Introduction
Gastric cancer commonly develops within the lining
of the stomach (1). The diagnosis
rate has recently greatly increased with ~28,000 new cases of
gastric cancer diagnosed in the USA during 2017 according to the
National Cancer Institute (2). The
mechanism for the development and progression of gastric cancer is
not fully understood which hampers the search for novel treatments
(3). Chemotherapy and radiotherapy
are the most commonly used approaches to treat gastric cancer
despite being time-consuming and susceptible to development of drug
resistance (4). The mechanisms of
gastric cancer were reported to involve the increasing platinum-DNA
adducts and apoptosis-regulating genes (5). It is of great value to understand the
underlying mechanisms to accelerate the search for a new
therapeutic target for gastric cancer.
MicroRNAs (miRs) are a class of small,
single-stranded and non-protein-coding RNAs. They are implicated in
the expression of mRNA (6) and
regulation of various biological activities such as tumor
development, cardiac diseases and hepatitis (7). Recent studies have demonstrated that
miRs facilitated cancer progression by targeting specific genes
(8,9)
and some functional miRs may be used to treat cancer. A common type
of miR, miR-454, has been identified to be involved in the
development of cancer (10).
Expression of miR-454 is downregulated in osteosarcoma and
glioblastoma and thus recognized as a potential tumor suppressor
(11,12). By contrast, miR-454 is overexpressed
in colorectal cancer, uveal melanoma and hepatocellular carcinoma
suggesting that it could also be an oncogene (13,14).
However, the relationship between miR-454 and gastric cancer
remains uninvestigated.
The present study demonstrated that miR-454 promoted
cellular proliferation and inhibited apoptosis of SGC-7901 gastric
carcinoma cells, through downregulation of CYLD. It was identified
that miR-454 could also induce oxaliplatin (OXA) resistance of
gastric carcinoma cell via targeting CYLD. Taken together, the
results suggested that miR-454 may be a potential novel therapeutic
target of gastric cancer.
Materials and methods
Cell culture and transfection
Human SGC-7901 gastric cancer cells were obtained
from the Cell Bank of the Guangzhou Institute of Biochemistry and
Cell Biology (Guangzhou, China). The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM), supplemented with fetal
bovine serum (FBS) and incubated at 37˚C in a humidified atmosphere
containing 5% CO2.
The control (scramble) and miR-454 mimics, and the
miR-454 inhibitor and its associated negative control, were
synthesized by Shanghai GenePharma Co., Ltd. and added to cells at
a concentration of 20 nmol/l per oligonucleotide. The sequences
were as follows: miR-454 mimic, 5'-UGGGAUAUUCGCUGGAAUCCUCAU-3';
miR-454 inhibitor, 5'-UCACAUAGGAAUAAAAAGCCAUA-3'; inhibitor
control, 5'-CAGUACUUUUGUGUAGUACAA-3'. Cells were collected 48 h
post-transfection for subsequent experimentation. DharmaFECT1
reagent (GE Healthcare Dharmacon, Inc.) was used to perform the
transfections according to manufacturer's protocol.
For the ectopic expression of CYLD, CYLD ORFs with
3'-UTR was amplified from the RNA samples of HepG2 or BEL-7402
cells using PCR and subcloned into the two restriction sites of
pEGFP-C1 (Clontech Laboratories, Inc.) and pGL3 vectors (Promega
Corporation). The primers used were as follows: CYLD-3'UTR-GFP
forward, 5'-GCCCTCGAGCTTGACTCCGTTCCCCTTCAGAC-3' and reverse,
5'-GCCGGATCCAACCAAGGGCAGTTGAGTC-3' and CYLD-3' UTR-luc forward,
5'-GCCCCGCGGCTCCGTTCCCCTTCAGAC-3' and reverse,
5'-GCCCTGCAGAACCAAGGGCAGTTGAGTC-3'. Reporter gene assay was
initiated 24 h post-transfection. A total of 5 µg plasmids were
used per transfection reaction. Lipofectamine® 3000
transfection reagent was used in this study (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol®
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was
synthesized using High-Capacity cDNA reverse transcriptase kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. The expression of mRNA and miRNA were quantified using
TaqMan™ Universal PCR Master Mix (Invitrogen; Thermo
Fisher Scientific, Inc.) by RT-qPCR in the ABI 7500 System (Bio-Rad
Laboratories, Inc.) according to manufacturer's protocols. The
primers selected are as follows: CYLD forward,
5'-TCCTCTCCAAAATGCCAGAG-3' and reverse, 5'-GGCGGATTGGAAATGAACTT-3';
miR-454 forward, 5'-TCAAGAGGCGAACACACAAC-3' and reverse,
5'-GGCCTTTTCATTGTTTTCCA-3'; GAPDH forward,
5'-GACTCATGACCACAGTCCATGC-3' and reverse,
3'-AGAGGCAGGGATGATGTTCTG-5' and U6 snRNA forward,
5'-ATACAGAGAAAGTTAGCACGG-3' and reverse,
3'-GGAATGCTTCAAAGAGTTGTG-5'. GAPDH and U6 snRNA were selected as
the internal control for mRNA and miRNAs, respectively. The
thermocycling conditions were as follows: Initial denaturation at
95˚C for 20 sec, followed by 45 cycles of 95˚C for 10 sec and 70˚C
for 5 sec. Relative expression levels were calculated using the
2-ΔΔCq method (15).
