Introduction
Liver cancer is a common malignant tumor type with
high morbidity and mortality (1).
Although various treatments have been improved, the mortality rate
of liver cancer is still high and the prognosis remains poor
(2,3). Therefore, prognostic biomarkers of
liver cancer have become one of the hotspots of current research
(4). The discovery of accurate
prognostic biomarkers may contribute to clinical guidance in order
to improve the evaluation system of liver cancer.
The family of eukaryotic translation initiation
factors (EIFs) participates in eukaryotic translation by regulating
the interaction between ribosomes and RNA. It is the rate-limiting
step of protein synthesis and participates in numerous processes
that are deregulated in cancer cells, including DNA repair and
proliferation, cell cycle and apoptosis (5). EIF 3 subunit B (EIF3B) is an important
member of the family of EIFs and has been observed to be
overexpressed in numerous cancer types, including clear cell renal
cell carcinoma (6), esophageal
squamous cell carcinoma (7),
glioblastoma (8), ovarian cancer
(9), osteosarcoma (10) and lung cancer (11), and has an important role in the
progression and prognosis of several cancer types (12-14).
In addition, Golob-Schwarzl et al (15) reported that EIF3B was upregulated in
hepatitis C virus (HCV)-associated hepatocellular carcinoma (HCC).
However, the precise role of EIF3B in liver cancer has remained
elusive.
To further evaluate the roles of EIF3B in patients
with liver cancer, the expression of EIF3B was examined in a
dataset from The Cancer Genome Atlas (TCGA) database. The
χ2 and Fisher's exact tests were used to assess the
association of EIF3B with clinicopathological parameters and
demographic features. Receiver operating characteristics (ROC)
curve analysis was used for evaluating the diagnostic value of
EIF3B. Kaplan-Meier overall survival and relapse-free survival
analysis were performed to determine the association between EIF3B
expression and survival. Univariate and multivariate Cox regression
analysis were performed to identify the factors affecting overall
survival and relapse-free survival. Furthermore, gene set
enrichment analysis (GSEA) was used to explore EIF3B-associated
signaling pathways.
Materials and methods
Data source
The clinical information of patients and their
RNAseq data were obtained from TCGA (https://cancergenome.nih.gov/). All patients from the
liver hepatocellular carcinoma (LIHC) cohort were screened based on
TCGA inclusion and exclusion pre-selection criteria.
Statistical analysis
R (version 3.6.1; The R Foundation) (16) was used for statistical analysis
(t-test, Kruskal-Wallis with Dunn's post-hoc test, Wilcoxon
sum-rank test) and generation of images. The ggplot2 package
(17) was used to draw boxplots of
the EIF3B expression in subgroups by clinical characteristics.
χ2 and Fisher's exact tests were applied to estimate the
significance of the association between EIF3B expression and
clinicopathological or demographic characteristics. The pROC
package (18) was used to plot the
ROC curves and assess the diagnostic ability of EIF3B, and patients
were divided into a high expression group and low expression group
according to the best operating system cut-off value determined by
the Youden index. The survival package (19) was used to draw survival curves. A
univariate Cox linear regression model was utilized to select
correlative variables affecting survival time. Multivariate Cox
regression analysis was employed to evaluate the independent
influencing factors of survival time.
GSEA
GSEA may be used to determine whether a predefined
set of genes is able to indicate significant, consistent
differences between two biological states (20). In the present study, GSEA was
performed in the ‘h.all.v6.2.symbols.gmt’ and ‘c2.
cp.biocarta.v6.2.symbols.gmt’ gene sets using GSEA3.0 software. The
standardized enrichment fraction was obtained by 1,000 permutation
analyses.
Results
Patient characteristics
In Table I, the
clinical features of the 373 patient cohort, including sex (female
121, male 252), age (16-90 years old, median 61, mean 59.47), use
of radiation therapy, residual tumor, relapse-free survival,
histological type, stage, vital status, survival data, T/N/M
classification and EIF3B expression are provided.
 | Table IDemographic and clinical
characteristics of the cohort from The Cancer Genome Atlas-liver
hepatocellular carcinoma dataset. |
Table I
Demographic and clinical
characteristics of the cohort from The Cancer Genome Atlas-liver
hepatocellular carcinoma dataset.
| Characteristics | N (373) |
|---|
| Age (years) | |
|
<55 | 117 (31.45) |
|
≥55 | 255 (68.55) |
|
NA | 1 (0.00) |
| Sex | |
|
Female | 121 (32.44) |
|
Male | 252 (67.56) |
| Histological
type | |
|
Fibrolamellar
carcinoma | 3 (0.80) |
|
Hepatocellular
carcinoma | 363 (97.32) |
|
Hepatocholangiocarcinoma
(mixed) | 7 (1.88) |
| Histologic
grade | |
|
NA | 5 (1.34) |
|
G1 | 55 (14.75) |
|
G2 | 178 (47.72) |
|
G3 | 123 (32.98) |
|
G4 | 12 (3.22) |
| Stage | |
|
NA | 24 (6.43) |
|
I | 172 (46.11) |
|
II | 87 (23.32) |
|
III | 85 (22.79) |
|
IV | 5 (1.34) |
| T
classification | |
|
NA | 2 (0.54) |
|
T1 | 182 (48.79) |
|
T2 | 95 (25.47) |
|
T3 | 80 (21.45) |
|
T4 | 13 (3.49) |
|
TX | 1 (0.27) |
| N
classification | |
|
NA | 1 (0.27) |
|
N0 | 253 (67.83) |
|
N1 | 4 (1.07) |
|
NX | 115 (30.83) |
| M
classification | |
|
M0 | 267 (71.58) |
|
M1 | 4 (1.07) |
|
MX | 102 (27.35) |
| Radiation
therapy | |
|
NA | 25 (6.70) |
|
No | 340 (91.15) |
|
Yes | 8 (2.14) |
| Residual tumor | |
|
NA | 7 (1.88) |
|
R0 | 326 (87.40) |
|
R1 | 17 (4.56) |
|
R2 | 1 (0.27) |
|
RX | 22 (5.90) |
| Vital status | |
|
Deceased | 130 (34.85) |
|
Alive | 243 (65.15) |
| Sample type | |
|
Primary
tumor | 371 (99.46) |
|
Recurrent
tumor | 2 (0.54) |
| Overall survival
(ten years) | |
|
No | 237 (64.58) |
|
Yes | 130 (35.42) |
| Relapse-free
survival (ten years) | |
|
No | 179 (55.94) |
|
Yes | 141 (44.06) |
| EIF3B | |
|
High | 109 (29.22) |
|
Low | 264 (70.78) |
Expression of EIF3B in liver
tissues
Boxplots revealed that EIF3B was significantly
upregulated in liver cancer compared with that in normal liver
tissues (Fig. 1A; P<0.001).
