P=0.16). Furthermore, low levels of both CCR9 and
CCL25 mRNA were indicative of a favorable OS outcome among patients
who smoked (Fig. 5E and F).
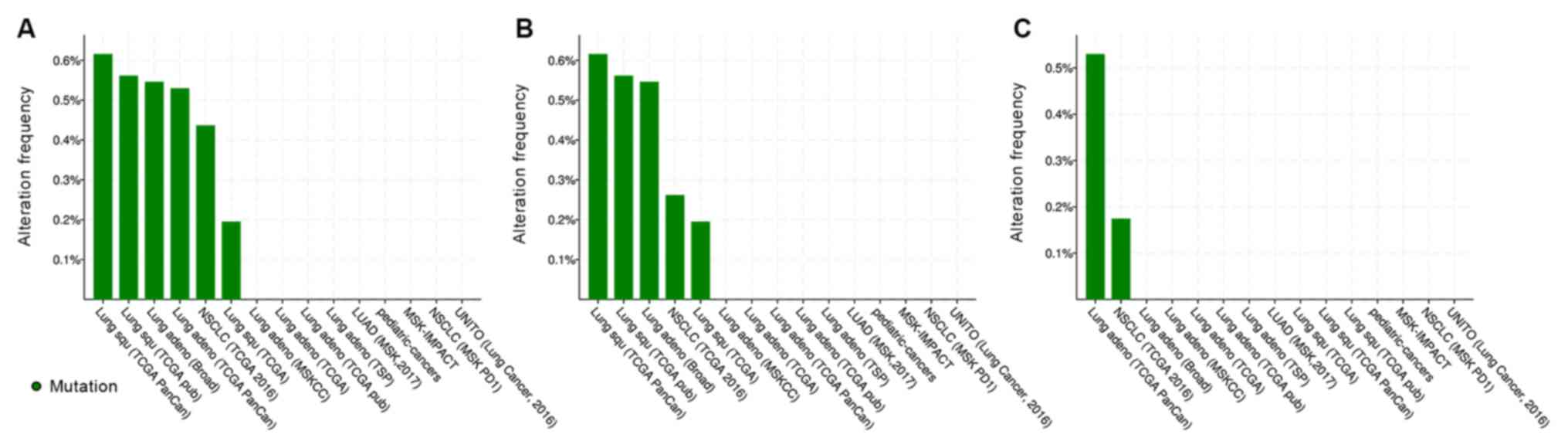
Mutation analysis
The mutation status of CCR9 and CCL25 in lung tumor
tissues was examined using the public database of cBioPortal for
Cancer Genomics (Fig. 6). As
presented in Fig. 6B, CCR9 had a
somatic mutation rate of 0.26% in NSCLC (3/1,144) in a TCGA 2016
study (18), 0.55% in lung AC
(1/183) in the Imielinski et al (19) study. For lung SCC, the CCR9
mutational rates were found to be 0.62 (3/487), 0.56 (1/178) and
0.2% (1/511) in TCGA PanCan (20),
TCGA pub (21) and TCGA studies
(http://gdac.broadinstitute.org/runs/stddata_2016_01_28/data/LUSC/20160128/gdac.broadinstitute.org_LUSC.Mutation_Packager_Calls.Level_3.2016012800.0.0.tar.gz),
respectively.
CCL25 had a somatic mutation rate of 0.17% (2/1,144)
in NSCLC data from TCGA 2016 study and 0.53% (3/566) in lung AC
data from TCGA PanCan study (Fig.
6C). Altogether, these results suggested that mutation of CCR9
and CCL25 is a rare occurrence in patients with NSCLC. This is in
stark contract with epidermal growth factor receptor (EGFR), which
has a mutational rate of ~10% in Caucasian patients with NSCLC and
≤50% of Asian patients with NSCLC (22-24),
or ALK receptor tyrosine kinase (ALK) with a 3-5% rate in patients
with NSCLC (25).
Discussion
In the present study, the CCL25/CCR9 signaling axis
was demonstrated to regulate the expression of VEGF-C, VEGF-D,
MMP-1 and MMP-7, and may promote the invasion and migration of the
lung cancer cells. Survival analysis demonstrated that patients
with lower expression of CCL25 or CCR9 in their tumors displayed
better prognosis.