Cell proliferation assay
SGC-7901 cancer cells were seeded in 96-well plates
(4x103 cells/well). CCK-8 solution (Dojindo Molecular
Technologies, Inc.) was added to each well at 24, 48, 72, 96 and
120 h following cell seeding. The number of viable cells was then
counted at a wavelength of 450 nm following the manufacturer's
protocol.
Colony formation assay
SGC-7901 human gastric cancer cells were seeded into
6-well plates at a density of 1,000 cells/well and incubated for 12
days. The colonies were fixed with formaldehyde for 20 min then
stained with 1% crystal violet (Sigma-Aldrich; Merck KGaA) for 30
sec. Colonies with over 50 cells were counted.
Flow cytometric analysis of
apoptosis
Cells were stained with Annexin V-fluorescein
isothiocyanate (Clontech Laboratories, Inc.) and
7-amino-actinomycin D (10 µg/ml; Sigma-Aldrich; Merck KGaA) at 4˚C
for 30 min and were measured using BD FACSCaliburTM
system (Becton-Dickinson; BD Biosciences). The experiment was
repeated three times and the data was analyzed with Multi Cycle
Flow System Software (V-321; Phoenix flow systems, Inc.). Cell
apoptosis was examined with Annexin V FITC/7-amino-actinomycin D
double staining.
Luciferase activity assay
Luciferase activity assay was performed with the
Dual-Luciferase Reporter Assay System (Promega Corporation). Cells
were seeded into 96-well plates at 4,000 cells/well. The
pGL3-luciferase reporter gene plasmids pGL3-CYLD-3'-UTR mutant
(Mut) or pGL3-CYLD-3'-UTR wild-type (WT) were purchased from
Guangzhou RiboBio Co., Ltd. They were co-transfected (5 µg) into
SGC-7901 human gastric cancer cells, along with Renilla
plasmid and miR-NC (negative control; 100 nmol/l) or miR-454 (100
nmol/l) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Firefly luciferase activity of the
transfection efficiency was normalized to Renilla luciferase
activity.
Bioinformatics analysis
The public web-based prediction site TargetScan 7.1
(http://www.targetscan.org) was used to
predict the potential miRNA-targeted gene transcripts.
Western blot analysis
Total proteins were extracted with
radioimmunoprecipitation assay buffer (1% NP-40, 0.5% sodium
deoxycholate, 50 mM Tris-HCl, 0.1% SDS, 150 mM NaCl, pH 7.5)
supplemented with 1% protease inhibitor. The solution was
centrifuged at 15,000 x g at 4˚C for 5 min, after which the
supernatant was collected. Protein concentrations were quantified
using Bio-Rad protein assay (Bio-Rad Laboratories, Inc.). The
target protein (30 µg) was separated by 10% sodium dodecyl sulfate
polyacrylamide gel electrophoresis and transferred onto the
nitrocellulose membranes (Bio-Rad Laboratories, Inc.). The membrane
was blocked with 5% non-fat milk solution for 1 h at 4˚C and
incubated with anti-GAPDH (1:500; cat. no. ab181602; Abcam) and
anti-CYLD (1:1,000; cat. no. 4495; Cell Signaling Technology, Inc.)
primary antibodies at 4˚C for 12 h, washed briefly and then
incubated with a horseradish peroxidase-conjugated secondary
antibody (1:1,000; cat. no. sc-2357; Santa Cruz Biotechnology,
Inc.) at 4˚C for 2 h. Intensity of bands was analyzed using
Image-Pro Plus 6.0 software (Media Cybernetics, Inc.) with
chemiluminescence (Amersham Pharmacia Biotech; GE Healthcare) with
GAPDH as the loading control.
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical significance was assessed using unpaired two-tailed
Student's t-test for comparisons between two groups or using
one-way analysis of variance followed by Newman-Keuls comparison
for more than three groups. Data were processed with GraphPad Prism
v.8.0 (GraphPad Software, Inc.). P<0.05 was considered to
indicate statistical significance.