Furthermore, EIF3B was also differentially expressed between
subgroups by vital status (P=0.005) and histologic grade
(P<0.001; Fig. 1).
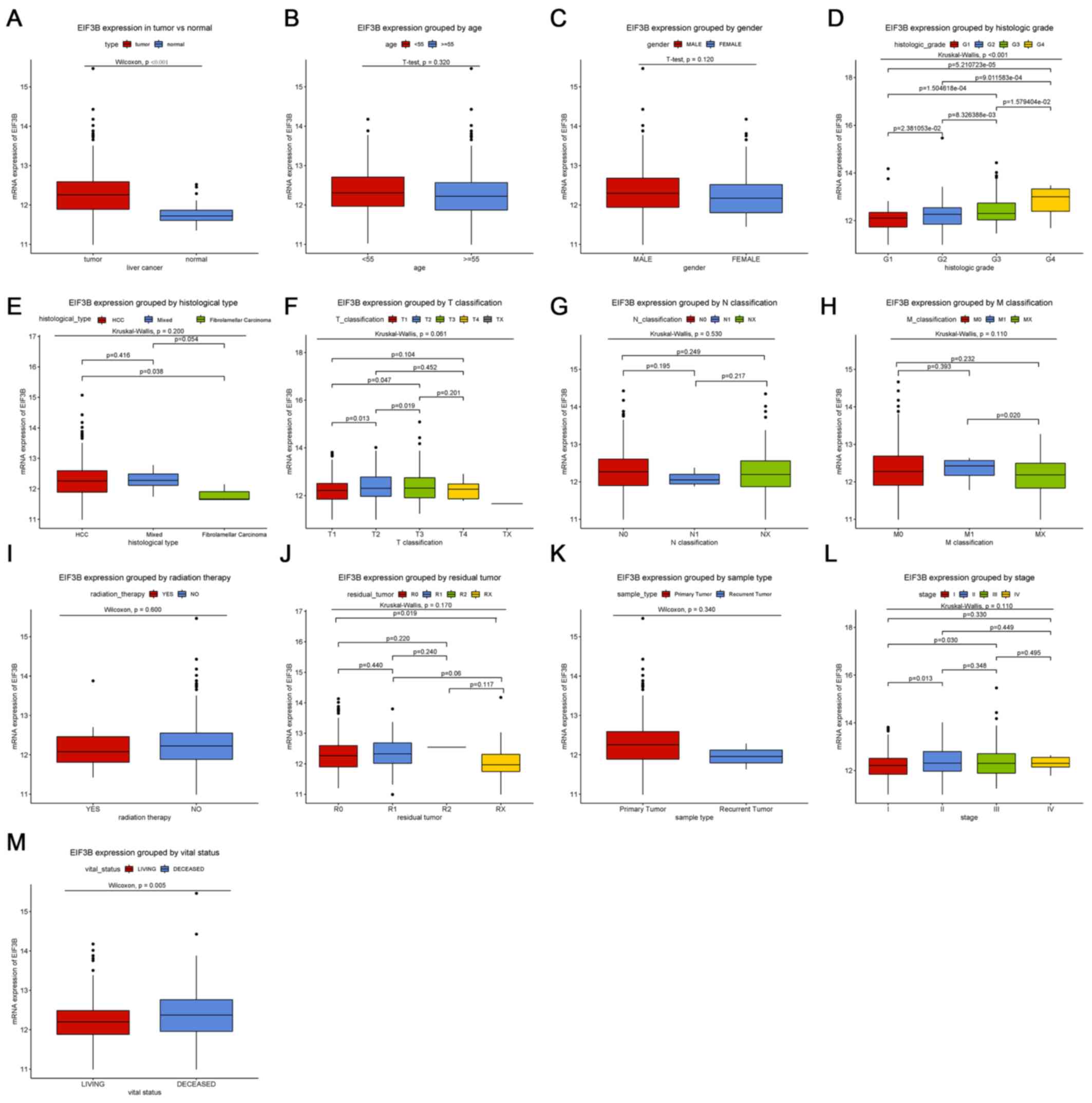 | Figure 1Differences in EIF3B expression among
subgroups of patients. Boxplots showing differences in EIF3B
expression according to (A) tissue type (P<0.001 vs. normal
tissue controls), (B) age, (C) sex, (D) histologic grade
(P<0.001), (E) histological type, (F) T classification, (G) N
classification, (H) M classification, (I) radiation therapy, (J)
residual tumor classification, (K) sample type, (L) clinical stage
and (M) vital status (P=0.005). EIF3B, eukaryotic translation
initiation factor 3 subunit B. G1-4, grade relating to degree of
differentiation; T1-4, size and or extension of the primary tumor;
TX, tumor could not be assessed; N0, no regional lymph node
metastasis; N1, regional lymph node metastasis present; NX, lymph
nodes could not be assessed; M0, no distant metastasis; M1,
metastasis to distant organs; MX, metastasis could not be assessed;
R0, no residual tumor visible under the microscope; R1, residual
tumor visible under the microscope; R2, residual tumor visible to
the naked eye; RX, residual tumor could not be assessed. |
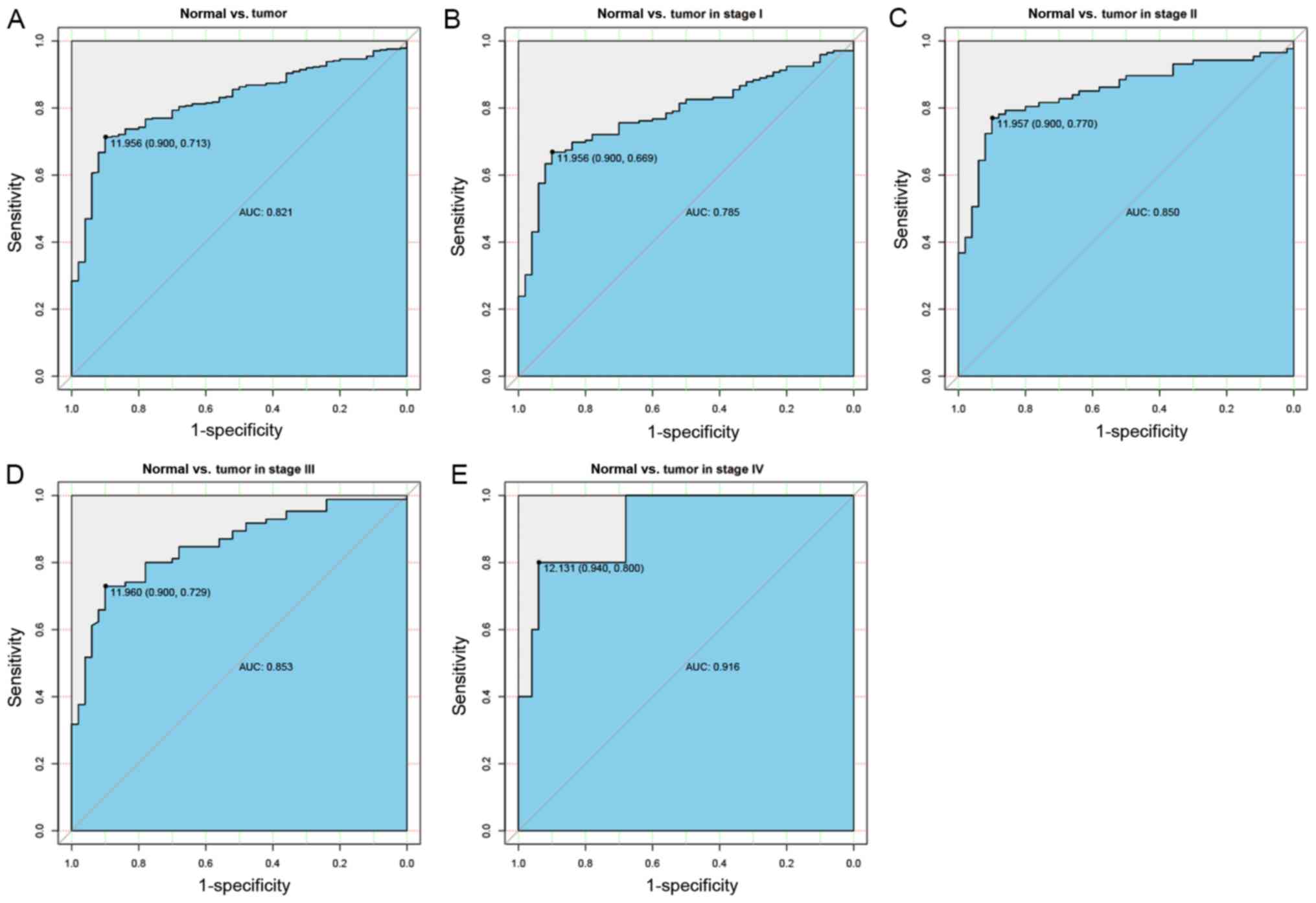
Diagnostic capability of EIF3B
To assess the diagnostic performance of EIF3B in
liver cancer, ROC curve analysis was used. The area under the curve
(AUC) was 0.821, indicating that EIF3B has moderate diagnostic
ability. In addition, similar AUCs were obtained for distinguishing
normal liver tissues from liver cancer at specific stages (stage I,
0.785; stage II, 0.850; stage III, 0.853; stage IV, 0.916; Fig. 2).
Association between EIF3B expression
and clinical features of patients with liver cancer
As indicated in Table
II, the vital status of the patients with liver cancer
(P<0.001), overall survival (P<0.001; duration ten years) and
the histologic grade (P<0.001) were associated with the
expression of EIF3B.
 | Table IIAssociation between the expression of
EIF3B and the clinicopathological characteristics of patients with
liver cancer. |
Table II
Association between the expression of
EIF3B and the clinicopathological characteristics of patients with
liver cancer.
| | EIF3B
expression | |
|---|
| Clinical
characteristics | No. of
patients | High | Low | χ2 | P-value |
|---|
| Age (years) | | | | 0.089 | 0.765 |
|
<55 | 117 | 36 (33.03) | 81 (30.80) | | |
|
≥55 | 255 | 73 (66.97) | 182 (69.20) | | |
| Sex | | | | 0.484 | 0.486 |
|
Female | 121 | 32 (29.36) | 89 (33.71) | | |
|
Male | 252 | 77 (70.64) | 175 (66.29) | | |