Chemokines are known mediators of leukocyte
trafficking and host defense (26),
Indeed, previous studies have determined that the involvement of
chemokine receptors is of importance in patient prognosis (27), apoptosis (28) and metastatic (29) signaling machinery in various cancer
types. Among all the chemokine receptors, studies on the role of
CXCR4 have been more extensive. These previous studies suggested
that CXCR4 (8,12,30-32)
was highly expressed in NSCLC, and functional blockade of this
interaction could inhibit metastasis to the bone marrow, lymph
nodes or pleural space. These findings highlight the effect of
chemokine/chemokine receptor signaling on the metastatic potential
of NSCLC.
Several steps are required to achieve metastasis,
including active migration, extracellular matrix degradation and
adhesion to vascular endothelial cells. Migration and invasion
associated with metastatic potential can be triggered by chemokine
binding to chemokine receptor on the cell surface (33,34).
Tumor lymphangiogenesis was previously found to be associated with
the VEGF-C/VEGF-D/VEGF receptor-3 (VEGFR-3) signaling axis
(35,36). Previous studies have determined
VEGF-C activates VEGFR-3, which in turn promotes proliferation
(37), migration (38) and apoptosis protection (39). In various cancer types, the tumor
cells produce VEGF-C and recruit monocytes or macrophages into
tumor tissue (40). These monocytes
and macrophages differentiate to M2-polarized tumor-associated
macrophages, which also produce VEGF-C, and further increase
lymphatic vessel development (41).
Moreover, lymph-angiogenic factors derived from normal lymphatic
cells can reprogram the gene expression profile of these cells and
convert them to tumor-derived lymphatic cells during tumor
development and progression (42).
Tumor-derived lymphatic cells express specific lymphatic markers,
such as VEGFR-3 and lymphatic vessel endothelial receptor 1, and
form a lymphatic system in vivo (43).
VEGF-C and VEGF-D, members of the VEGF family, have
been demonstrated to stimulate the proliferation of lymphatic
endothelial cells, and to promote lymphatic invasion and lymph node
metastasis through VEGFR-3 signaling, which is critical for the
growth of lymphatic vessels (44,45).
These findings provided theoretical evidence for the role of VEGF-C
and VEGF-D in promoting cancer metastasis (46). In the present study, VEGF expression
was higher in NSCLC tumors compared with the tumor-adjacent normal
tissues, and CCL25 treatment upregulated the expression of VEGF-C
and -D in lung cancer cell lines, which could be blocked by the
anti-CCR9 antibody. The aforementioned results were consistent with
previous cancer-related studies on VEGF (47,48).
In addition to VEGF-C and -D, the metalloproteinases
MMP-1 and MMP-7, which serve as predominant components in the
degradation of collagenous extracellular matrix, play a critical
role in cancer biology, including cell migration, invasion and
angiogenesis (49,50). Previous studies (51-53)
have demonstrated an important role for MMP-1 and MMP-7 in the
development of various cancer types; however, the effect of MMP-1
and MMP-7 have not been well characterized in NSCLC. In the present
study, it was observed that the migration and invasion of lung
cancer cells was enhanced by CCL25 treatment. This provides insight
on a potential association between the CCL25/CCR9 interaction, and
the migration and invasion of lung cancer cells. CCL25 was
identified to influence the secretion of VEGF-C, VEGF-D, MMP-1 and
MMP-7 in NSCLC cells, which influenced migratory and invasive
potential in vitro. In addition, low expression of CCR9 and
CCL25 is associated with good OS in NSCLC. In consideration of our
previous study (14) and the present
study, the CCL25/CCR9 axis may promote the invasion and migration
of cancer cells through modulation of VEGF-C, VEGF-D, MMP-1 and
MMP-7.
In the present study, another notable result was the
low mutation of CCR9 and CCL25, compared with EGFR. For clinical
therapy, the effectiveness of either ALK or EGFR inhibitors is
restricted by the eventual occurrence of drug resistance despite
the initial promising responses. Considering the low mutation rates
of CCL25 and CCR9 in NSCLC, future research should focus on
targeting the CCL25/CCR9 axis.
There are several limitations in the present study.
Firstly, only a blocking antibody was used to validate the
expression of CCL25 and CCR9 on lung cancer cell lines. Additional
methods of altering the signaling pathway, such as with gene
expression manipulation with lentivirus, for example, would
additionally be informative. In vivo animal experiments
would also be valuable in helping to understand this signaling
pathway in lung cancer cells. Investigating the effect of
CCL25/CCR9 signaling on transcription factor interactions using
chromatin immunoprecipitation analysis could also improve the
understanding of the pathway in lung cancer cells. Finally, the OS
benefit of CCL25 may be re-evaluated by expanding the number of
samples, and to examine the potential reasons for the different OS
benefits observed between CCR9 and CCL25 in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Research
Projects of Shanghai Health and Planning Commission (grant no.