Results
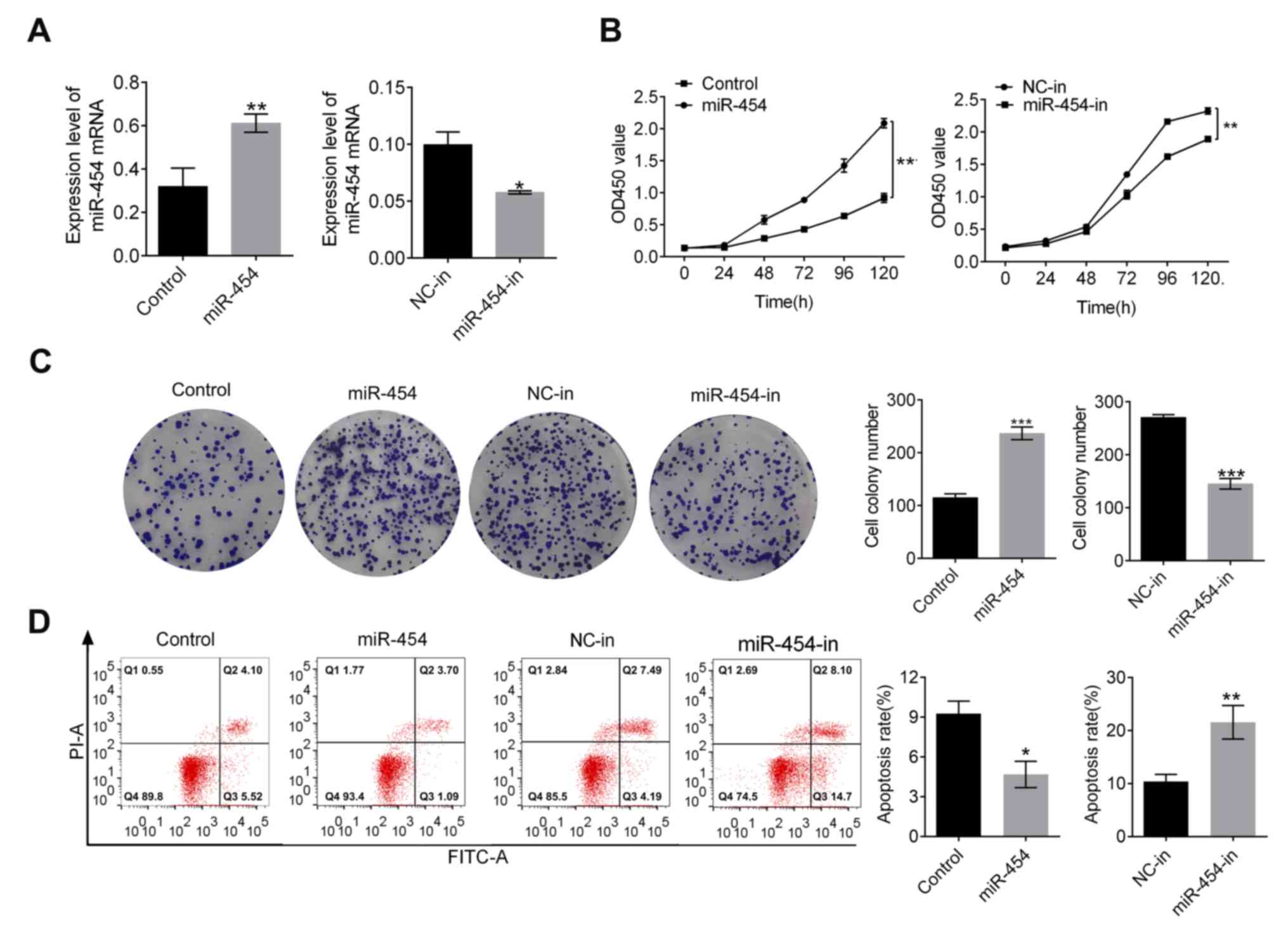
miR-454 promotes proliferation and
inhibits apoptosis in gastric cancer cells
Expression of miR-454 in SGC-7901 gastric cancer
cells was measured post-transfection (Fig. 1A). Compared with the control group,
miR-454 expression significantly increased upon transfection
(P<0.01; Fig. 1A) but
significantly decreased upon transfection with miR-454 inhibitor
(P<0.05; Fig. 1A). At 120 h,
proliferation of the miR-454 groups was significantly higher
compared with the control group. Proliferation of the
inhibitor-transfected group was significantly lower than the NC-in
group (P<0.01; Fig. 1B). The
number of colonies of SGC-7901 increased with the expression of
miR-454 (P<0.001, Fig. 1C) but
decreased in the miR-454-in group compared with the NC-in group
(P<0.001, Fig. 1C). In addition,
miR-454 expression was related to SGC-7901 cell apoptosis rate
(Fig. 1D) where the control group
demonstrated much higher apoptosis rate than the miR-454 group
(P<0.05; Fig. 1D) whilst the
NC-in group demonstrated a decreased apoptosis rate compared with
the miR-454-in group (P<0.01; Fig.
1D). Taken together, these findings suggested that miR-454
suppressed apoptosis in SGC-7901 cells.