| Histological
type | | | | 1.251 | 0.534 |
|
Fibrolamellar
carcinoma | 3 | 0 (0.00) | 3 (1.14) | | |
|
Hepatocellular
carcinoma | 363 | 107 (98.17) | 256 (96.97) | | |
|
Hepatocholangiocarcinoma
(mixed) | 7 | 2 (1.83) | 5 (1.89) | | |
| Histologic
grade | | | | 17.796 | <0.001 |
|
G1 | 55 | 9 (8.33) | 46 (17.69) | | |
|
G2 | 178 | 45 (41.67) | 133 (51.15) | | |
|
G3 | 123 | 46 (42.59) | 77 (29.62) | | |
|
G4 | 12 | 8 (7.41) | 4 (1.54) | | |
| Stage | | | | 4.532 | 0.209 |
|
I | 172 | 43 (40.95) | 129 (52.87) | | |
|
II | 87 | 32 (30.48) | 55 (22.54) | | |
|
III | 85 | 28 (26.67) | 57 (23.36) | | |
|
IV | 5 | 2 (1.9) | 3 (1.23) | | |
| T
classification | | | | 7.720 | 0.102 |
|
T1 | 182 | 44 (40.37) | 138 (52.67) | | |
|
T2 | 95 | 34 (31.19) | 61 (23.28) | | |
|
T3 | 80 | 29 (26.61) | 51 (19.47) | | |
|
T4 | 13 | 2 (1.83) | 11 (4.2) | | |
|
TX | 1 | 0 (0.00) | 1 (0.38) | | |
| N
classification | | | | 1.936 | 0.379 |
|
N0 | 253 | 77 (70.64) | 176 (66.92) | | |
|
N1 | 4 | 0 (0.00) | 4 (1.52) | | |
|
NX | 115 | 32 (29.36) | 83 (31.56) | | |
| M
classification | | | | 2.882 | 0.236 |
|
M0 | 267 | 83 (76.15) | 184 (69.70) | | |
|
M1 | 4 | 2 (1.83) | 2 (0.76) | | |
|
MX | 102 | 24 (22.02) | 78 (29.55) | | |
| Radiation
therapy | | | | <0.001 | >0.999 |
|
No | 340 | 92 (97.87) | 248 (97.64) | | |
|
Yes | 8 | 2 (2.13) | 6 (2.36) | | |
| Residual tumor | | | | 5.116 | 0.163 |
|
R0 | 326 | 98 (91.59) | 228 (88.03) | | |
|
R1 | 17 | 5 (4.67) | 12 (4.63) | | |
|
R2 | 1 | 1 (0.93) | 0 (0.00) | | |
|
RX | 22 | 3 (2.80) | 19 (7.34) | | |
| Vital status | | | | 17.505 | <0.001 |
|
Deceased | 130 | 56 (51.38) | 74 (28.03) | | |
|
Alive | 243 | 53 (48.62) | 190 (71.97) | | |
| Sample type | | | | 0.017 | 0.895 |
|
Primary
tumor | 371 | 109(100) | 262 (99.24) | | |
|
Recurrent
tumor | 2 | 0 (0.00) | 2 (0.76) | | |
| Overall survival
(ten years) | | | | 18.690 | <0.001 |
|
No | 237 | 50 (47.17) | 187 (71.65) | | |
|
Yes | 130 | 56 (52.83) | 74 (28.35) | | |
| Relapse-free
survival (ten years) | | | | 1.018 | 0.312 |
|
No | 179 | 42 (50.6) | 137 (57.81) | | |
|
Yes | 141 | 41 (49.4) | 100 (42.19) | | |
High expression of EIF3B is associated
with poor overall survival of patients with liver cancer
Kaplan-Meier analysis indicated that high expression
of EIF3B was significantly associated with poor overall survival
(P<0.001; Fig. 3). Subgroup
analysis provided similar results, particularly in female
(P<0.001), younger (<55; P=0.008) and older subjects (≥55;
P<0.001), and in patients with R0 (P<0.001), G2 (P=0.003), G3
(P<0.0001), stage I (P<0.001) and stage III (P<0.001).