201740015); Shanghai Municipal Natural Science Foundation (grant
no. 18ZR1413000); and Natural Science Foundation of China (grant
no. 81560383).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YN and DT wrote the manuscript and analyzed the
data. YN and LF performed the experiments. YN contributed to the
conceptual idea for the manuscript. WG and HL contributed to study
design and manuscript editing. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study protocol was approved by The
Ethics Committee Review Board of The Huadong Hospital Affiliated
with Fudan University (approval no. 20180046), and written informed
consent was obtained from every participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Fidler IJ: Critical factors in the biology
of human cancer metastasis: Twenty-eighth GHA Clowes memorial award
lecture. Cancer Res. 50:6130–6138. 1990.PubMed/NCBI
|
|
3
|
Fidler IJ: Critical determinants of cancer
metastasis: Rationale for therapy. Cancer Chemother Pharmacol. 43
(43 Suppl):S3–S10. 1999.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Strieter RM: Chemokines: Not just
leukocyte chemoattractants in the promotion of cancer. Nat Immunol.
2:285–286. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Stein JV and Nombela-Arrieta C: Chemokine
control of lymphocyte trafficking: A general overview. Immunology.
116:1–12. 2005.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Olson TS and Ley K: Chemokines and
chemokine receptors in leukocyte trafficking. Am J Physiol
Regulatory Integrative Comparative Physiol. 283:R7–R28.
2002.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Saintigny P, Massarelli E, Lin S, Ahn YH,
Chen Y, Goswami S and Zhang L: CXCR2 expression in tumor cells is a
poor prognostic factor and promotes invasion and metastasis in lung
adenocarcinoma. Cancer Res. 73:571–582. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Su L, Zhang J, Xu H, Wang Y, Chu Y, Liu R
and Xiong S: Differential expression of CXCR4 is associated with
the metastatic potential of human non-small cell lung cancer cells.
Clin Cancer Res. 11:8273–8280. 2005.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hald SM, Kiselev Y, Al-Saad S, Richardsen
E, Johannessen C, Eilertsen M and Bremnes RM: Prognostic impact of
CXCL16 and CXCR6 in non-small cell lung cancer: Combined high
CXCL16 expression in tumor stroma and cancer cells yields improved
survival. BMC Cancer. 15(441)2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mukaida N, Sasaki SI and Baba T:
Chemokines in cancer development and progression and their
potential as targeting molecules for cancer treatment. Mediators
Inflamm. 2014(170381)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhu YM, Webster SJ, Flower D and Woll PJ:
Interleukin-8/CXCL8 is a growth factor for human lung cancer cells.
Br J Cancer. 91(1970)2004.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Phillips RJ, Burdick MD, Lutz M, Belperio
JA, Keane MP and Strieter RM: The stromal derived
factor-1/CXCL12–CXC chemokine receptor 4 biological axis in
non-small cell lung cancer metastases. Am J Respir Crit Care Med.
167:1676–1686. 2003.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gupta P, Sharma PK, Mir H, Singh R, Singh
N, Kloecker GH and Singh S: CCR9/CCL25 expression in non-small cell
lung cancer correlates with aggressive disease and mediates key
steps of metastasis. Oncotarget. 5(10170)2014.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li B, Wang Z, Zhong Y, Lan J, Li X and Lin
H: CCR9-CCL25 interaction suppresses apoptosis of lung cancer cells
by activating the PI3K/Akt pathway. Med Oncol.
32(66)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhong Y, Jiang L, Lin H, Li B, Lan J,
Liang S, Shen B, Lei Z and Zheng W: Expression of CC chemokine
receptor 9 predicts poor prognosis in patients with lung
adenocarcinoma. Diagn Pathol. 10(101)2015.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8(e82241)2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Campbell JD, Alexandrov A, Kim J, Wala J,
Berger AH, Pedamallu CS and Imielinski M: Distinct patterns of
somatic genome alterations in lung adenocarcinomas and squamous
cell carcinomas. Nat Genet. 48(607)2016.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E and Sougnez C: Mapping the hallmarks
of lung adenocarcinoma with massively parallel sequencing. Cell.