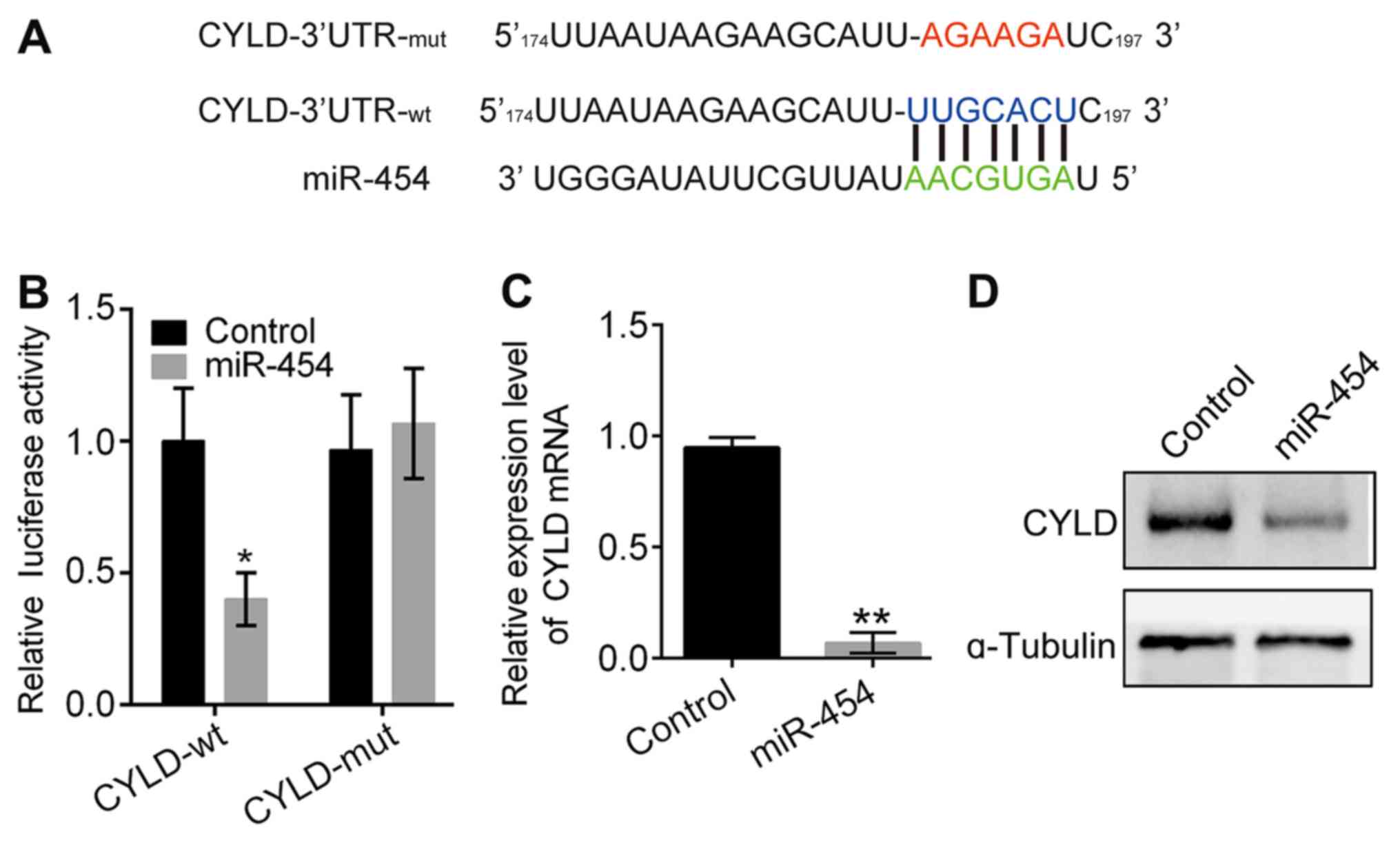
miR-454 directly downregulates
CYLD
CYLD was predicted to be a miR-454 target gene,
based on analysis with TargetScan (http://www.targetscan.org/vert_72/) (Fig. 2A). The relationship between CYLD and
miR-454 was examined by the luciferase reporter assay (Fig. 2B) Results determined that luciferase
activity of the wild-type construct was significantly reduced by
miR-454 (P<0.05; Fig. 2B), but
there was no significant effect observed with the mutant construct.
These results were further confirmed by data demonstrating that the
expression of CYLD mRNA and protein was inhibited by miR-454
overexpression (Fig. 2C and D).