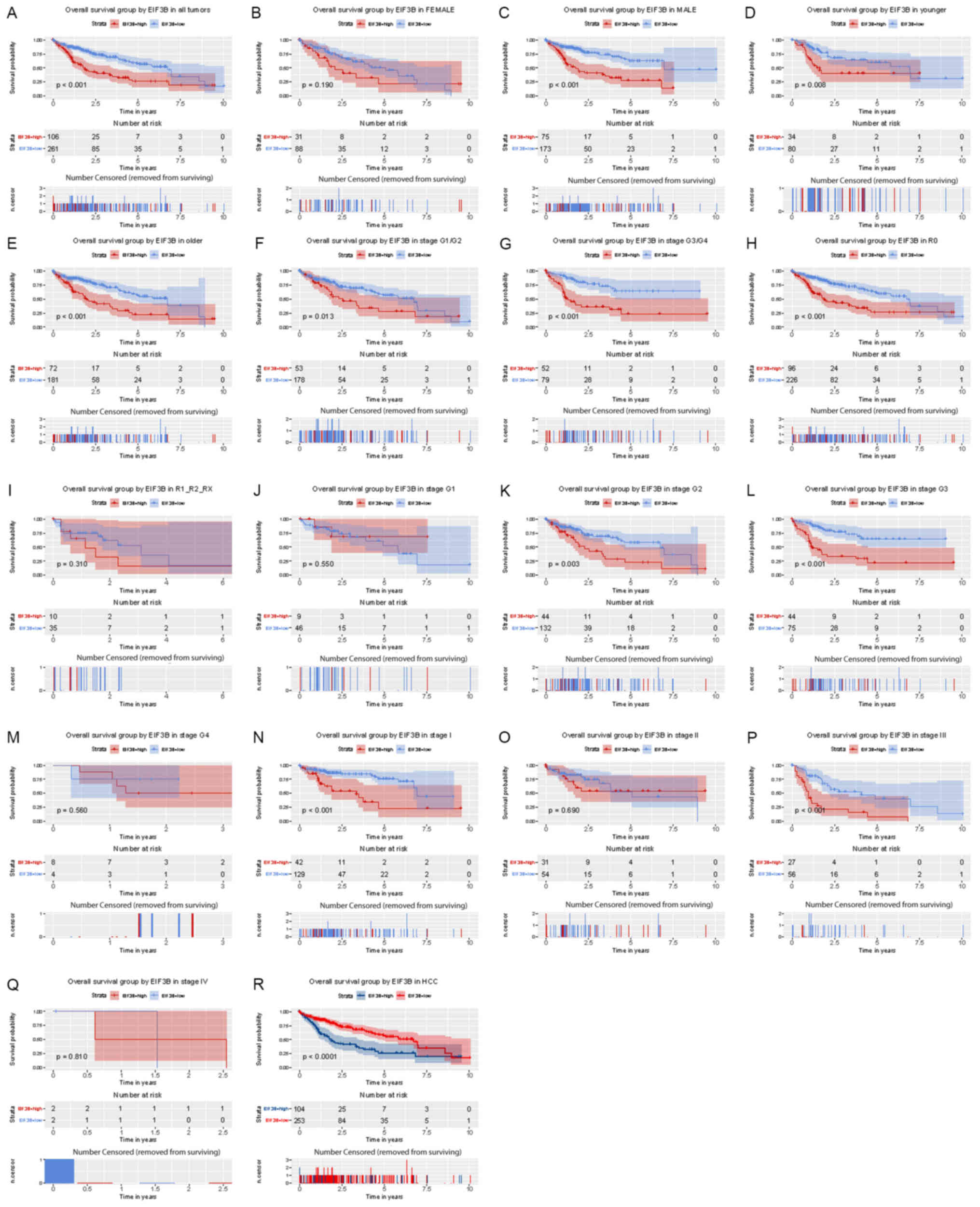 | Figure 3Kaplan-Meier analysis of the influence
of EIF3B expression on overall survival. (A) All patients. Subgroup
analysis for (B) females, (C) males, (D) younger patients (<55),
(E) older patients (≥55), (F) no lymph node dissection (R0), (G)
lymph node dissection (R1/R2/RX), (H-M) histological grade, (H)
G1/G2, (I) G3/G4, (J) G1, (K) G2, (L) G3, (M) G4, (N-Q) clinical
stage (N) I, (O) II, (P) III, (Q) IV and (R) HCC. EIF3B, eukaryotic
translation initiation factor 3 subunit B; HCC, hepatocellular
carcinoma; G1-4, grade relating to degree of differentiation; T1-4,
size and or extension of the primary tumor; TX, tumor could not be
assessed; N0, no regional lymph node metastasis; N1, regional lymph
node metastasis present; NX, lymph nodes could not be assessed; M0,
no distant metastasis; M1, metastasis to distant organs; MX,
metastasis could not be assessed; R0, no residual tumor visible
under the microscope; R1, residual tumor visible under the
microscope; R2, residual tumor visible to the naked eye; RX,
residual tumor could not be assessed. The number of results
censored (removed from surviving) is indicated below the survival
curve. |
As presented in Table
III, T classification, stage, residual tumor and EIF3B
expression were variables associated with overall survival
according to univariate Cox regression analysis. In addition,
multivariate Cox regression indicated that high EIF3B expression, T
classification and residual tumor were independent risk factors for
overall survival of patients with liver cancer [hazard ratio
(HR)=2.44, 95% CI=1.71-3.47, P<0.001].
 | Table IIISummary of univariate and
multivariate Cox regression analyses for overall survival duration
(ten years). |
Table III
Summary of univariate and
multivariate Cox regression analyses for overall survival duration
(ten years).
