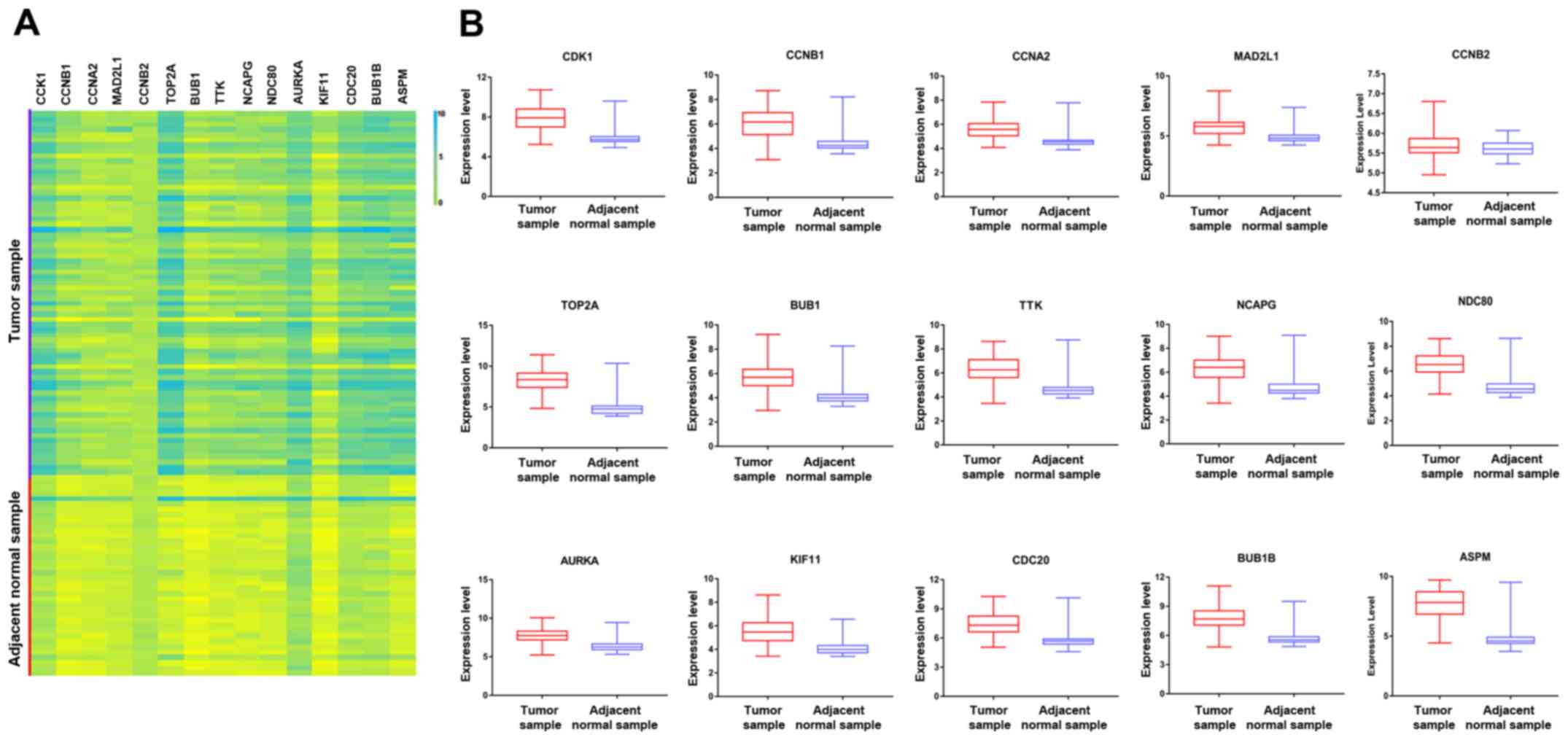
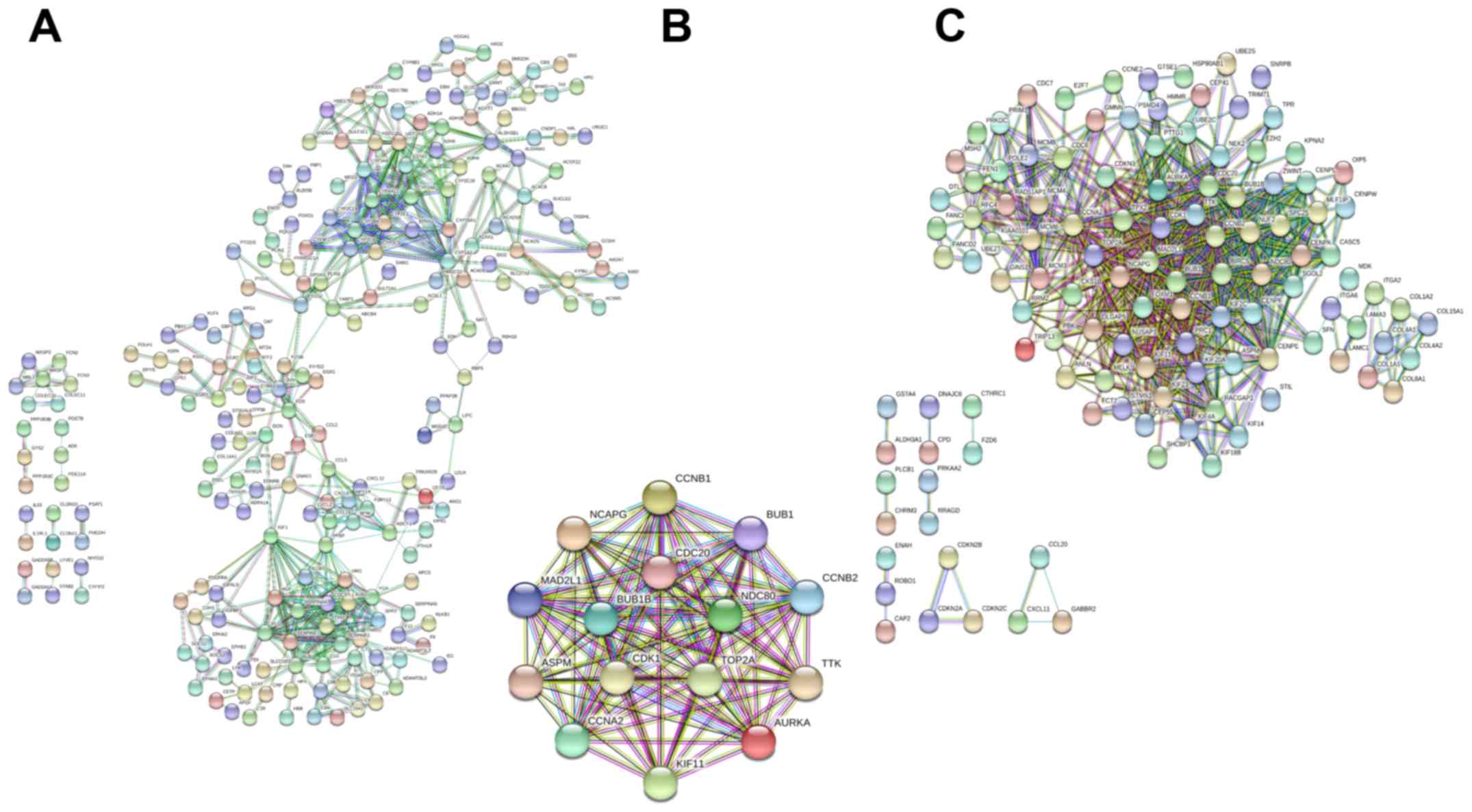
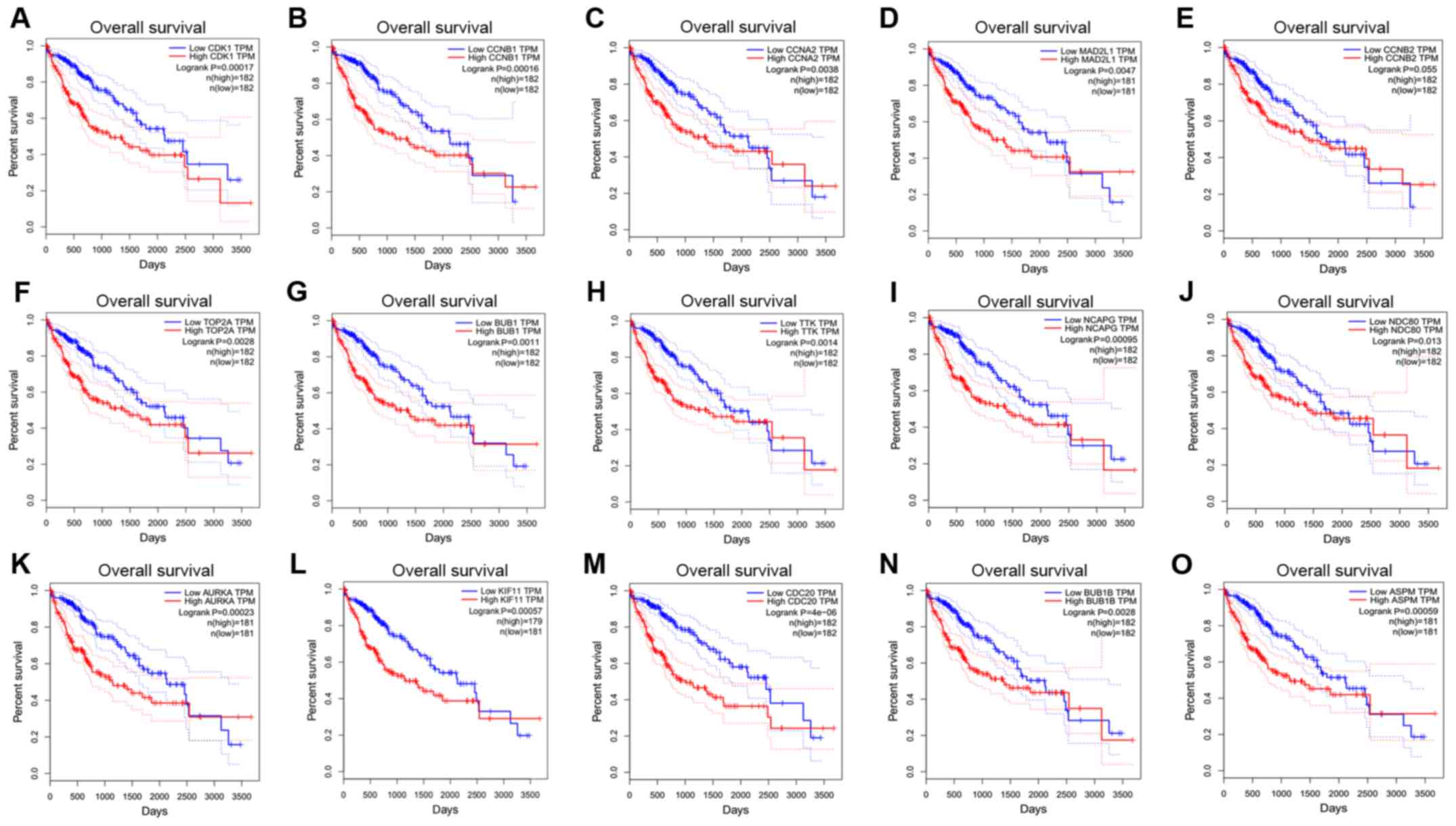
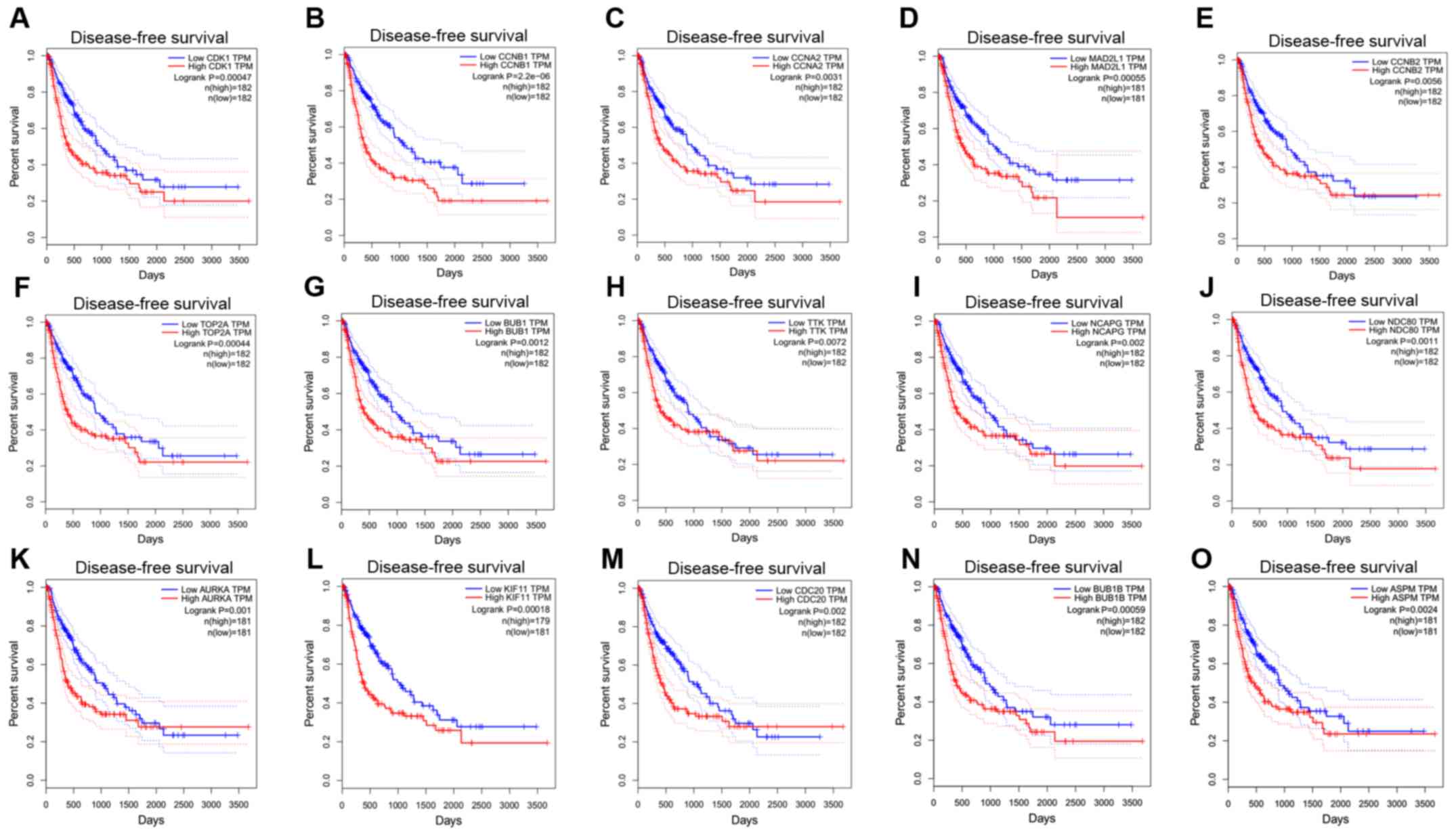
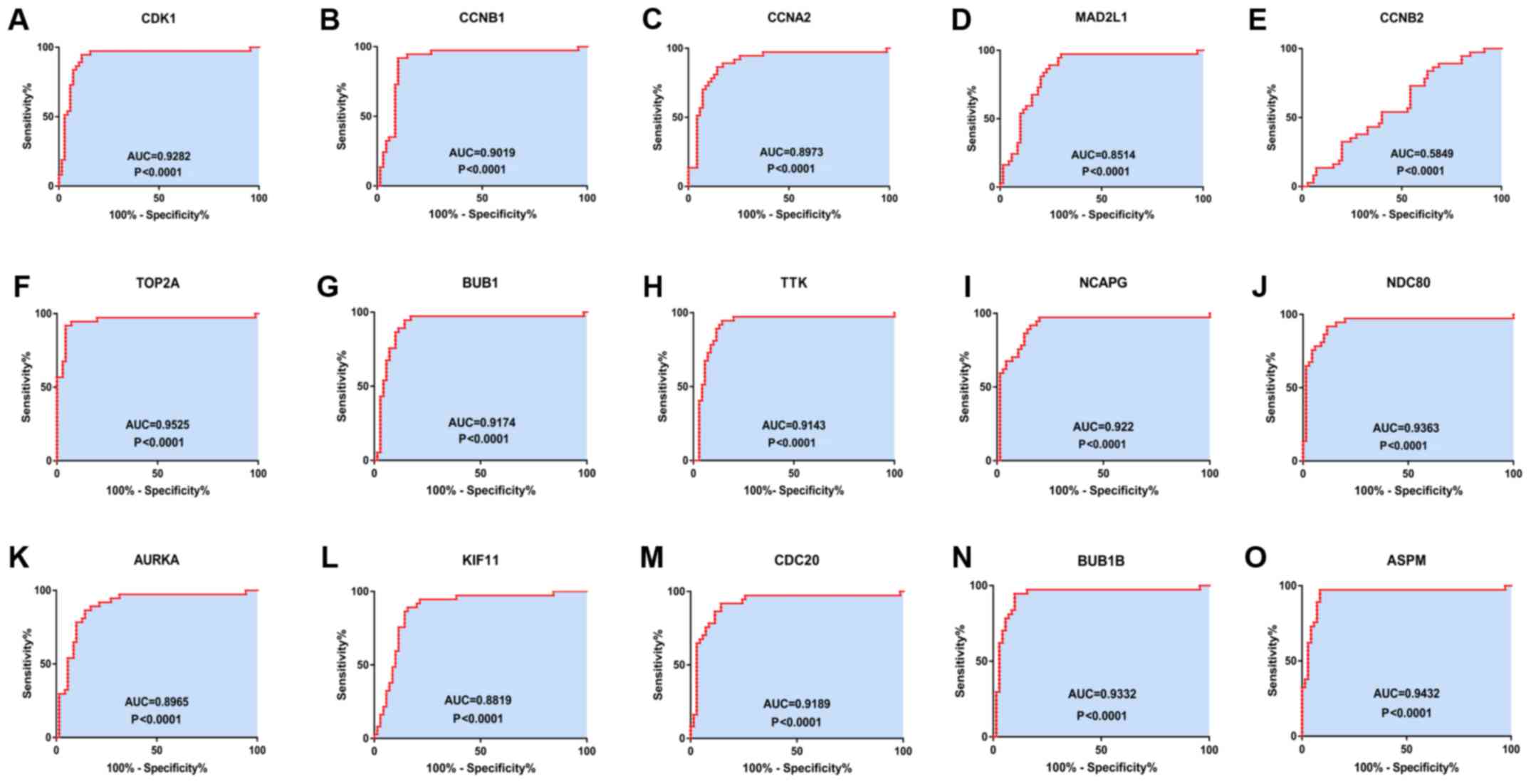
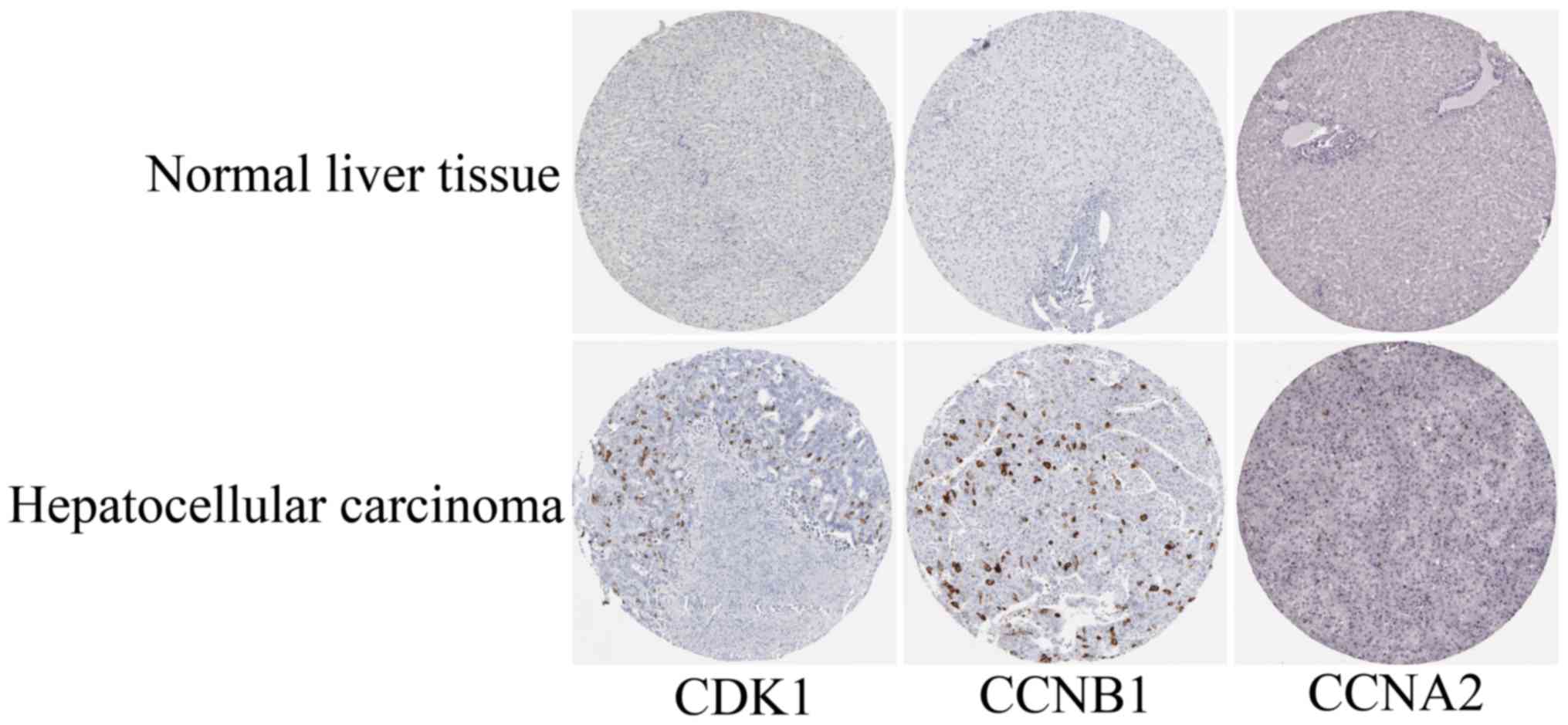
|
1
|
Ghouri YA, Mian I and Rowe JH: Review of
hepatocellular carcinoma: Epidemiology, etiology, and
carcinogenesis. J Carcinog. 16(1)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Geier A, Gartung C and Dietrich CG:
Hepatitis B e antigen and the risk of hepatocellular carcinoma. N