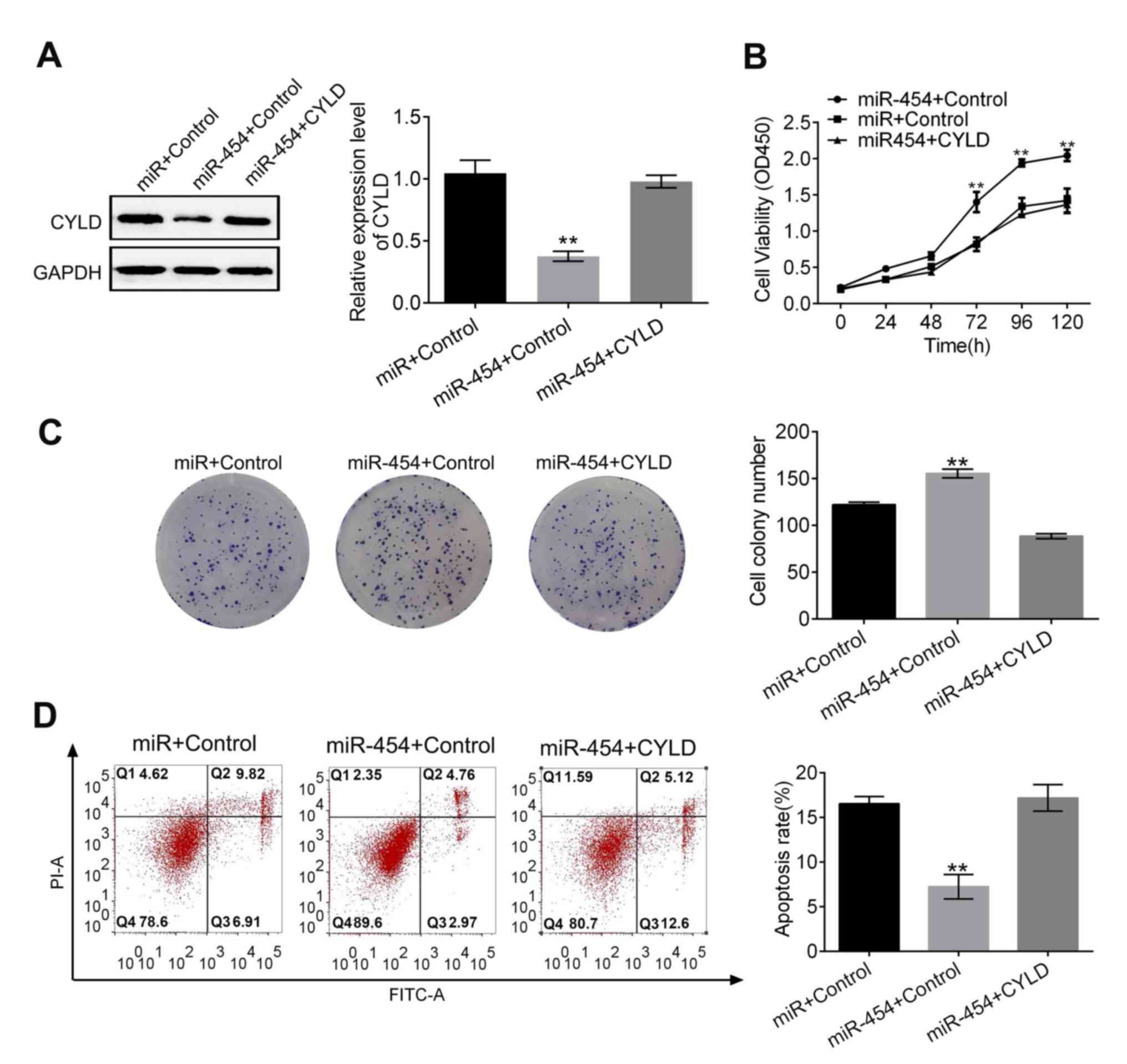
miR-454 promoted proliferation and
inhibited apoptosis in gastric cancer cells via targeting CYLD
Western blot analysis determined that the
miR-454-induced inhibition of CYLD was reversed upon the
transfection of pCDNA CYLD into the SGC-7901 cells (Fig. 3A). Results for the proliferation
assays, colony formation and apoptosis assays demonstrated that
co-transfection of miR-454 mimic and pCDNA CYLD counteracted the
effects of miR-454 on cell proliferation and apoptosis (Fig. 3B-D). These results suggested that the
effects of miR-454 on gastric cancer cells were mediated by
downregulating CYLD.