| | Univariate
analysis | Multivariate
analysis |
|---|
| Parameters | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥55/<55
years) | 1.00 | 0.69-1.45 | 0.997 | | | |
| Sex
(male/female) | 0.80 | 0.56-1.14 | 0.220 | | | |
| Histological type
(hepatocholangiocarcinoma/Hepatocellular, Hepatocellular
/fibrolamellar) | 0.99 | 0.27-3.66 | 0.986 | | | |
| Histologic grade
(G4/G3/G2/G1) | 1.04 | 0.84-1.30 | 0.698 | | | |
| Stage
(IV/III/II/I) | 1.38 | 1.15-1.66 | 0.001 | 0.81 | 0.65-1.01 | 0.060 |
| T classification
(T4/T3/T2/T1/NX) | 1.66 | 1.39-1.99 | <0.001 | 1.91 | 1.51-2.42 | <0.001 |
| N classification
(N1/N0/NX) | 0.73 | 0.51-1.05 | 0.086 | | | |
| M classification
(M1/M0/MX) | 0.72 | 0.49-1.04 | 0.077 | | | |
| Radiation therapy
(yes/no) | 0.51 | 0.26-1.03 | 0.060 | | | |
| Residual tumor
classification (RX/R2/R1/R0) | 1.42 | 1.13-1.80 | 0.003 | 1.45 | 1.13-1.87 | 0.004 |
| EIF3B
(high/low) | 2.41 | 1.70-3.42 | <0.001 | 2.44 | 1.71-3.47 | <0.001 |
High expression of EIF3B is associated
with poor relapse-free survival of patients with liver cancer
Kaplan-Meier analysis indicated that patients with
high expression of EIF3B had significantly poorer relapse-free
survival (P=0.013; Fig. 4). Subgroup
analysis provided similar results, particularly in female (P=0.001)
and older (≥55; P=0.032) subjects and in patients with R0
(P=0.020), G2 (P<0.001) and stage III (P=0.004).
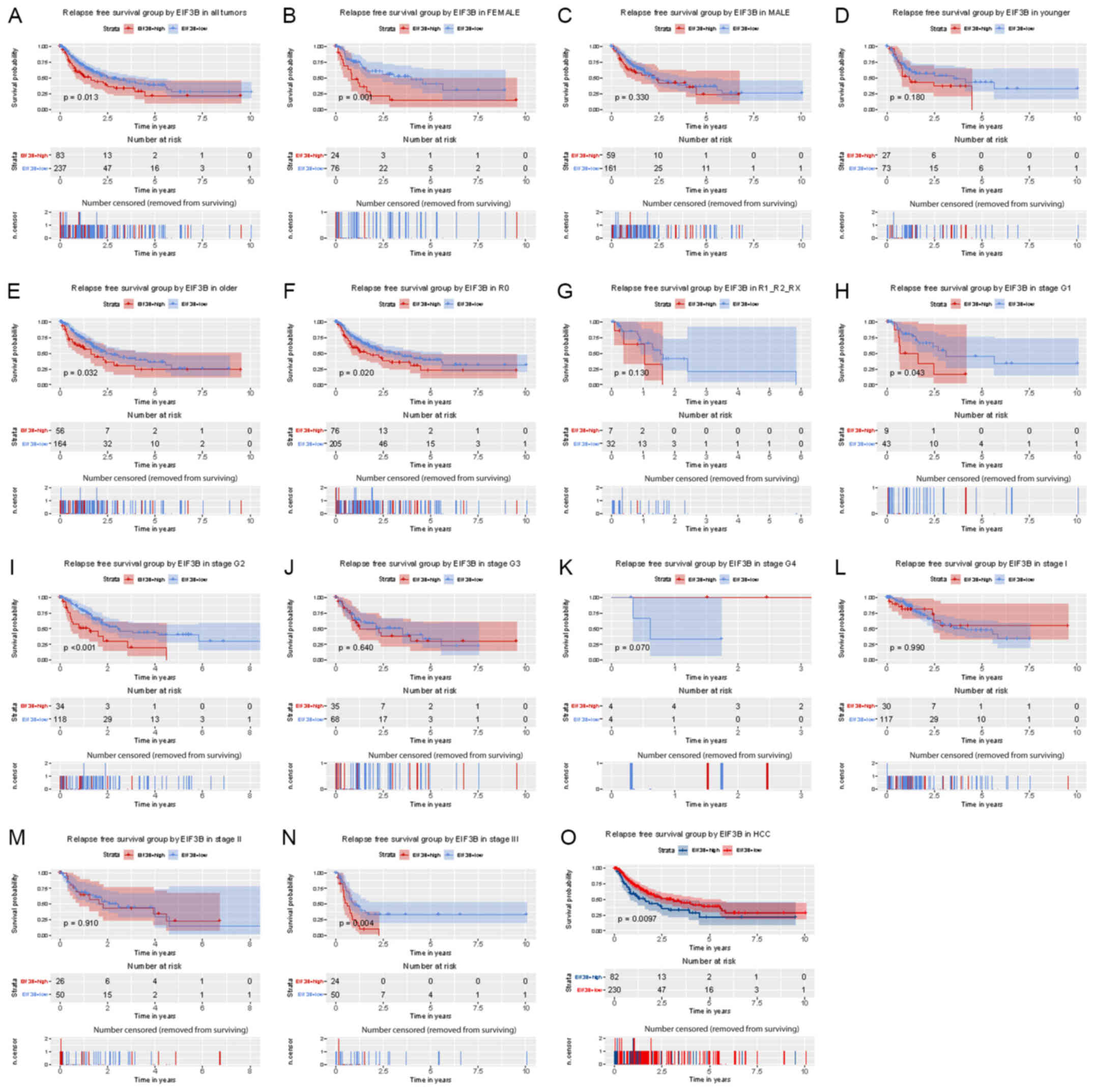 | Figure 4Kaplan-Meier analysis of the influence
of EIF3B expression on relapse-free survival. (A) All patients.
(B-R) Subgroup analysis for (B) females, (C) males, (D) younger
patients (<55), (E) older patients (≥55), (F) no lymph node
dissection (R0), (G) lymph node dissection (R1/R2/RX), (H-K)
histological grade (H) G1, (I) G2, (J) G3, (K) G4, (L-N) clinical
stage (L) I, (M) II, (N) III and (O) HCC. EIF3B, eukaryotic
translation initiation factor 3 subunit B; HCC, hepatocellular
carcinoma; G1-4, grade relating to degree of differentiation; T1-4,
size and or extension of the primary tumor; TX, tumor could not be
assessed; N0, no regional lymph node metastasis; N1, regional lymph
node metastasis present; NX, lymph nodes could not be assessed; M0,
no distant metastasis; M1, metastasis to distant organs; MX,
metastasis could not be assessed; R0, no residual tumor visible
under the microscope; R1, residual tumor visible under the
microscope; R2, residual tumor visible to the naked eye; RX,
residual tumor could not be assessed. The number of results
censored (removed from surviving) is indicated below the survival
curve. |
As presented in Table
IV, T classification, stage, residual tumor and EIF3B
expression were variables associated with relapse-free survival
according to the univariate Cox regression analysis. In addition,
high EIF3B expression, T classification and residual tumor were
independent risk factors for relapse-free survival of patients with
liver cancer in the multivariate Cox regression analysis (HR=1.54,
95% CI=1.06-2.23, P=0.022).