Engl J Med. 347:1721–1722. 2002.PubMed/NCBI View Article : Google Scholar
|
|
4
|
European Association for the Study of the
Liver. Electronic address: easloffice@easloffice.eu; European
Association for the Study of the Liver. EASL Clinical Practice
Guidelines: Management of hepatocellular carcinoma. J Hepatol.
69:182–236. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Fu PY, Hu B, Ma XL, Yang ZF, Yu MC, Sun
HX, Huang A, Zhang X, Wang J, Hu ZQ, et al: New insight into BIRC3:
A novel prognostic indicator and a potential therapeutic target for
liver cancer. J Cell Biochem. 120:6035–6045. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Long J, Zhang L, Wan X, Lin J, Bai Y, Xu
W, Xiong J and Zhao H: A four-gene-based prognostic model predicts
overall survival in patients with hepatocellular carcinoma. J Cell
Mol Med. 22:5928–5938. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sauzay C, Petit A, Bourgeois AM, Barbare
JC, Chauffert B, Galmiche A and Houessinon A: Alpha-foetoprotein
(AFP): A multi-purpose marker in hepatocellular carcinoma. Clin
Chim Acta. 463:39–44. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tsuchiya N, Sawada Y, Endo I, Saito K,
Uemura Y and Nakatsura T: Biomarkers for the early diagnosis of
hepatocellular carcinoma. World J Gastroenterol. 21:10573–10583.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Yan P, He Y, Xie K, Kong S and Zhao W: In
silico analyses for potential key genes associated with gastric
cancer. PeerJ. 6(e6092)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Teufel A: Bioinformatics and database
resources in hepatology. J Hepatol. 62:712–719. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Jiao Y, Fu Z, Li Y, Meng L and Liu Y: High
EIF2B5 mRNA expression and its prognostic significance in liver
cancer: A study based on the TCGA and GEO database. Cancer Manag
Res. 10:6003–6014. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shen S, Kong J, Qiu Y, Yang X, Wang W and
Yan L: Identification of core genes and outcomes in hepatocellular
carcinoma by bioinformatics analysis. J Cell Biochem.
120:10069–10081. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhou SL, Hu ZQ, Zhou ZJ, Dai Z, Wang Z,
Cao Y, Fan J, Huang XW and Zhou J: miR-28-5p-IL-34-macrophage
feedback loop modulates hepatocellular carcinoma metastasis.
Hepatology. 63:1560–1575. 2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Gaudet P, Skunca N, Hu JC and Dessimoz C:
Primer on the gene ontology. Methods Mol Biol. 1446:25–37.