150:1107–1120. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ellrott K, Bailey MH, Saksena G, Covington
KR, Kandoth C, Stewart C and Sofia HJ: Scalable open science
approach for mutation calling of tumor exomes using multiple
genomic pipelines. Cell Systems. 6:271–281. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cancer Genome Atlas Research Network:
Comprehensive genomic characterization of squamous cell lung
cancers. Nature 489: 519, 2012.
|
|
22
|
Skov BG, Høgdall E, Clementsen P, Krasnik
M, Larsen KR, Sørensen JB and Mellemgaard A: The prevalence of EGFR
mutations in non-small cell lung cancer in an unselected Caucasian
population. APMIS. 123:108–115. 2015.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shi Y, Li J, Zhang S, Wang M, Yang S, Li N
and Zheng M: Molecular epidemiology of EGFR mutations in Asian
patients with advanced non-small-cell lung cancer of adenocarcinoma
histology-Mainland China subset analysis of the PIONEER study. PLoS
One. 10(e0143515)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hirsch FR and Bunn PA Jr: EGFR testing in
lung cancer is ready for prime time. Lancet Oncol. 5:432–433.
2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Haratake N, Seto T, Takamori S, Toyozawa
R, Nosaki K, Miura N, Ohba T, Toyokawa G, Taguchi K, Yamaguchi M,
et al: Short progression-free survival of ALK inhibitors sensitive
to secondary mutations in ALK-positive NSCLC patients. Thorac
Cancer. 10:1779–1787. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zlotnik A and Yoshie O: Chemokines: A new
classification system and their role in immunity. Immunity.
12:121–127. 2000.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Nishikawa G, Kawada K, Nakagawa J, Toda K,
Ogawa R, Inamoto S and Sakai Y: Bone marrow-derived mesenchymal
stem cells promote colorectal cancer progression via CCR5. Cell
Death Dis. 10(264)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Zhu F, Liu Y, Liu L, Zhou Y, Zhou P, Yan Q
and Ding S: CKLF1 enhances Inflammation-mediated carcinogenesis and
prevents Doxorubicin-induced apoptosis via IL-6/STAT3 signaling in
HCC. Clin Cancer Res. 25:4141–4154. 2019.
|
|
29
|
Steele CW, Karim SA, Leach JD, Bailey P,
Upstill-Goddard R, Rishi L and Eberlein C: CXCR2 inhibition
profoundly suppresses metastases and augments immunotherapy in
pancreatic ductal adenocarcinoma. Cancer Cell. 29:832–845.
2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Oonakahara KI, Matsuyama W, Higashimoto I,
Kawabata M, Arimura K and Osame M: Stromal-derived
factor-1alpha/CXCL12-CXCR 4 axis is involved in the dissemination
of NSCLC cells into pleural space. Am J Respir Cell Mol Biol.
30:671–677. 2004.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yusen W, Xia W, Shengjun Y, Shaohui Z and
Hongzhen Z: The expression and significance of tumor associated
macrophages and CXCR4 in non-small cell lung cancer. J BUON.
23:398–402. 2018.PubMed/NCBI
|
|
32
|
Choi YH, Burdick MD, Strieter BA, Mehrad B
and Strieter RM: CXCR4, but not CXCR7, discriminates metastatic
behavior in non-small cell lung cancer cells. Mol Cancer Res.
12:38–47. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Siegel G, Malmsten M and Klüssendorf D:
Tumor cell locomotion and metastatic spread. Microsc Res Tech.
43:276–282. 1998.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Navolotski A, Rumjnzev A, Lü H, Proft D,
Bartholmes P and Zänker KS: Migration and gap junctional
intercellular communication determine the metastatic phenotype of
human tumor cell lines. Cancer Lett. 118:181–187. 1997.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Achen MG and Stacker SA: Molecular control
of lymphatic metastasis. Ann N Y Acad Sci. 1131:225–234.
2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yonemura Y, Endo Y, Tabata K, Kawamura T,
Yun HY, Bandou E, Sasaki T and Miura M: Role of VEGF-C and VEGF-D
in lymphangiogenesis in gastric cancer. Int J Clin Oncol.
10:318–327. 2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Marchiò S, Primo L, Pagano M, Palestro G,
Albini A, Veikkola T, Cascone I, Alitalo K and Bussolino F:
Vascular endothelial growth factor-C stimulates the migration and
proliferation of Kaposi's sarcoma cells. J Biol Chem.