 | Table IVSummary of univariate and
multivariate Cox regression analyses or relapse-free survival
duration. |
Table IV
Summary of univariate and
multivariate Cox regression analyses or relapse-free survival
duration.
| | Univariate
analysis | Multivariate
analysis |
|---|
| Parameters | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥55/<55
years) | 0.90 | 0.63-1.28 | 0.550 | | | |
| Sex
(male/female) | 0.99 | 0.70-1.41 | 0.966 | | | |
| Histological type
(hepatocholangiocarcinoma/hepatocellular, hepatocellular
/fibrolamellar) | 2.02 | 0.66-6.24 | 0.220 | | | |
| Histologic grade
(G4/G3/G2/G1) | 0.98 | 0.80-1.21 | 0.883 | | | |
| Stage
(IV/III/II/I) | 1.66 | 1.38-1.99 | <0.001 | 1.10 | 0.85-1.42 | 0.473 |
| T classification
(T4/T3/T2/T1/TX) | 1.78 | 1.49-2.12 | <0.001 | 1.67 | 1.28-2.18 | <0.001 |
| N classification
(N1/N0/NX) | 0.97 | 0.67-1.40 | 0.874 | | | |
| M classification
(M1/M0/MX) | 1.17 | 0.79-1.74 | 0.432 | | | |
| Radiation therapy
(yes/no) | 0.74 | 0.26-2.16 | 0.584 | | | |
| Residual tumor
classification (RX/R2/R1/R0) | 1.28 | 1.01-1.61 | 0.042 | 1.36 | 1.07-1.73 | 0.012 |
| EIF3B
(high/low) | 1.58 | 1.10-2.28 | 0.014 | 1.54 | 1.06-2.23 | 0.022 |
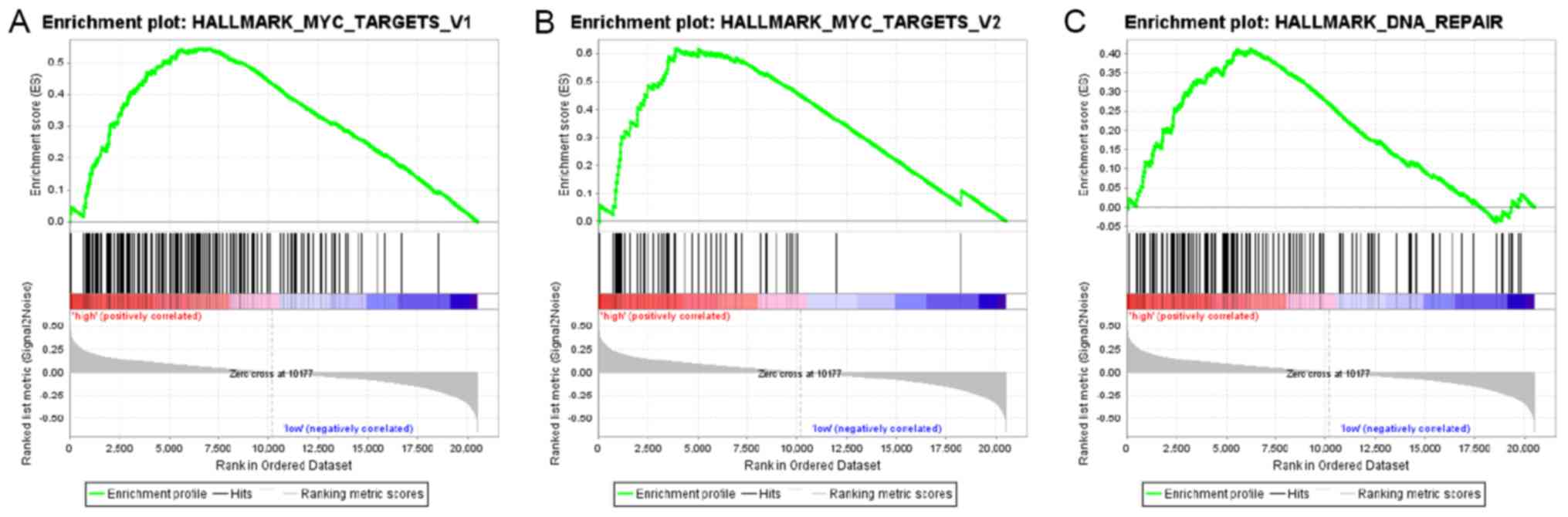
Signaling pathways associated with
EIF3B
To identify the signaling pathways associated with
EIF3B in liver cancer, GSEA was performed between the low EIF3B
expression dataset and the high EIF3B expression dataset. The
enrichment of the molecular signatures database (MSigDB) determined
by GSEA was significantly different (nominal P-value <0.050,
false discovery rate <0.250; Table
V). As presented in Fig. 5 and
Table V, GSEA indicated that MYC-V1
(HALLMARK_MYC_TARGETS_V1 geneset; P=0.009), MYC-V2
(HALLMARK_MYC_TARGETS_V2 geneset; P=0.004) and DNA repair pathways
(HALLMARK_DNA_REPAIR geneset; P<0.001) were differentially
enriched in high EIF3B expression and low EIF3B expression
groups.
 | Table VGene sets enriched in phenotype
high. |
Table V
Gene sets enriched in phenotype
high.
| Molecular
signatures database collection | Gene set name | NES | NOM P-value | FDR q-value |
|---|
|
h.all.v6.2.symbols.gmt |
HALLMARK_MYC_TARGETS_V2 | 2.154 | 0.004 | 0.006 |
|
h.all.v6.2.symbols.gmt |
HALLMARK_MYC_TARGETS_V1 | 2.028 | 0.009 | 0.010 |
|
h.all.v6.2.symbols.gmt |
HALLMARK_DNA_REPAIR | 2.006 | <0.001 | 0.009 |
Discussion
Liver cancer is associated with a high mortality
rate worldwide; the development of this cancer type may be
influenced by viral infection, diet and environmental factors
(21-24).
In recent years, with the continuous progression of molecular
biology and treatments, including chemotherapeutic drugs and
surgical technology, the understanding of cancer biology and the
treatment of liver cancer have made great progress. However, the
prognosis of liver cancer remains poor. The World Health
Organization/International Classification of Diseases-10 classifies
diseases according to their etiology, pathology, clinical
manifestations and anatomical location. Cancer is a gene-associated
disease and molecular typing is required to deepen our
understanding of the underlying mechanisms of disease development
(25). Therefore, novel biomarkers
are urgently required. Our research group has been exploring novel
biomarkers for a number of years (26-38).
The present study focused on EIF3B and indicated that EIF3B is a
potential and independent prognostic biomarker for liver
cancer.