2017.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wixon J and Kell D: The Kyoto encyclopedia
of genes and genomes-KEGG. Yeast. 17:48–55. 2000.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Baryshnikova A: Exploratory analysis of
biological networks through visualization, clustering, and
functional annotation in cytoscape. Cold Spring Harb Protoc 2016,
2016.
|
|
18
|
McClinton KJ, Aliani M, Kuny S, Sauvé Y
and Suh M: Differential effect of a carotenoid-rich diet on retina
function in non-diabetic and diabetic rats. Nutr Neurosci. 11:1–11.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Prevo R, Pirovano G, Puliyadi R, Herbert
KJ, Rodriguez-Berriguete G, O'Docherty A, Greaves W, McKenna WG and
Higgins GS: CDK1 inhibition sensitizes normal cells to DNA damage
in a cell cycle dependent manner. Cell Cycle. 17:1513–1523.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Haider C, Grubinger M, Řezníčková E, Weiss
TS, Rotheneder H, Miklos W, Berger W, Jorda R, Zatloukal M, Gucky
T, et al: Novel inhibitors of cyclin-dependent kinases combat
hepatocellular carcinoma without inducing chemoresistance. Mol
Cancer Ther. 12:1947–1957. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhou Z, Li Y, Hao H, Wang Y, Zhou Z, Wang
Z and Chu X: Screening Hub genes as prognostic biomarkers of
hepatocellular carcinoma by bioinformatics analysis. Cell
Transplant. 11(963689719893950)2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Nakayama Y and Yamaguchi N: Role of cyclin
B1 levels in DNA damage and DNA damage-induced senescence. Int Rev
Cell Mol Biol. 305:303–337. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chai N, Xie HH, Yin JP, Sa KD, Guo Y, Wang
M, Liu J, Zhang XF, Zhang X, Yin H, et al: FOXM1 promotes
proliferation in human hepatocellular carcinoma cells by
transcriptional activation of CCNB1. Biochem Biophys Res Commun.
500:924–929. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Weng L, Du J, Zhou Q, Cheng B, Li J, Zhang
D and Ling C: Identification of cyclin B1 and Sec62 as biomarkers
for recurrence in patients with HBV-related hepatocellular
carcinoma after surgical resection. Mol Cancer.
11(39)2012.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chiu YT, Wong JK, Choi SW, Sze KM, Ho DW,
Chan LK, Lee JM, Man K, Cherny S, Yang W, et al: Novel pre-mRNA
splicing of intronically integrated HBV generates oncogenic chimera
in hepatocellular carcinoma. J Hepatol. 64:1256–1264.
2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Jin Q, Dai Y, Wang Y, Zhang S and Liu G:
High kinesin family member 11 expression predicts poor prognosis in
patients with clear cell renal cell carcinoma. J Clin Pathol.
72:354–362. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen J, Li S, Zhou S, Cao S, Lou Y, Shen
H, Yin J and Li G: Kinesin superfamily protein expression and its
association with progression and prognosis in hepatocellular
carcinoma. J Cancer Res Ther. 13:651–659. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li R, Jiang X, Zhang Y, Wang S, Chen X, Yu
X, Ma J and Huang X: Cyclin B2 overexpression in human
hepatocellular carcinoma is associated with poorprognosis. Arch Med
Res. 50:10–17. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li Y, Bai W and Zhang J: MiR-200c-5p
suppresses proliferation and metastasis of human hepatocellular
carcinoma (HCC) via suppressing MAD2L1. Biomed Pharmacother.
92:1038–1044. 2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Yun MY, Kim SB, Park S, Han CJ, Han YH,
Yoon SH, Kim SH, Kim CM, Choi DW, Cho MH, et al: Mutation analysis
of p31comet gene, a negative regulator of Mad2, in human
hepatocellular carcinoma. ExperimeExp Mol Med. 39:508–513.
2007.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Wong N, Yeo W, Wong WL, Wong NL, Chan KY,
Mo FK, Koh J, Chan SL, Chan AT, Lai PB, et al: TOP2A overexpression
in hepatocellular carcinoma correlates with early age onset,
shorter patients survival and chemoresistance. Int J Cancer.
124:644–652. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xu B, Xu T, Liu H, Min Q, Wang S and Song
Q: MiR-490-5p suppresses cell proliferation and invasion by
targeting BUB1 in hepatocellular carcinoma cells. Pharmacology.