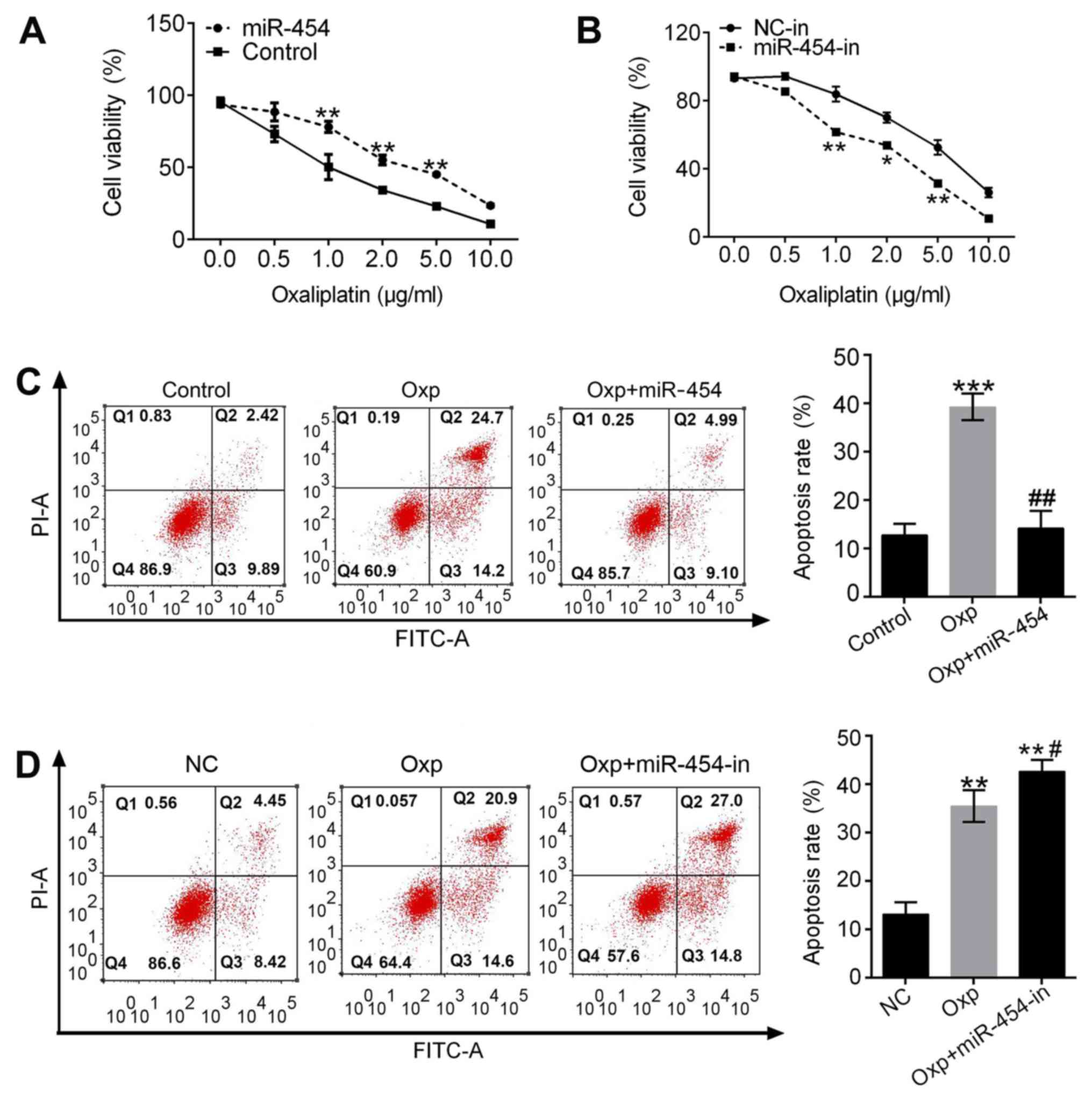
miR-454 enhances oxaliplatin
resistance in gastric cancer cells
SGC-7901 cells in the miR-454 group exhibited higher
cell viability than the control group following treatment with
oxaliplatin (Fig. 4A). Cell
viability of the miR-454-in group was lower than the NC-in group
following oxaliplatin treatment (Fig.
4B). Apoptosis rate was significantly higher for the
oxaliplatin group compared with the controls (P<0.001; Fig. 4C). Overexpression of miR-454 reversed
the effect of oxaliplatin (Fig. 4C).
Apoptosis rates of the oxaliplatin and Oxp + miR-454-in groups
increased (P<0.01; Fig. 4D) with
the Oxp+miR-454-in group higher than the oxaliplatin group
(P<0.05, Fig. 4D). These results
suggested that drug resistance was reversed by inhibiting miR-454
and that miR-454 decreased the sensitivity of SGC-7901 cells to
oxaliplatin.
Discussion
The present study evaluated the effects of miR-454
on gastric cancer cell proliferation and explored the corresponding
mechanism. The effect of miR-454-induced oxaliplatin resistance was
also observed. Overexpression of miR-454 increased the number of
gastric cancer cell colonies formed and decreased apoptosis rate,
which indicated that miR-454 promoted proliferation. CYLD was
directly downregulated by miR-454 thus increasing the survival rate
of the SGC-7901 cells. These results suggested that the influence
of miR-454 on SGC-7901 cells was achieved via downregulating CYLD.
It was also determined that miR-454 reduced the sensitivity of
SGC-7901 cells to oxaliplatin, which may further influence the
therapeutic effects of oxaliplatin on gastric cancer. The present
study provided preliminary evidence for the effect of miR-454 on
gastric carcinoma cells and could be used to develop a novel
approach for gastric cancer treatment.