EIF3B is closely linked to cancer progression.
Consistent with previous studies, it was indicated that EIF3B was
highly expressed in patients with liver cancer. Although
Golob-Schwarzl et al (15)
reported that EIF3B was upregulated in HCV-associated HCC, all of
their patients were Asians. The patients assessed in the present
study were from all over the world and covered other types of liver
cancer that may be related to HCV. All of these results indicate
that EIF3B has an important role in cancer-associated processes. A
previous study suggested that EIF3B is involved in the
proliferation and metastasis of gastric cancer (39). In addition, the present study further
determined that EIF3B is associated with the histologic grade and
survival status of patients with liver cancer. Therefore, in-depth
studies using experimental and bioinformatics methods are
required.
EIF3B has been indicated to have a marked influence
on the prognosis of patients with cancer. In the present study, it
was observed that upregulation of EIF3B was associated with poor
overall/relapse-free survival of patients with liver cancer. In
addition, patients with high EIF3B expression in clear cell renal
cell carcinoma, esophageal squamous cell carcinoma and non-small
cell lung cancer had a shorter survival time (6,7,11). In order to further explore the
association between EIF3B and clinical characteristics of patients
with liver cancer, a subgroup analysis was performed. As the
TCGA-LIHC dataset does not have a Barcelona Clinic Liver Cancer
staging system, TNM staging was used. Kaplan-Meier subgroup
analysis indicated that high expression of EIF3B was associated
with poor overall survival in the subgroups of females, younger
(<55) or older (≥55) patients, R0, G2, G3, stage I and stage
III. Furthermore, high expression of EIF3B was associated with poor
relapse-free survival in the subgroups of females, older patients,
R0, G2 and stage III. However, Tian et al (11) reported that upregulation of EIF3B was
associated with tumor depth, TNM stage and lymph node metastasis in
patients with esophageal squamous cell carcinoma. However, the
results of the present study indicated that EIF3B was related to
histological grade and survival status. This may be linked to the
heterogeneity of tumor types and individual differences, which may
help to select personalized treatments. Owing to the TCGA-LIHC data
not including the body mass index and the presence of diabetes
mellitus as variables, it is not possible to calculate their
association with the prognosis of patients.
In the GSEA analysis, high EIF3B expression was
indicated to be associated with MYC-V1, MYC-V2 and DNA repair in
liver cancer. The MYC oncogene is an important regulator of liver
cancer progression. Previous studies have indicated that MYC is
able to promote the proliferation, metastasis and metabolism of
liver cancer by regulating signaling pathways including AKT/mTOR
and RAS/mitogen-activated protein kinase (40-42).
In addition, each replication of DNA in cancer cells may cause a
large amount of damage, including DNA substitutions or deletions
(43). Therefore, DNA repair
mechanisms (damage induction, signal transduction, signal response)
are particularly important. This may explain why EIF3B may promote
the progression of liver cancer through MYC-V1/V2 and DNA repair
pathways.
The present study mainly uncovered the prognostic
value of the EIF3B mRNA expression in liver cancer. Along with
other studies on EIF3B, the present study contributed to a better
understanding of the role of EIF3B, as well as the great
possibility for precise prognostication. However, the underlying
mechanisms remain to be fully elucidated and require further
exploration by scientific research. In the future, the mechanisms
of EIF3B will be studied at a deeper level.
In conclusion, the present study investigated the
prognostic value of EIF3B in patients with liver cancer. High EIF3B
expression was proved to be a potential and independent prognostic
biomarker for liver cancer. Future work will include in vivo
and in vitro experiments to explore the biological functions
of EIF3B and the underlying mechanisms.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Patient information was obtained from the open TCGA
database (https://portal.gdc.cancer.gov/). The datasets used
and/or analyzed during the current study are available from the
corresponding author on reasonable request.
Authors' contributions
WH designed the study. QY and LM were responsible
for extracting data, conducted data analysis and wrote the first
draft of the manuscript. BJ participated in the analysis of data
and critical modification of important knowledge content. WH and BJ
critically revised the manuscript and gave final approval for
submission. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ryerson AB, Eheman CR, Altekruse SF, Ward
JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM,
et al: Annual Report to the Nation on the Status of Cancer,
1975-2012, Featuring the Increasing Incidence of Liver Cancer.
Cancer. 122:1312–1337. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li L and Wang H: Heterogeneity of liver
cancer and personalized therapy. Cancer Lett. 379:191–197.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Dawkins J and Webster RM: The
hepatocellular carcinoma market. Nat Rev Drug Discov. 18:13–14.
2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Smith MD, Arake-Tacca L, Nitido A,
Montabana E, Park A and Cate JH: Assembly of eIF3 mediated by
mutually dependent subunit insertion. Structure. 24:886–896.
2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zang Y, Zhang X, Yan L, Gu G, Li D, Zhang
Y, Fang L, Fu S, Ren J and Xu Z: Eukaryotic translation initiation
factor 3b is both a promising prognostic biomarker and a potential
therapeutic target for patients with clear cell renal cell
carcinoma. J Cancer. 8:3049–3061. 2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Xu F, Xu CZ, Gu J, Liu X, Liu R, Huang E,
Yuan Y, Zhao G, Jiang J, Xu C, et al: Eukaryotic translation
initiation factor 3B accelerates the progression of esophageal
squamous cell carcinoma by activating β-catenin signaling pathway.
Oncotarget. 7:43401–43411. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liang H, Ding X, Zhou C, Zhang Y, Xu M,
Zhang C and Xu L: Knockdown of eukaryotic translation initiation
factors 3B (EIF3B) inhibits proliferation and promotes apoptosis in
glioblastoma cells. Neurol Sci. 33:1057–1062. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang L and Ouyang L: Effects of EIF3B gene
downregulation on apoptosis and proliferation of human ovarian
cancer SKOV3 and HO-8910 cells. Biomed Pharmacother. 109:831–837.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Choi YJ, Lee YS, Lee HW, Shim DM and Seo
SW: Silencing of translation initiation factor eIF3b promotes
apoptosis in osteosarcoma cells. Bone Joint Res. 6:186–193.
2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Tian Y, Zhao K, Yuan L, Li J, Feng S, Feng
Y, Fang Z, Li H and Deng R: EIF3B correlates with advanced disease
stages and poor prognosis, and it promotes proliferation and
inhibits apoptosis in non-small cell lung cancer. Cancer Biomark.