100:269–282. 2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ricke RM, Jeganathan KB and van Deursen
JM: Bub1 overexpression induces aneuploidy and tumor formation
through Aurora B kinase hyperactivation. J Cell Biol.
193:1049–1064. 2011.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Chen QF, Xia JG, Li W, Shen LJ, Huang T
and Wu P: Examining the key genes and pathways in hepatocellular
carcinoma development from hepatitis B virus-positive cirrhosis.
Mol Med Rep. 18:4940–4950. 2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Liu X, Liao W, Yuan Q, Ou Y and Huang J:
TTK activates Akt and promotes proliferation and migration of
hepatocellular carcinoma cells. Oncotarget. 6:34309–34320.
2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Baffy G: Decoding multifocal
hepatocellular carcinoma: An opportune pursuit. Hepatobiliary Surg
Nutr. 4:206–210. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zaman GJR, de Roos JADM, Libouban MAA,
Prinsen MBW, de Man J, Buijsman RC and Uitdehaag JCM: TTK
inhibitors as a targeted therapy for CTNNB1 (β-catenin) mutant
cancers. Mol Cancer Ther. 16:2609–2617. 2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu W, Liang B, Liu H, Huang Y, Yin X,
Zhou F, Yu X, Feng Q, Li E, Zou Z and Wu L: Overexpression of
non-SMC condensin I complex subunit G serves as a promising
prognostic marker and therapeutic target for hepatocellular
carcinoma. Int J Mol Med. 40:731–738. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Liu K, Li Y, Yu B, Wang F, Mi T and Zhao
Y: Silencing non-SMC chromosome-associated polypeptide G inhibits
proliferation and induces apoptosis in hepatocellular carcinoma
cells. Can J Physiol Pharmacol. 96:1246–1254. 2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ju LL, Chen L, Li JH, Wang YF, Lu RJ, Bian
ZL and Shao JG: Effect of NDC80 in human hepatocellular carcinoma.
World J Gastroenterol. 23:3675–3683. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu B, Yao Z, Hu K, Huang H, Xu S, Wang Q,
Yang Y and Ren J: ShRNA-mediated silencing of the Ndc80 gene
suppress cell proliferation and affected hepatitis B virus-related
hepatocellular carcinoma. Clin Res Hepatol Gastroenterol.
40:297–303. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhang L, Huang Y, Ling J, Zhuo W, Yu Z,
Shao M, Luo Y and Zhu Y: Screening and function analysis of hub
genes and pathways in hepatocellular carcinoma via bioinformatics
approaches. Cancer Biomark. 22:511–521. 2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Bao Z, Lu L, Liu X, Guo B, Zhai Y, Li Y,
Wang Y, Xie B, Ren Q, Cao P, et al: Association between the
functional polymorphism Ile31Phe in the AURKA gene and
susceptibility of hepatocellular carcinoma in chronic hepatitis B
virus carriers. Oncotarget. 8:54904–54912. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Li J, Gao JZ, Du JL, Huang ZX and Wei LX:
Increased CDC20 expression is associated with development and
progression of hepatocellular carcinoma. Int J Oncol. 45:1547–1555.
2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Chen PF, Li QH, Zeng LR, Yang XY, Peng PL,
He JH and Fan B: A 4-gene prognostic signature predicting survival
in hepatocellular carcinoma. J Cell Biochem. 120:9117–9124.
2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wen DY, Lin P, Pang YY, Chen G, He Y, Dang
YW and Yang H: Expression of the long intergenic non-protein coding
RNA 665 (LINC00665) gene and the cell cycle in hepatocellular
carcinoma using the cancer genome atlas, the gene expression
omnibus, and quantitative real-time polymerase chain reaction. Med
Sci Monit. 24:2786–2808. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lin SY, Pan HW, Liu SH, Jeng YM, Hu FC,
Peng SY, Lai PL and Hsu HC: ASPM is a novel marker for vascular
invasion, early recurrence, and poor prognosis of hepatocellular
carcinoma. Clin Cancer Res. 14:4814–4820. 2008.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Okuda K, Nakashima T, Sakamoto K, Ikari T,
Hidaka H, Kubo Y, Sakuma K, Motoike Y, Okuda H and Obata H:
Hepatocellular carcinoma arising in noncirrhotic and highly
cirrhotic livers: A comparative study of histopathology and
frequency of hepatitis B markers. Cancer. 49:450–455.
1982.PubMed/NCBI View Article : Google Scholar
|