Regulation of miRNA expression has recently been
used for the treatment of various types of tumor (16,17). The
regulatory function of miRNAs in gastric cancer remains not fully
understood therefore it would be beneficial to study the role of
miR-454 in regards to this condition. Overexpression of miR-454
promoted proliferation of SGC-7901 cells whilst inhibition of
miR-454 decreased it which suggested that inhibiting miR-454 may be
a potential treatment approach to gastric cancer.
Effects of miR-454 on the progression of various
cancers have been comprehensively reported. Inhibition of miR-454
suppresses the invasion and proliferation of hepatoma cells by
inhibiting chromodomain helicase DNA binding protein 5(14). Tumorigenesis of lung cancer and uveal
melanoma is accelerated by miR-454 via targeting the phosphatase
and tensin homolog deleted on chromosome ten (13,10). The
tumor-suppressing functions of miR-454 have been demonstrated
including the inhibition of glioma cell proliferation by
miR-454(11); inhibition of
osteosarcoma invasion and proliferation by miR-454 via inhibiting
c-Met (12); and cellular radiated
susceptibility of renal carcinoma cells promoted by miR-454 via
targeting the cell translocation gene (18). The present study demonstrated that
miR-454 could regulate the survival of gastric cancer cells.
CYLD is a deubiquitination enzyme that functions as
a regulator in the NF-κB and transforming growth factor-β signaling
pathways (19), and also as a tumor
suppressor (20). It has been
reported that CYLD inhibited c-Jun N-terminal kinase and
uncontrolled activation of TGF-β activated kinase 1, which, in turn
regulated hepatocyte homeostasis (21,22). The
present study determined that CYLD was a target gene of miR-454.
The results demonstrated that miR-454 downregulated CYLD mRNA and
protein expression which increased the apoptosis rate of cancer
cells.
The overexpression of miR454 in gastric cancer
tissues has been previously reported (23) where it was hypothesized that
overexpression of miR-454 was associated with the advancement of
the clinical stage, metastases of the lymph node and a poor
prognosis for patients. CYLD acts as a suppressor in many types of
tumor including breast cancer and non-melanoma skin cancer
(24,25). Both miR-425-5p and miR-20a promote
the progression of gastric cancer via regulating CYLD (26,27).
Oxaliplatin is an effective chemotherapeutic drug
commonly used for treating gastric carcinoma (28). Despite the high remission rate, most
patients will ultimately develop resistance to the
oxaliplatin-based chemotherapy treatment (29) with the underlying mechanism not fully
understood. The present study demonstrated that downregulation of
miR-454 could improve the outcome of oxaliplatin treatment on
SGC-7901 cells. The in vitro study suggested miR-454
increased stomach cell proliferation and induced drug resistance to
oxaliplatin by directly targeting CYLD. It would be of use to carry
out in vivo studies to confirm the results, as the in
vitro study (i.e., cancer cell lines) might not accurately
reflect the in vivo environment however findings
demonstrated that miR-454 might be a novel therapeutic target for
gastric cancer.
Acknowledgements
Not applicable.
Funding
This work was supported by the Science Fund of the
National Natural Science Foundation of China (grant no. 81502113)
and Wuhan Clinical Research Center for Peritoneal Carcinomatosis
(grant no. 2015060911020462).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CQH and JYL performed the experiments and wrote the
manuscript. XKP collected and analyzed the experimental data. CWP
and BX analyzed the experimental data and revised the manuscript.
MHF and XJY designed the experiments and approved the final version
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Oliveira C, Pinheiro H, Figueiredo J,
Seruca R and Carneiro F: Familial gastric cancer: Genetic
susceptibility, pathology, and implications for management. Lancet
Oncol. 16:e60–e70. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wadhwa R, Song S, Lee JS, Yao Y, Wei Q and
Ajani JA: Gastric cancer-molecular and clinical dimensions. Nat Rev
Clin Oncol. 10:643–655. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Goetze OT, Al-Batran SE, Chevallay M and
Mönig SP: Multimodal treatment in locally advanced gastric cancer.
Updates Surg. 70:173–179. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bi J, Bai Z, Ma X, Song J, Guo Y, Zhao J,
Yi X, Han S and Zhang Z: Txr1: An important factor in oxaliplatin
resistance in gastric cancer. Med Oncol. 31(807)2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Peng B, Chen Y and Leong KW: MicroRNA
delivery for regenerative medicine. Adv Drug Deliv Rev. 88:108–122.