274:27617–27622. 1999.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Timoshenko AV, Rastogi S and Lala PK:
Migration-promoting role of VEGF-C and VEGF-C binding receptors in
human breast cancer cells. Br J Cancer. 97:1090–1098.
2007.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhao L, Zhu Z, Yao C, Huang Y, Zhi E, Chen
H, Tian R, Li P, Yuan Q, Xue Y, et al: VEGFC/VEGFR3 signaling
regulates mouse Spermatogonial cell proliferation via the
activation of AKT/MAPK and cyclin D1 pathway and mediates the
apoptosis by affecting caspase 3/9 and Bcl-2. Cell Cycle.
17:225–239. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Schmid MC and Varner JA: Myeloid cell
trafficking and tumor angiogenesis. Cancer Lett. 250:1–8.
2007.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Movahedi K, Laoui D, Gysemans C, Baeten M,
Stangé G, Van den Bossche, Mack M, Pipeleers D, In't Veld P, De
Baetselier P and Van Ginderachter JA: Different tumor
microenvironments contain functionally distinct subsets of
macrophages derived from Ly6C(high) monocytes. Cancer Res.
70:5728–5739. 2010.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Volk-Draper LD, Hall KL, Wilber AC and Ran
S: Lymphatic endothelial progenitors originate from plastic myeloid
cells activated by toll-like receptor-4. PLoS One.
12(e0179257)2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Alitalo A and Detmar M: Interaction of
tumor cells and lymphatic vessels in cancer progression. Oncogene.
31(4499)2012.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Achen MG and Stacker SA: Vascular
endothelial growth factor-D: Signaling mechanisms, biology, and
clinical relevance. Growth Factors. 30:283–296. 2012.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Stacker SA, Achen MG, Jussila L, Baldwin
ME and Alitalo K: Metastasis: Lymphangiogenesis and cancer
metastasis. Nature Rev Cancer. 2(573)2002.PubMed/NCBI View
Article : Google Scholar
|
|
47
|
Chen Y, Liu Y, Wang Y, Li W, Wang X, Liu
X, Chen Y, Ouyang C and Wang J: Quantification of STAT3 and VEGF
expression for molecular diagnosis of lymph node metastasis in
breast cancer. Medicine (Baltimore). 96(e8488)2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Guo J, Lou W, Ji Y and Zhang S: Effect of
CCR7, CXCR4 and VEGF-C on the lymph node metastasis of human
pancreatic ductal adenocarcinoma. Oncol Lett. 5:1572–1578.
2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Sánchez-Lorencio MI, Saenz L, Ramirez P,
Villalba-López F, de la Orden V, Mediero-Valeros B, Revilla Nuin B,
Gonzalez MR, Cascales-Campos PA, Ferreras-Martínez D, et al: Matrix
metalloproteinase 1 as a novel biomarker for monitoring
hepatocellular carcinoma in liver transplant patients. Transplant
Proc. 50:623–627. 2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Gao Y, Nan X, Shi X, Mu X, Liu B, Zhu H,
Yao B, Liu X, Yang T, Hu Y and Liu S: SREBP1 promotes the invasion
of colorectal cancer accompanied upregulation of MMP7 expression
and NF-κB pathway activation. BMC Cancer. 19(685)2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Yokoi A, Yoshioka Y, Yamamoto Y, Ishikawa
M, Ikeda SI, Kato T, Kiyono T, Takeshita F, Kajiyama H, Kikkawa F
and Ochiya T: Malignant extracellular vesicles carrying MMP1 mRNA
facilitate peritoneal dissemination in ovarian cancer. Nat Commun.
8(14470)2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sizemore ST, Sizemore GM, Booth CN,
Thompson CL, Silverman P, Bebek G, Abdul-Karim FW, Avril S and Keri
RA: Hypomethylation of the MMP7 promoter and increased expression
of MMP7 distinguishes the basal-like breast cancer subtype from
other triple-negative tumors. Breast Cancer Res Treat. 146:25–40.
2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Yuan S, Lin LS, Gan RH, Huang L, Wu XT,
Zhao Y, Su BH, Zheng D and Lu YG: Elevated matrix metalloproteinase
7 expression promotes the proliferation, motility and metastasis of
tongue squamous cell carcinoma. BMC Cancer. 20(33)2020.PubMed/NCBI View Article : Google Scholar
|