23:291–300. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Spilka R, Ernst C, Mehta AK and Haybaeck
J: Eukaryotic translation initiation factors in cancer development
and progression. Cancer Lett. 340:9–21. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang H, Ru Y, Sanchez-Carbayo M, Wang X,
Kieft JS and Theodorescu D: Translation initiation factor eIF3b
expression in human cancer and its role in tumor growth and lung
colonization. Clin Cancer Res. 19:2850–2860. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lin L, Holbro T, Alonso G, Gerosa D and
Burger MM: Molecular interaction between human tumor marker protein
p150, the largest subunit of eIF3, and intermediate filament
protein K7. J Cell Biochem. 80:483–490. 2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Golob-Schwarzl N, Krassnig S, Toeglhofer
AM, Park YN, Gogg-Kamerer M, Vierlinger K, Schröder F, Rhee H,
Schicho R, Fickert P and Haybaeck J: New liver cancer biomarkers:
PI3K/AKT/mTOR pathway members and eukaryotic translation initiation
factors. Eur J Cancer. 83:56–70. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Team RDCJCR. A language and environment
for statistical computing. R Foundation for Statistical Computing,
Vienna, Austria. 14:12–21. 2009. View Article : Google Scholar
|
|
17
|
Wickham H: Ggplot2: Elegant graphics for
data analysis. J R Stat Soc. 174:245–246. 2011.
|
|
18
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12(77)2011.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Therneau TM and Grambsch PM: Modeling
survival data: Extending the Cox model. Springer, New York,
2000.
|
|
20
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Cronin KA, Lake AJ, Scott S, Sherman RL,
Noone AM, Howlader N, Henley SJ, Anderson RN, Firth AU, Ma J, et
al: Annual report to the nation on the status of cancer, part I:
National cancer statistics. Cancer. 124:2785–2800. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Xu N, Liu YN, Yin P, Wang LJ, Dou YS, Yang
WJ and Zhou MG: Impact of liver cancer deaths on life expectancy in
14 counties (districts) from the Huai River Basin, 2013:
Relationship between the water environment and liver cancer.
Zhonghua Yu Fang Yi Xue Za Zhi. 50:629–633. 2016.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
23
|
Tu T, Bühler S and Bartenschlager R:
Chronic viral hepatitis and its association with liver cancer. Biol
Chem. 398:817–837. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Chen YJ, Wallig MA and Jeffery EH: Dietary
broccoli lessens development of fatty liver and liver cancer in
mice given diethylnitrosamine and fed a western or control diet. J
Nutr. 146:542–550. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Global Burden of Disease Cancer
Collaboration. Fitzmaurice C, Akinyemiju TF, Al Lami FH, Alam T,
Alizadeh-Navaei R, Allen C, Alsharif U, Alvis-Guzman N, Amini E, et
al: Global, regional, and national cancer incidence, mortality,
years of life lost, years lived with disability, and
disability-adjusted life-years for 29 cancer groups, 1990 to 2016:
A systematic analysis for the global burden of disease study. JAMA
Oncol. 4:1553–1568. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jiao Y, Fu Z, Li Y, Meng L and Liu Y: High
EIF2B5 mRNA expression and its prognostic significance in liver
cancer: A study based on the TCGA and GEO database. Cancer Manag
Res. 10:6003–6014. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Jiao Y, Fu Z, Li Y, Zhang W and Liu Y:
Aberrant FAM64A mRNA expression is an independent predictor of poor
survival in pancreatic cancer. PLoS One.
14(e0211291)2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jiao Y, Li Y, Lu Z and Liu Y: High
trophinin-associated protein expression is an independent predictor
of poor survival in liver cancer. Dig Dis Sci. 64:137–143.
2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Jiao Y, Li Y, Fu Z, Hou L, Chen Q, Cai Y,
Jiang P, He M and Yang Z: OGDHL expression as a prognostic
biomarker for liver cancer patients. Dis Markers.
2019(9037131)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jiao Y, Li Y, Jiang P, Han W and Liu Y:
PGM5: A novel diagnostic and prognostic biomarker for liver cancer.
PeerJ. 7(e7070)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jiao Y, Li Y, Liu S, Chen Q and Liu Y:
ITGA3 serves as a diagnostic and prognostic biomarker for
pancreatic cancer. Onco Targets Ther. 12:4141–4152. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Li Y, Jiao Y, Fu Z, Luo Z, Su J and Li Y:
High miR-454-3p expression predicts poor prognosis in
hepatocellular carcinoma. Cancer Manag Res. 11:2795–2802.
2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li Y, Jiao Y, Li Y and Liu Y: Expression
of La ribonucleoprotein domain family member 4B (LARP4B) in liver
cancer and their clinical and prognostic significance. Dis Markers.
2019(1569049)2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li Y, Jiao Y, Luo Z, Li Y and Liu Y: High
peroxidasin-like expression is a potential and independent
prognostic biomarker in breast cancer. Medicine (Baltimore).
98(e17703)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang X, Cui Y, He M, Jiao Y and Yang Z:
Lipocalin-1 expression as a prognosticator marker of survival in
breast cancer patients. Breast Care, 2019.
|
|
36
|
Cui Y, Jiao Y, Wang K, He M and Yang Z: A
new prognostic factor of breast cancer: High carboxyl ester lipase
expression related to poor survival. Cancer Genet. 239:54–61.
2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hou L, Zhang X, Jiao Y, Li Y, Zhao Y, Guan
Y and Liu Z: ATP binding cassette subfamily B member 9 (ABCB9) is a
prognostic indicator of overall survival in ovarian cancer.
Medicine (Baltimore). 98(e15698)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Cai H, Jiao Y, Li Y, Yang Z, He M and Liu
Y: Low CYp24A1 mRNA expression and its role in prognosis of breast
cancer. Sci Rep. 9(13714)2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Ma F, Li X, Ren J, Guo R, Li Y, Liu J, Sun
Y, Liu Z, Jia J and Li W: Downregulation of eukaryotic translation
initiation factor 3b inhibited proliferation and metastasis of
gastric cancer. Cell Death Dis. 10(623)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ladu S, Calvisi DF, Conner EA, Farina M,
Factor VM and Thorgeirsson SS: E2F1 inhibits c-Myc-driven apoptosis
via pIK3CA/Akt/mTOR and COX-2 in a mouse model of human liver
cancer. Gastroenterology. 135:1322–1332. 2008.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xin B, Yamamoto M, Fujii K, Ooshio T, Chen
X, Okada Y, Watanabe K, Miyokawa N, Furukawa H and Nishikawa Y:
Critical role of Myc activation in mouse hepatocarcinogenesis
induced by the activation of AKT and RAS pathways. Oncogene.
36:5087–5097. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li X, Wu Q, Bu M, Hu L, Du WW, Jiao C, Pan
H, Sdiri M, Wu N, Xie Y and Yang BB: Ergosterol peroxide activates
Foxo3-mediated cell death signaling by inhibiting AKT and c-Myc in
human hepatocellular carcinoma cells. Oncotarget. 7:33948–33959.
2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Barnes JL, Zubair M, John K, Poirier MC
and Martin FL: Carcinogens and DNA damage. Biochem Soc Trans.
46:1213–1224. 2018.PubMed/NCBI View Article : Google Scholar
|