2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
El Bezawy R, Cominetti D, Fenderico N,
Zuco V, Beretta GL, Dugo M, Arrighetti N, Stucchi C, Rancati T,
Valdagni R, et al: miR-875-5p counteracts epithelial-to-mesenchymal
transition and enhances radiation response in prostate cancer
through repression of the EGFR-ZEB1 axis. Cancer Lett. 395:53–62.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Qiu X and Dou Y: miR-1307 promotes the
proliferation of prostate cancer by targeting FOXO3A. Biomed
Pharmacother. 88:430–435. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhu DY, Li XN, Qi Y, Liu DL, Yang Y, Zhao
J, Zhang CY, Wu K and Zhao S: MiR-454 promotes the progression of
human non-small cell lung cancer and directly targets PTEN. Biomed
Pharmacother. 81:79–85. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fang B, Zhu J, Wang Y, Geng F and Li G:
MiR-454 inhibited cell proliferation of human glioblastoma cells by
suppressing PDK1 expression. Biomed Pharmacother. 75:148–152.
2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Niu G, Li B, Sun J and Sun L: miR-454 is
down-regulated in osteosarcomas and suppresses cell proliferation
and invasion by directly targeting c-Met. Cell Prolif. 48:348–355.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sun L, Wang QL, Gao XC, Shi DJ, Mi SY and
Han Q: MicroRNA-454 functions as an oncogene by regulating PTEN in
uveal melanoma. FEBS Lett. 589:2791–2796. 2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yu L, Gong XJ, Sun L, Yao H, Lu BL and Zhu
LY: miR-454 functions as an oncogene by inhibiting CHD5 in
hepatocellular carcinoma. Oncotarget. 6:39225–39234.
2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene exression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lee HK, Finniss S, Cazacu S, Bucris E,
Ziv-Av A, Xiang C, Bobbitt K, Rempel SA, Hasselbach L, Mikkelsen T,
et al: Mesenchymal stem cells deliver synthetic microRNA mimics to
glioma cells and glioma stem cells and inhibit their cell migration
and self-renewal. Oncotarget. 4:346–361. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yu X, Li Z and Liu J: MiRNAs in primary
cutaneous lymphomas. Cell Prolif. 48:271–277. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wu X, Ding N, Hu WT, He J, Xu S, Pei H,
Hua J, Zhou G and Wang J: Down-regulation of BTG1 by miR-454-3p
enhances cellular radiosensitivity in renal carcinoma cells.
Radiation Oncol. 9(179)2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Huang DH, Wang GY, Zhang JW, Li Y, Zeng XC
and Jiang N: MiR-501-5p regulates CYLD expression and promotes cell
proliferation in human hepatocellular carcinoma. Jpn J Clin Oncol.
45:738–744. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Massoumi R: CYLD: A deubiquitination
enzyme with multiple roles in cancer. Future Oncol. 7:285–297.
2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Gautheron J and Luedde T: A novel player
in inflammation and cancer: The deubiquitinase CYLD controls HCC
development. J Hepatol. 57:937–939. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Nikolaou K, Tsagaratou A, Eftychi C,
Kollias G, Mosialos G and Talianidis I: Inactivation of the
deubiquitinase CYLD in hepatocytes causes apoptosis, inflammation,
fibrosis, and cancer. Cancer Cell. 21:738–750. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Xu G, Zhu H, Zhang M and Xu J: Histone
deacetylase 3 is associated with gastric cancer cell growth via the
miR-454-mediated targeting of CHD5. Int J Mol Med. 41:155–163.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Masoumi KC, Shaw-Hallgren G and Massoumi
R: Tumor suppressor function of CYLD in nonmelanoma skin cancer. J
Skin Cancer. 2011(614097)2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Orfanidou T, Xanthopoulos K, Dafou D,
Pseftogas A, Hadweh P, Psyllaki C, Hatzivassiliou E and Mosialos G:
Down-regulation of the tumor suppressor CYLD enhances the
transformed phenotype of human breast cancer cells. Anticancer Res.
37:3493–3503. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yan YF, Gong FM, Wang BS and Zheng W:
MiR-425-5p promotes tumor progression via modulation of CYLD in
gastric cancer. Eur Rev Med Pharmacol Sci. 21:2130–2136.
2017.PubMed/NCBI
|
|
27
|
Zhu M, Zhou X, Du Y, Huang Z, Zhu J, Xu J,
Cheng G, Shu Y, Liu P, Zhu W and Wang T: miR-20a induces cisplatin
resistance of a human gastric cancer cell line via targeting CYLD.
Mol Med Rep. 14:1742–1750. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Niu J and Mims MP: Oxaliplatin-induced
thrombotic thrombocytopenic purpura: Case report and literature
review. J Clin Oncol. 30:E312–E314. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tsimberidou AM, Said R, Culotta K, Wistuba
I, Jelinek J, Fu S, Falchook G, Naing A, Piha-Paul S, Zinner R, et
al: Phase I study of azacitidine and oxaliplatin in patients with
advanced cancers that have relapsed or are refractory to any
platinum therapy. Clin Epigenetics. 7(29)2015.PubMed/NCBI View Article : Google Scholar
|